Abstract
Chitinase Chi1 of Aeromonas caviae CB101 possesses chitin binding sites at both its N and C termini. Four putative exposed residues aligned in a line on the surface of the N-terminal domains of Chi1 were found to contribute to the enzyme-chitin binding and hydrolysis via site-directed mutagenesis. Also, it was found that Chi1 requires the cooperation of the N- and C-terminal domains to bind fully with crystalline and colloidal chitin.
Chitinases (EC 3.2.1.14) are essential in chitin recycling in nature (5). So far, the three-dimensional (3D) structures of the following three bacterial chitinases have been resolved: ChiA and ChiB from Serratia marcescens (13, 18) and ChiA1 from Bacillus circulans (9, 10, 17). all of these enzymes belong to family 18 subfamily A (7, 8, 15). Excluding the catalytic domains, all three bacterial chitinases whose structures have been resolved have substrate binding domains either at their N (ChiA) or C (ChiB, ChiA1) termini, and this modular organizing feature is common for most of the known bacterial chitinases (2, 11, 12, 14, 15, 16).
From the offshore sediment of the Xiamen coast, various chitinolytic bacteria were isolated in our laboratory. Strain CB101 was chosen for further analysis because it exhibited the highest chitinolytic activity (21). CB101 was determined to be an Aeromonas caviae strain (21) by cellular fatty acid analysis, carbon source utilization tests (BIOLOG, Inc., Hayward, Calif.), and 16S rRNA gene sequence comparison (98% identity) (unpublished data). Chitinase Chi1 of A. caviae CB101 has at least four domains: an N-terminal all-β-sheet domain (ChiN), a catalytic domain, an A region, and a C-terminal chitin binding domain (ChBD) (19, 22). Chi1 thus serves as a good model to study the functions of the discrete domains in the chitinase reaction. The functions of the C-terminal A region and the ChBD have been studied previously (19), and it was found that the C-terminal ChBD of Chi1 contained the major binding sites for crystalline chitin; the A region was not involved in chitin binding, but it had a function in soluble substrate hydrolysis. Also, it was suggested that the N-terminal domains may contribute more than the ChBD to colloidal chitin binding (19). The N-terminal 563-amino-acid segment of Chi1 (Chi1ΔAΔChBD) exhibited 74% identity with the corresponding segment of ChiA from S. marcescens. By using the 3D structure modeling method with the Swiss-Pdb viewer version 3.7 program (6), the 3D structure of the N-terminal 563 amino acids of Chi1 was constructed based on the structure of ChiA. One hydroxyl containing amino acid Ser33 and three aromatic residues (Trp70, Trp232, and Trp245) of Chi1 were aligned in a line on the surface of the N-terminal domains, corresponding to the exposed aromatic residues Trp33, Trp69, Phe232, and Trp245 of ChiA (17; photo not shown). In this study, the possible functions of these four putative exposed residues of Chi1 in chitin binding and hydrolysis were studied to determine the functions of the N-terminal domains. Also, the contributions of the N-terminal and C-terminal domains to enzyme-substrate binding and hydrolysis were compared and analyzed further.
Site-directed mutagenesis of the aromatic residues Trp70, Trp232, and Trp245 to alanine and of Ser33 to alanine or tryptophan was carried out by PCR using pQE86 containing the complete chi1 sequence (19) as the template and the procedure recommended for a Quikchange site-directed mutagenesis kit (Stratagene). The primers used were 5′-CCCACCATTGGCTGGGGCCCCACCAAG for S33W, 5′-CCCACCATTGGCGCAGGCCCCACCAAG for S33A, 5′-ACCCTGGAACCTTGCGTCCGGTGACGTC for W70A, 5′-ATCCATGATCCCGCGGCGGCGATCCAG for W232A, and 5′-AACCTCAGCGCCGCAGATGAGCCCTAC for W245A. The mutant clones were verified by sequencing (Sangon, Shanghai, People's Republic of China). Protein Chi1 and five mutants of this protein (W70A, S33A, S33W, W232A, and W245A) with a C-terminal six-His tag were purified using an Ni-nitrilotriacetic acid matrix column as recommended by the manufacturer (QIAGEN).The purified proteins were checked by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and were found to be very pure (photo not shown).
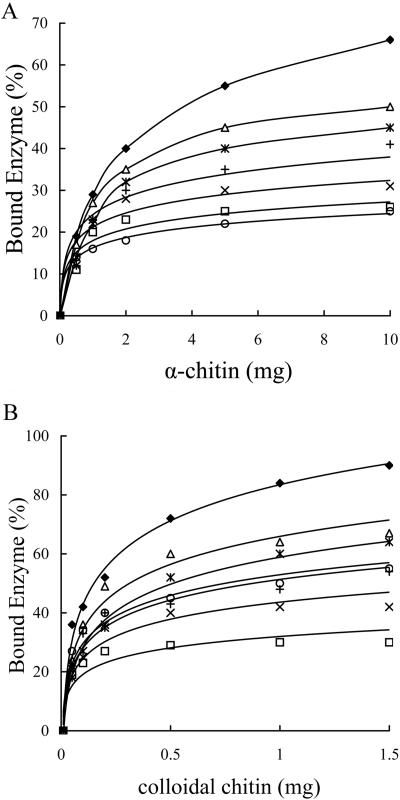
Binding assays for Chi1 and five site-directed mutants of this enzyme with various amounts of crystalline chitin (α-chitin; Sigma) and colloidal chitin prepared from crab shells were carried out as follows. Twenty micrograms of protein was mixed thoroughly with 1 to 10 mg of chitin or 0.1 to 1.5 mg of colloidal chitin in 200 μl of 0.1-mol/liter sodium phosphate buffer (pH 6). The mixtures were kept on ice with rotary shaking for 1 h. After centrifugation at 13,000 × g for 5 min, the supernatants containing unabsorbed proteins were collected, and the concentrations of the unbound proteins were determined by the Bradford method (3). The amounts of the bound proteins were calculated by determining the difference between the amount of the total protein and the amount of the unbound protein. The percentages of bound enzymes were calculated. All tests were performed in triplicate. There was no significant difference among the data obtained from three repeated tests as determined at a P value of <0.05. The mean values for the three repeated tests were used, and the results are shown in Fig. 1. It was found that all four putative exposed amino acids contributed to both crystalline chitin and colloidal chitin binding, to different extents. Trp70 contributed most significantly to the enzyme-chitin binding, followed by Trp245, Ser33, and finally Trp232 (Fig. 1A and B). Changing Trp70 to alanine caused the enzyme to lose the majority of its binding capacity for crystalline chitin, an effect similar to that observed for the Chi1 truncate Chi1ΔChBD (without the C-terminal chitin binding domain) (Fig. 1A). Chi1ΔChBD still retained most of the ability to bind colloidal chitin; however, the site-directed mutant W70A lost most of the ability to bind colloidal chitin (Fig. 1B). Replacement of Ser30 with Trp resulted in slight decreases in the binding activities of the enzyme with crystalline and colloidal chitin, whereas replacement of Ser30 with Ala resulted in much more severe defects in the enzyme-chitin binding reaction (Fig. 1A and B). Previously, Wu et al. (20) and Chang et al. (4) indicated that the C-terminal ChBD of Chi92 of Aeromonas hydrophila, which is 100% identical to the ChBD of Chi1, exhibited efficient chitin binding activity when it was expressed separately in vitro. Our data revealed that the C-terminal ChBD of Chi1 did not play a role alone in the enzyme; the enzyme needed the cooperation of the N- and C-terminal domains to confer full chitin binding activity.
FIG. 1.
Chitin binding assays for Chi1 and mutants of this enzyme. The binding activities of Chi1 and the derived mutants were compared. Symbols: ⧫, Chi1; □, W70A; ▵, S33W; ×, W245A; *, W232A; +, S33A; ○, Chi1ΔChBD.
The abilities of Chi1 and the five site-directed mutants to hydrolyze different substrates, including crystalline chitin, colloidal chitin, soluble chitin (acetylation degree, 61.2%; average molecular weight, 200,000 to 300,000; Yaizu Suisan Chemiacal Co., Ltd., Shizuoka, Japan), and p-nitrophenyl (pNP)-chitobiose, were compared using the methods described previously (19). In brief, 50 pmol of chitinase was incubated with 1.5 mg of substrate in 200 μl of 0.1-mol/liter sodium phosphate buffer (pH 6) at 45°C for various times, or 20 pmol of chitinase was mixed with 0.5 mmol/liter pNP-chitobiose in 0.1 ml of 0.1-mol/liter sodium phosphate buffer (pH 6) at 37°C for various times. One unit of enzyme activity was defined as the amount of enzyme that released 1 μmol of reducing sugar or pNP per min. Each activity test was repeated three times independently. As shown in Table 1, all five site-directed mutants exhibited significantly reduced hydrolyzing activities with crystalline chitin (range, approximately 37% to 58%). Replacement of Trp70, Trp245, and Ser33 with Ala decreased the enzyme-digesting activity with crystalline chitin most severely, as Chi1ΔChBD did (around 37% of the activity was left), whereas the enzyme exhibited more than one-half the wild-type digestion activity with crystalline chitin when Trp232 was replaced with Ala or Ser33 was replaced with Trp. Also, the abilities of the five mutants to hydrolyze colloidal chitin were slightly higher than their abilities to hydrolyze crystalline chitin; around 44% to 63% of the activity was retained. W70A, in which the colloidal chitin binding activity was reduced most severely, exhibited the highest colloidal chitin-hydrolyzing activity among the five mutants. Mutant W245A had the lowest hydrolyzing activity with colloidal chitin. All five site-directed mutants exhibited much lower digestion activity with colloidal chitin than the truncate Chi1ΔChBD. On the other hand, all five site-directed mutants studied, together with Chi1ΔChBD, exhibited full hydrolyzing activities with the water-soluble substrates tested, soluble chitin and pNP-chitobiose; some even exhibited higher levels of digestion activity than the wild-type Chi1 (Table 1). This was probably because the soluble substrates do not need to be bound and guided into the catalytic cleft through interactions with the putative exposed residues; therefore, mutations in these residues had no effect on hydrolysis of the soluble substrates.
TABLE 1.
Hydrolyzing activities of Chi1 and mutants of this enzyme
| Enzyme | Specific hydrolyzing activity (U/μmol protein)
|
|||
|---|---|---|---|---|
| Crystalline chitin | Colloidal chitin | Soluble chitin | pNP-chitobiose | |
| Chi1 | 5.24 ± 0.22 (100)a | 68.55 ± 2.83 (100) | 1,315.0 ± 54.2 (100) | 48.66 ± 5.91 (100) |
| W70A | 1.91 ± 0.13 (37) | 42.84 ± 2.72 (62) | 1,328.2 ± 50.1 (101) | 53.52 ± 6.82 (110) |
| S33A | 1.92 ± 0.11 (37) | 40.44 ± 2.06 (59) | 1,354.5 ± 57.8 (103) | 49.15 ± 5.73 (101) |
| W245A | 1.93 ± 0.15 (37) | 30.02 ± 2.31 (44) | 1,275.6 ± 55.7 (97) | 45.74 ± 5.88 (94) |
| W232A | 3.02 ± 0.18 (58) | 40.78 ± 1.78 (60) | 1,341.3 ± 60.5 (102) | 58.39 ± 4.91 (120) |
| S33W | 2.86 ± 0.19 (55) | 40.44 ± 2.51 (59) | 1,354.5 ± 50.4 (103) | 62.28 ± 7.12 (128) |
| Chi1ΔChBD | 1.73 ± 0.09 (33) | 51.41 ± 2.56 (75) | 1,301.9 ± 48.1 (99) | 48.64 ± 5.56 (100) |
The values are means ± standard deviations for three independent experiments. The values in parentheses are relative specific activities (expressed as percentages).
The results presented here strongly support the computer structure modeling results for Chi1ΔAΔChBD and suggest that Chi1 may have crystalline chitin-hydrolyzing activity similar to that of ChiAsm (1, 17). The reaction mode of Chi1 can be hypothesized: the chitin chain is bound and guided into the catalytic cleft through the N terminus of the enzyme by interaction of the exposed aromatic and hydroxyl residues with the chitin chain; the C-terminal chitin binding domain cooperates with the N terminus for full binding; and the C-terminal ChBD may contribute more to distortion of the highly crystallized chitin to facilitate attachment of the chitin chain to the enzyme.
Acknowledgments
This work was supported in part by the Chinese High Tech (863) program (2002AA625010) and by the Fujian Science Fund (2002F006).
REFERENCES
- 1.Aronson, N. A., Jr., B. A. Halloran, M. F. Alexyv, L. Amable, J. D. Madura, L. Pasuplati, C. Worth, and P. van Roey. 2003. Family 18 chitinase-oligosaccharide substrate interaction: subsite preference and anomer selectivity of Serratia marcescens chitinase A. Biochem. J. 376:87-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blaak, H., J. Schnellmann, S. Walter, B. Henrissat, and H. Schrempf. 1993. Characteristics of an exochitinase from Streptomyces olivaceoviridis, its corresponding gene, putative protein domains and relationship to other chitinase. Eur. J. Biochem. 214:659-669. [DOI] [PubMed] [Google Scholar]
- 3.Bradford, M. M. 1976. A rapid and sensitive method for quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 4.Chang, M. C., P. L. Lai, and M. L. Wu. 2004. Biochemical characterization and site-directed mutational analysis of the double chitin-binding domain from chitinase 92 of Aeromonas hydrophila JP101. FEMS Microbiol. Lett. 232:61-66. [DOI] [PubMed] [Google Scholar]
- 5.Cohen-Kupiec, R., and I. Chert. 1998. The molecular biology of chitin digestion. Curr. Opin. Biotechnol. 9:270-277. [DOI] [PubMed] [Google Scholar]
- 6.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-Pdb Viewer: an environment for comparative protein modeling. Electrophoresis 18:2714-2723. [DOI] [PubMed] [Google Scholar]
- 7.Henrissat, B. 1991. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 280:309-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Henrissat, B., and A. Bairoch. 1993. New families in the classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 293:781-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ikegami, T., T. Okada, M. Hashimoto, S. Seino, T. Watanabe, and M. Shirakawa. 2000. Solution structure of the chitin-binding domain of Bacillus circulans WL-12 chitinase A1. J. Biol. Chem. 275:13654-13661. [DOI] [PubMed] [Google Scholar]
- 10.Jee, J. G., T. Ikegami, M. Hashimoto, T. Kawabata, M. Ikeguchi, T. Watanabe, and M. Shirakawa. 2002. Solution structure of the fibronectin type III domain from Bacillus circulans WL-12 chitinase A1. J. Biol. Chem. 277:1388-1397. [DOI] [PubMed] [Google Scholar]
- 11.Kobayashi, D. Y., R. M. Reedv, J. A. Bick, and P. V. Oudemans. 2002. Characterization of a chitinase gene from Stenotrophomonas maltophilia strain 34S1 and its involvement in biological control. Appl. Environ. Microbiol. 68:1047-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee, H. S., D. S. Han, S. J. Choi, S. W. Choi, D. S. Kim, D. H. Bai, and J. H. Yu. 2000. Purification, characterization, and primary structure of a chitinase from Pseudomonas sp. YHS-A2. Appl. Microbiol. Biotechnol. 54:397-405. [DOI] [PubMed] [Google Scholar]
- 13.Perrakis, A., I. Tews, Z. Dauter, A. B. Oppenheim, I. Chet, K. S. Wilson, and C. E. Vorgias. 1994. Crystal structure of a bacterial chitinase at 2.3 Å resolution. Structure 2:1169-1180. [DOI] [PubMed] [Google Scholar]
- 14.Robbins, P. W., C. Albright, and B. Benfield. 1988. Cloning and expression of a Streptomyces plicatus chitinase (chitinase-63) in Escherichia coli. J. Biol. Chem. 263:443-447. [PubMed] [Google Scholar]
- 15.Suzuki, K., M. Taiyoji, N. Sugawara, N. Nikaidou, B. Henrissat, and T. Watanabe. 1999. The third chitinase gene (chiC) of Serratia marcescens 2170 and the relationship of its product to other bacterial chitinases. Biochem. J. 343:587-596. [PMC free article] [PubMed] [Google Scholar]
- 16.Tsujibo, H., H. Orikoshi, K. Shiotani, M. Hayashi, J. Umeda, K. Miyamoto, C. Imada, Y. Okami, and Y. Inamori. 1998. Characterization of chitinase C from a marine bacterium, Alteromonas sp. strain O-7, and its corresponding gene and domain structure. Appl. Environ. Microbiol. 64:472-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Uchiyama, T., F. Katouno, N. Nikaidou, T. Nonaka, J. Sugiyama, and T. Watanabe. 2001. Roles of the exposed aromatic residues in crystalline chitin hydrolysis by chitinase A from Serratia marcescens 2170. J. Biol. Chem. 276:41343-41349. [DOI] [PubMed] [Google Scholar]
- 18.van Aalten, D. M. F., B. Synstad, M. B. Brurberg, E. Hough, B. W. Riise, V. G. H. Eijsink, and R. K. Wierenga. 2000. Structure of a two-domain chitotriosidase from Serratia marcescens at 1.9 Å resolution. Proc. Natl. Acad. Sci. USA 97:5842-5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang, F. P., Q. Li, Y. Zhou, M. G. Li, and X. Xiao. 2003. The C-terminal module of Chi1 from Aeromonas caviae CB101 has function in substrate binding and hydrolysis. Proteins 53:908-916. [DOI] [PubMed] [Google Scholar]
- 20.Wu, M. L., Y. C. Chuang, J. P. Chen, C. S. Chen, and M. C. Chang. 2001. Identification and characterization of the three chitin-binding domains within the multidomain chitinase Chi92 from Aeromonas hydrophila JP101. Appl. Environ. Microbiol. 67:5100-5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiao, X., Y. Zhou, and F. P. Wang. 2003. Isolation and identification of a high-efficient chitin-degrading marine bacterium CB101 and studies on its chitinases system. Acta Oceanol. Sin. 25:38-142. (In Chinese with English abstract.) [Google Scholar]
- 22.Zhou, Y., F. P. Wang, and X. Xiao. 2002. Structures of the cloned chitinase and its truncate from A. caviae. Prog. Nat. Sci. 12:587-591. [Google Scholar]