Abstract
The Geobacteraceae citrate synthase is phylogenetically distinct from those of other prokaryotes and is a key enzyme in the central metabolism of Geobacteraceae. Therefore, the potential for using levels of citrate synthase mRNA to estimate rates of Geobacter metabolism was evaluated in pure culture studies and in four different Geobacteraceae-dominated environments. Quantitative reverse transcription-PCR studies with mRNA extracted from cultures of Geobacter sulfurreducens grown in chemostats with Fe(III) as the electron acceptor or in batch with electrodes as the electron acceptor indicated that transcript levels of the citrate synthase gene, gltA, increased with increased rates of growth/Fe(III) reduction or current production, whereas the expression of the constitutively expressed housekeeping genes recA, rpoD, and proC remained relatively constant. Analysis of mRNA extracted from groundwater collected from a U(VI)-contaminated site undergoing in situ uranium bioremediation revealed a remarkable correspondence between acetate levels in the groundwater and levels of transcripts of gltA. The expression of gltA was also significantly greater in RNA extracted from groundwater beneath a highway runoff recharge pool that was exposed to calcium magnesium acetate in June, when acetate concentrations were high, than in October, when the levels had significantly decreased. It was also possible to detect gltA transcripts on current-harvesting anodes deployed in freshwater sediments. These results suggest that it is possible to monitor the in situ metabolic rate of Geobacteraceae by tracking the expression of the citrate synthase gene.
The ability to estimate in situ rates of microbial metabolism can provide invaluable insight into microbial processes in the environment. Yet the metabolic rates of some of these processes can be difficult to measure. For example, with current technology, it is difficult to measure the in situ rates of Fe(III) reduction in most sedimentary environments, in part because of the insoluble nature not only of Fe(III) oxides but also of the Fe(II) produced. In addition, suitable tracer methods are currently unavailable (13). The development of tools to assess in situ rates of Fe(III) reduction would be particularly beneficial, as dissimilatory Fe(III)-reducing bacteria are known to play an essential role in a number of ecologically significant microbial processes. For example, Fe(III) reducers are able to degrade natural and contaminant organic matter in sedimentary environments (28, 31), they are useful agents for the bioremediation of groundwater contaminated with uranium and other metals (5, 35, 37), and they can use electrodes as alternative electron acceptors to harvest electricity from sediment organic matter (9, 20, 43).
An alternative approach might be to correlate levels of mRNA transcripts for key genes involved in Fe(III) reduction with rates of Fe(III) reduction and then measure their transcript levels in the environment (13). This is a possibility because even though there is a wide phylogenetic diversity of dissimilatory Fe(III)-reducing microorganisms with different strategies for Fe(III) reduction (31), molecular studies have demonstrated that Geobacteraceae are the predominant microorganisms in many sedimentary environments in which Fe(III) reduction is an important process (4, 21, 39-41), as well as on the surfaces of electrodes harvesting electricity from aquatic sediments (9, 20, 43). Thus, if it were possible to identify a gene that was highly conserved within the Geobacteraceae family with expression patterns that correlated with rates of Fe(III) reduction, it should theoretically be possible to estimate Fe(III) reduction rates by monitoring the expression of this gene.
Previous studies have suggested that it is possible to monitor the gene expression of Geobacteraceae in subsurface environments (23). For example, Geobacteraceae nifD transcript levels could be measured in Fe(III)-reducing subsurface sediments (23). These nitrogen fixation studies were facilitated by the fact that the genes involved in the N2 fixation pathway are well understood for a diversity of microorganisms. However, Fe(III) respiration is not as well understood, making it difficult to identify the appropriate gene(s) to monitor in the environment. Initial studies of Fe(III) reduction rates focused on Geobacter sulfurreducens, a heavily studied member of the Geobacteraceae. This organism was selected because it is closely related to the Geobacter species that are most prevalent in many Fe(III)-reducing environments (31) and because both this organism's complete genome sequence (32) and a genetic system (15) are available.
Analysis of chemostat cultures of G. sulfurreducens demonstrated that there was a direct correlation between levels of transcripts for fumarate reductase and the rate of fumarate respiration (13). More importantly, levels of transcripts for omcB, a gene which encodes an outer membrane c-type cytochrome required for Fe(III) reduction (27), were directly correlated with rates of Fe(III) reduction (13). However, the level of omcB transcripts was also significantly influenced by whether metabolism was limited by electron donor or Fe(III). These results suggest that environmental conditions could also have an important influence on the expression of omcB (13). Furthermore, although omcB is an essential gene for Fe(III) reduction in G. sulfurreducens, this gene has not been detected in the genome of the closely related species Geobacter metallireducens (J. Butler, D. R. Lovley, et al., unpublished data) or in genomic DNAs extracted from Geobacteraceae-dominated environments. Thus, monitoring levels ofomcB transcripts is unlikely to be a useful strategy for evaluating rates of Fe(III) reduction in microbial communities comprised of multiple Geobacter species.
An alternative to monitoring the expression of genes directly involved in Fe(III) respiration would be to evaluate rates of central metabolism, which provides the electrons that are required for dissimilatory Fe(III) reduction. Acetate is expected to be the primary electron donor for Fe(III) reduction in a variety of sedimentary environments (30) as well as during in situ bioremediation of uranium (5). Citrate synthase is a key gene in the incorporation of acetate into the tricarboxylic acid (TCA) cycle (19, 47), and the genome of G. sulfurreducens was found to contain a novel putative citrate synthase that is more closely related to the citrate synthases of eukaryotic microorganisms than to those of prokaryotes (32). The gene product was purified and found to possess the only citrate synthase activity in G. sulfurreducens (11). All members of the Geobacteraceae examined contained a similar citrate synthase gene (11), designated gltA. We report that the levels of transcripts for gltA correspond with the rates of acetate metabolism and Fe(III) reduction in chemostat cultures of G. sulfurreducens and of current production when G. sulfurreducens is growing with an electrode serving as the sole electron acceptor. Furthermore, Geobacteraceae-specific gltA genes can be detected in a variety of environments in which Geobacteraceae are important members of the microbial community, and levels of gltA transcripts in the environment are related to the availability of acetate to support Fe(III) reduction.
MATERIALS AND METHODS
Source of organisms and culture conditions.
Geobacter sulfurreducens (ATCC 51573), Geobacter psychrophilus strain P35T (JCM 12644), and Pelobacter massiliensisT (DSM 6233) were obtained from our laboratory culture collection and grown under previously described conditions (22, 33). Standard anaerobic techniques were used throughout (8, 34). All media were sterilized by autoclaving, and all incubations were performed at 30°C.
G. sulfurreducens grown under chemostat conditions.
G. sulfurreducens was grown in electron donor-limited chemostats, with acetate (5 mM) provided as the electron donor and Fe(III) citrate (55 mM) provided as the electron acceptor, at 30°C as previously described (18). G. sulfurreducens was grown in triplicate chemostats at dilution rates of 0.03 h−1 and 0.07 h−1. Analyses of Fe(II), acetate, and protein were performed as previously described (18).
Growth of G. sulfurreducens on an electrode.
G. sulfurreducens cells that had been grown with acetate (10 mM) as the electron donor and Fe(III) citrate (55 mM) as the electron acceptor were harvested by centrifugation at 3,150 × g at 4°C and resuspended in anoxic medium lacking an electron donor or acceptor. This cell suspension served as an inoculum for the anaerobic anodic chamber (200 ml of medium) of a two-chambered electrode system constructed as previously described (9, 10). The electron acceptor provided for growth in the anode chamber consisted of a graphite electrode poised with a potentiostat (AMEL Instruments, Milan, Italy) at a constant potential of +0.52 V (in reference to a standard H2 electrode), and acetate (10 mM) was provided as the electron donor.
A Power Lab 4SP unit connected to a Power Macintosh computer collected current and voltage measurements directly from potentiostat outputs every 10 seconds. The data were logged with Chart 4.0 software (ADInstruments, Mountain View, CA) as levels of current (mA) production over time.
Geobacteraceae-dominated environment 1: U(VI)-contaminated aquifer.
A small-scale in situ bioremediation experiment was conducted on the grounds of a former uranium ore-processing facility in Rifle, Colorado (5), during the months of August and September 2004. This site, designated the Old Rifle site, is part of the Uranium Mill Tailings Remedial Action program of the U.S. Department of Energy. Using previously described procedures (5), acetate (100 mM) and a bromide tracer (10 mM) were injected into the subsurface to provide <10 mM acetate in the groundwater over the course of 28 days via an injection gallery composed of five injection wells which was installed up-gradient from a series of four monitoring wells (each 1.83 m down-gradient of the injection gallery/previous monitoring well). The test plot was adjacent to the 2002 Rifle experiment plot and was one-fourth the size and similar in design (5).
Geobacteraceae-dominated environment 2: site exposed to high concentrations of CMA.
The second site consisted of a highway runoff recharge pool located adjacent to State Route 25 in Plymouth, Massachusetts. The pool was constructed to collect runoff generated by State Route 25, which opened in August 1987 (14). The Massachusetts Department of Environmental Protection enacted restrictions on this area requiring the use of nonchloride deicing agents along a 1,900-m section of highway impacting nearby cranberry bogs. Since the road opened, the primary road deicing agent used on this stretch of highway has been calcium magnesium acetate (CMA). The unconfined aquifer underlying the study site is part of the Wareham Outwash Plain consisting of fine to coarse-grained sand. The concentration of acetate in the groundwater varies between 0 and 5 mM (36).
Geobacteraceae-dominated environment 3: Fe(III) reduction zone of petroleum-contaminated aquifer.
Aquifer sediments at the USGS Groundwater Toxics Site in Bemidji, Minnesota, have been contaminated with crude oil for 18 years as a result of a break in an oil pipeline (7, 16, 24). Fe(III) reduction is an important terminal electron-accepting process in portions of the aquifer (3, 29, 40). Sediments were collected from the Fe(III) reduction zone of the contaminant plume in 2004 by split-spoon sample cores and transported immediately to the laboratory as previously described (23).
Geobacteraceae-dominated environment 4: fuel cell deployed in freshwater pond.
Sediment fuel cells were deployed at the University of Massachusetts Field Station in Nantucket, Massachusetts, as previously described (38, 43). A graphite electrode with a surface area of 189.58 in2 was submerged in the sediment and connected via a 560-Ω resistor to a cathode of the same size in the overlying aerobic water. Two control electrodes were not connected to a cathode and did not harvest electricity. Current was produced by the sediment fuel cells for 16.3 weeks, and the voltage was logged hourly by a Campbell scientific logger with cellular phone access for remote monitoring. Electrodes were deployed in May and harvested in October, and current generated by the freshwater sediment fuel cell ranged from 0.05 to 1.45 mA (25).
Extraction of mRNA from pure cultures.
Solutions for RNA extraction were made with diethyl pyrocarbonate-treated water (Ambion). As previously described (23), chemostat cultures (200 ml) at a steady state were transferred to prechilled 50-ml conical tubes and centrifuged at 4,000 rpm for 15 min at 4°C. The supernatant was discarded, and pellets were flash frozen in an ethanol-dry ice bath and stored at −80°C. Prior to RNA extraction, the pellets were resuspended in 1.5 ml TPE buffer (100 mM Tris-HCl, 100 mM KH2PO4, 10 mM EDTA; pH 8.0) and divided into aliquots in eight separate 2-ml screw-cap tubes.
Cells were harvested from current-harvesting electrodes at six different rates of current, i.e., 0.5 mA, 0.75 mA, 1.0 mA, 1.2 mA, 1.4 mA, and 2.0 mA. Once the desired current was obtained, electrodes were removed from the anodic chamber, rinsed with RNA Protect (QIAGEN), and vigorously scraped with a sterile razor blade into 100 ml of RNA Protect, producing a graphite slurry. Graphite suspensions were then transferred to prechilled 50-ml conical tubes and centrifuged at 4,000 rpm for 15 min at 4°C. The supernatant was discarded, and pellets were flash frozen in an ethanol-dry ice bath and stored at −80°C. Prior to RNA extraction, each pellet was resuspended in 3 ml TPE buffer and divided into aliquots in eight separate 2-ml screw-cap tubes. RNAs were extracted from chemostat and electrode pellets as previously described (23).
Extraction of mRNA from environmental samples.
Groundwater samples were collected from the same well (well M16) at the U(VI)-contaminated site in Rifle, Colorado, every other day over the course of 27 days in the months of August and September 2004. At the site exposed to CMA in Plymouth, Massachusetts, groundwater samples were collected from well AY in June and October 2004. Groundwater from both sites was concentrated by impact filtration on 293-mm-diameter Supor membrane disk filters (Pall Life Sciences). The filters were placed into Whirl-Pack bags, flash frozen in a dry ice-ethanol bath, and shipped back to the laboratory, where they were stored at −80°C. RNA could only be extracted from one-half of a filter at a time. The frozen filter halves were first crushed into a fine powder, dispensed into eight different 2-ml screw-cap tubes, and suspended in 800 μl of TPE buffer. RNA was extracted from this filter suspension by a previously described acetone precipitation protocol (23).
In order to extract RNA from the surfaces of current-harvesting anodes, the electrodes were first removed from the sediment, placed into Whirl-Pack bags, and flash frozen in liquid nitrogen. Each frozen graphite electrode was then fragmented into smaller pieces and allocated into 15-ml conical tubes. Four milliliters of TE-sucrose buffer (10 mM Tris-HCl, 1 mM EDTA, 6.7% sucrose, pH 8.0), 60 μl 10% sodium dodecyl sulfate, 20 μl lysozyme (50 mg/ml), and 6 μl proteinase K (20 mg/ml) were added to each tube. Samples were then incubated at 37°C for 10 min and centrifuged at 4,000 rpm for 20 min. The supernatants were allocated into 2-ml screw-cap tubes, and a hot acidic phenol-chloroform extraction followed by isopropanol precipitation at −30°C was done as previously described (23).
RNA extracted from all pure culture and environmental RNA samples was further purified with an RNeasy RNA cleanup kit (QIAGEN) and treated with DNA-free DNase (Ambion) according to the manufacturer's instructions. All samples had A260/A280 ratios of 1.8 to 2.0, indicating that they were of high purity (6). In order to confirm the fact that PCR products generated from cDNA templates did not result from the amplification of contaminating DNA, all PCR analyses included negative controls with RNA that had not been subjected to reverse transcription-PCR (RT-PCR).
Testing and design of primers.
Degenerate primers targeting Geobacteraceae gltA genes were designed from nucleotide sequences from the G. sulfurreducens (32), G. metallireducens, Desulfuromonas palmitatis, Pelobacter carbinolicus, and Pelobacter propionicus genomes. Preliminary sequence data from the G. metallireducens, D. palmitatis, P. carbinolicus, and P. propionicus genomes were obtained from the DOE Joint Genome Institute website (www.jgi.doe.gov). These nucleotide sequences were initially aligned in Clustal X (44) and imported into the Genetic Computer Group sequence editor (Wisconsin Package, version 1.0; Madison, WI). This alignment was then examined, and conserved regions were targeted for primer design.
The following primer set was used to amplify a 750-bp fragment of the gltA gene from genomic DNA extracted from pure cultures of P. massiliensis and G. psychrophilus, Plymouth groundwater, Bemidji sediment, and the current-harvesting anode: CS100F (5′ TCARTSTATTGGCGGTGC 3′) and CS850R (5′ CGGAGTCTTSAIGCAGAACTC 3′). The gltA gene was amplified from DNA extracted from Rifle groundwater with CS18F (5′ CTCGCGACATCC GGAGTCT 3′) and CS821R (5′ TGTCCGGCGTTCAGGGTAT 3′).
Primers were designed for TaqMan PCR analysis according to the manufacturer's instructions (amplicon size, 100 to 250 bp), and representative products from each of these primer sets were verified by sequencing. For pure-culture quantitative RT-PCR analyses of G. sulfurreducens, the following TaqMan primers that targeted G. sulfurreducens gltA, rpoD, recA, and proC transcripts were designed from the G. sulfurreducens genome sequence (32): CS418F (5′ TGC TCTCCGTCGGTATCCTT 3′) and CS552R (5′ AGATGAATGCGGCGATGAC 3′) amplified the citrate synthase gene, and the constitutively expressed housekeeping genes recA, proC, and rpoD were amplified with recA660F (5′ GTGAAGGTGGTCAAGAACAAGGT 3′) and recA737R (5′ GGAAAT GCCCTCACCGTAGTAA 3′), proC2F (5′ CATGCTGAAGGGAAGCACTCT 3′) and proC77R (5′ GGCCAGCAGCCCTTTGAT 3′), and rpoD1132F (5′ TCATGAAGGCGGTGGACAA 3′) and rpoD1210R (5′ GCCTGTCGA ATCCACCAAGT 3′), respectively.
Before TaqMan primers could be designed to quantify environmental RNA transcripts, it was necessary to construct cDNA libraries using primer sets that targeted a larger region of the gene (400 to 800 bp). The gltA gene was amplified from cDNA generated from Plymouth groundwater, Bemidji sediment, and the current-harvesting anode with CS100F/CS850R, and the primer set CS18F/CS821R was used to amplify cDNA from Rifle groundwater. The proC gene was also amplified from cDNA generated from RNAs extracted from Rifle groundwater with proC75f (5′ ATWGGIGGIGGIAATATGGC 3′) and proC471r (5′ TCCCCACCAGGTCGAACA 3′). The dominant sequences within each cDNA library constructed from these sites were then used to design specific Geobacteraceae primers suitable for quantitative RT-PCR analyses.
TaqMan primers specific for Geobacteraceae gltA genes in Rifle and Plymouth groundwater were amplified with CS375F (5′ AACAAGATGACCGCCTGG G 3′) and CS598R (5′ TCGTGGTCGGAGTGGAGAAT 3′). Primers specific for Geobacteraceae gltA mRNA on the freshwater electrode included nanfwcs352F (5′ ACCACCTACGAGGATGCCTG 3′) and nanfwcs452R (5′ TCGGGAGCGATGATGTTTC 3′), and proC cDNA from Rifle groundwater was amplified with proC100F (5′ AGATACGGCATACAGGTTAC 3′) and proC250R (5′ ACGCCCGCCATGATGGAGATG 3′).
Quantification of gene expression by TaqMan PCR.
A DuraScript enhanced avian RT single-strand synthesis kit (Sigma) was used to generate cDNA from gltA, recA, proC, and rpoD transcripts as previously described (23). Once the appropriate cDNA fragments were generated by RT-PCR, TaqMan PCR amplification and detection were performed with the GeneAmp 5700 sequence detection system (PE Biosystems, Foster City, CA). Optimal TaqMan PCR conditions were determined by using the manufacturer's guidelines. Each PCR mixture consisted of a total volume of 50 μl and contained 3 to 9 μl of the appropriate primers (stock concentration, 5 μM), 5 μl 10× SYBR green PCR buffer (PE Biosystems, Foster City, CA), 6 μl MgCl2 solution (25 mM; PE Biosystems), 4 μl deoxynucleoside triphosphate mix (2.5 mM; PE Biosystems), 0.5 U AmpErase uracil-N-glycosylase (UNG; PE Biosystems), and 0.25 U AmpliTaq Gold (PE Biosystems). The thermal cycling parameters consisted of a UNG activation step at 50°C for 2 min, a denaturation step at 95°C for 10 min, and 40 cycles of 95°C for 15 s and 63°C for 1 min. Standard curves were constructed as previously described (23) and covered a range of ∼8 orders of magnitude. To verify amplification and correct amplicon sizes, aliquots from real-time PCR were examined in an ethidium bromide-stained 1% agarose gel.
Amplification of gltA and 16S rRNA RT-PCR products and construction of cDNA libraries.
The optimal amplification conditions for CS100F/850R and CS18F/821R were determined in a gradient thermal cycler (MJ Research Inc., Waltham, MA). The amplification parameters included an initial denaturation step of 95°C for 5 min; 20 cycles of 95°C for 1 min, 55°C (−0.5°C per cycle) for 1.5 min, and 72°C for 1.5 min; another 20 cycles of 95°C for 1 min, 50°C for 1.5 min, and 72°C for 1.5 min; and a final extension at 72°C for 10 min.
In an attempt to reduce PCR bias and to provide the most reliable evaluation of microbial community dynamics, more than one bacterial primer set targeting different regions of 16S rRNA was used. The primer sets included 8F (17) with 519R (26) and 338F (2) with 907R (26). Once the appropriate 16S cDNA gene fragments were generated by RT-PCR, the following PCR parameters were used: an initial denaturation step at 95°C for 5 min, followed by 35 cycles of 95°C (45 s), 50°C (1 min), and 72°C (1 min) and a final extension step at 72°C for 7 min. To ensure sterility, the PCR mixtures were exposed to UV radiation for 8 min prior to the addition of cDNA template and Taq polymerase.
16S rRNA and gltA PCR products were purified with a gel extraction kit (QIAGEN), and clone libraries were constructed with a TOPO TA cloning kit, version M (Invitrogen, Carlsbad, CA), according to the manufacturer's instructions. One hundred plasmid inserts from each cDNA clone library were then sequenced with the M13F primer at the University of Massachusetts Sequencing Facility.
Phylogenetic analysis.
16S rRNA and gltA gene sequences were compared to the GenBank nucleotide and protein databases using the BLASTn and BLASTx algorithms (1). Nucleotide and amino acid sequences for each gene were initially aligned in Clustal X (44) and imported into the Genetic Computer Group sequence editor (Wisconsin Package, version 1.0; Madison, WI). These alignments were then imported into Clustal W (45) and Mview (12), where similarity and identity matrices were generated.
Aligned sequences were imported into PAUP 4.0b10 (42), where phylogenetic trees were inferred. Distances and the branching order were determined and compared using maximum parsimony and distance-based algorithms (HKY85 and Jukes-Cantor). Bootstrap values were obtained from 100 replicates.
RESULTS AND DISCUSSION
gltA gene expression by Geobacter sulfurreducens in chemostats and on current-harvesting electrodes.
It has been proposed that Geobacteraceae species oxidize acetate via a complete citric acid (TCA) cycle and that the citrate synthase protein exerts significant control over flux into this process. Biochemical studies have demonstrated that citrate synthase activity levels change at various stages of cell growth and when cells are grown under different medium conditions (11). For example, the specific activity of citrate synthase was greatest when cells were grown with acetate provided as the electron donor, with fumarate as the electron acceptor, and was lowest when hydrogen was provided as the electron donor, with fumarate as the electron acceptor. Citrate synthase activity levels were also greatest during the mid-logarithmic phase of growth and started to decrease as growth rates decreased.
In order to evaluate whether transcript levels for gltA might be affected by changes in the rate of metabolism, G. sulfurreducens was grown in chemostats at dilution rates near the lowest (0.03 h−1) and highest (0.07 h−1) rates at which G. sulfurreducens can be maintained in such systems (18). Acetate served as the electron donor and was the growth-limiting factor in order to mimic conditions that might typically be found in subsurface environments. Fe(III) was the electron acceptor. As expected, the steady-state cell yields (0.166 ± 0.029 mg protein/ml [mean ± standard error; n = 3] for 0.03 h−1 and 0.171 ± 0.014 mg protein/ml for 0.07 h−1) and Fe(II) concentrations (47.03 ± 8.48 mM for 0.03 h−1 and 45.92 ± 3.46 mM for 0.07 h−1) were comparable, and steady-state acetate concentrations were below the detection limit of 5 μM.
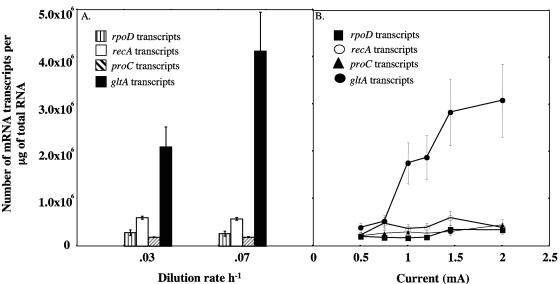
Quantitative real-time PCR studies demonstrated that the number of gltA transcripts was two times higher in the culture that was grown at 0.07 h−1 than in the 0.03 h−1 culture (Fig. 1A). In contrast, the levels of transcripts for the constitutively expressed housekeeping genes proC, recA, and rpoD remained relatively constant at both growth rates. These results suggest that levels of transcripts for gltA are related to the rates of growth and metabolism of G. sulfurreducens, and presumably other Geobacteraceae species.
FIG. 1.
Numbers of gltA, recA, rpoD, and proC transcripts expressed by G. sulfurreducens grown in chemostats under Fe(III)-reducing conditions (A) and on a current-harvesting electrode poised at +500 mV (in reference to a standard H2 electrode) (B). Each chemostat point is the average of five replicates from three separate chemostat cultures. RNAs were extracted from the electrode at 0.5 mA, 0.75 mA, 1.0 mA, 1.2 mA, 1.45 mA, and 2 mA of current, and each point is the average of triplicate determinations.
To further examine the expression of the gltA gene, G. sulfurreducens was also grown with a graphite electrode poised at +500 mV as the sole electron acceptor and with acetate (10 mM) as the electron donor. Quantitative real-time PCR indicated that gltA expression was proportional to the rate of current produced by the current-harvesting electrode; as the current went up, the transcription of the gltA gene increased (Fig. 1B). At 2 mA of current, the number of gltA transcripts/μg of total RNA was 7.8 times greater than that at 0.5 mA, whereas the levels of transcripts for the housekeeping control genes remained constant.
Phylogenetic analysis of 16S and gltA cDNA clone libraries.
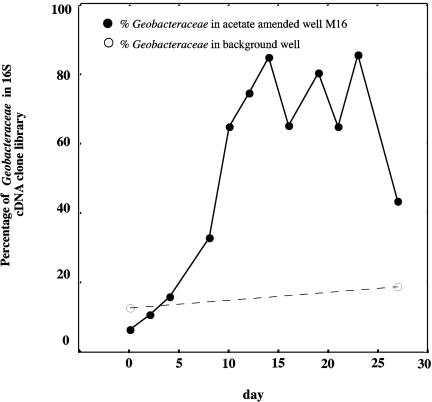
The phylogeny of the metabolically active members of the microbial community, as determined from analyses of 16S rRNA transcripts, as well as the presence and phylogeny of gltA transcripts, was evaluated in four environments in which Geobacteraceae were expected to be important components of the microbial community. The addition of acetate to a U(VI)-contaminated groundwater site in Rifle, Colorado, resulted in a dramatic increase in the number of Geobacteraceae 16S rRNA transcripts expressed in the groundwater, i.e., only 6.5% of the 16S rRNA bacterial transcripts were most similar to Geobacteraceae sequences on day 0, compared to 85% of the transcripts on day 14 (Fig. 2). It took approximately 7 days for acetate to reach well M16; the acetate concentration was 3.88 mM on day 7. Exposure to acetate resulted in a significant increase in the proportion of Geobacteraceae in the bacterial community; on day 8, Geobacteraceae accounted for 33% of the community. As a control, RNA was extracted from groundwater collected from a well that had not been exposed to acetate at the beginning (day 0) and end (day 27) of the experiment. The increase in Geobacteraceae sequences over time that was associated with the acetate-amended wells was not observed in these nonamended control samples, i.e., Geobacteraceae accounted for only 12.3% and 18.9% of the bacterial population on days 0 and 27, respectively.
FIG. 2.
Geobacteraceae 16S rRNA transcripts in Rifle groundwater expressed as percentages of the total 16S rRNA transcripts for all bacteria.
At the Plymouth, Massachusetts, site, where the groundwater is impacted by the CMA road deicer, Geobacteraceae 16S rRNA transcripts accounted for 77.6% of the community in the groundwater when groundwater acetate levels were at their peak (68.31 μM) in June, compared to only 23.8% of the community in October, when the amount of acetate in the groundwater had dramatically decreased (2.99 μM) (Table 1). Geobacteraceae 16S rRNA transcripts were also abundant in sediments collected from the Fe(III) reduction zone of a petroleum-contaminated aquifer. Geobacteraceae transcripts accounted for 26% of the transcripts in the Fe(III) reduction zone, in contrast to only 2.5% of the sequences from the Fe(III) reduction zone of nearby pristine sediments (data not shown). Furthermore, 16S cDNA analysis of the surface of a current-harvesting electrode from a sediment fuel cell deployed in freshwater sediments indicated that Geobacteraceae were the most metabolically active members of the microbial community; Geobacteraceae 16S rRNA transcripts accounted for 61.3% of the bacterial 16S rRNA on the current-harvesting anode, which generated up to 1.45 mA of current, compared to only 12% on the surface of a noncurrent control electrode (Table 1).
TABLE 1.
Number of gltA transcripts and percentage of Geobacteraceae 16S rRNA transcripts in cDNA clone libraries assembled from RNAs extracted from groundwater samples collected from the CMA-exposed site in Plymouth, Mass., and current-harvesting electrode deployed in a freshwater pond in Nantucket, Mass.
| Source | No. of gltA transcripts per μg of total RNAa | % Geobacteraceae in bacterial 16S rRNA clone library | Acetate concn (μM) |
|---|---|---|---|
| CMA-exposed site (Plymouth), June 7 | 2.02 × 106 ± 2.31 × 105 | 77.6 | 68.31 |
| CMA-exposed site (Plymouth), October 6 | 7.2 × 103 ± 1.65 × 102 | 23.8 | 2.99 |
| Current-harvesting freshwater electrode | 1.57 × 106 ± 6.55 × 105 | 61.3 | NAb |
| Noncurrent control freshwater electrode | 0 | 12.0 | NAb |
The numbers of gltA transcripts were determined by triplicate experiments (n = 3). Data are means ± standard errors of the means.
NA, not applicable.
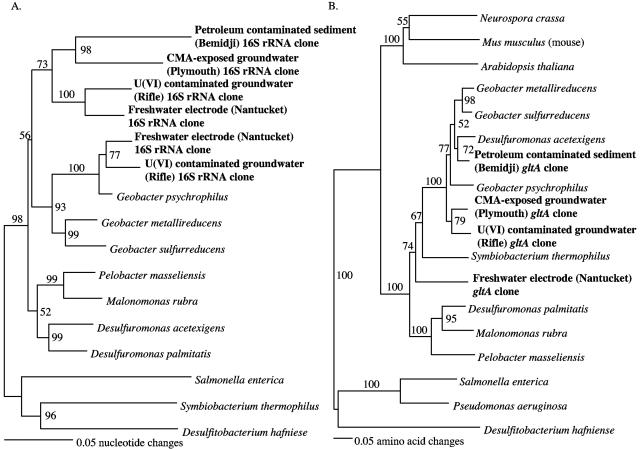
The Geobacteraceae 16S rRNA transcripts detected at all four of these sites clustered within the freshwater Geobacter clade of the family (Fig. 3A). Geobacteraceae 16S rRNA sequences detected in the U(VI)-contaminated groundwater were 86 to 100% identical to each other. The diversity among the Geobacteraceae 16S rRNA sequences detected in the CMA-exposed groundwater, petroleum-contaminated sediment, and freshwater anode was similar to that for the Geobacteraceae detected in the U(VI)-contaminated groundwater, with 89 to 100%, 80 to 99%, and 81 to 98% identities for the CMA, petroleum, and electrode studies, respectively. When comparisons were made across all four sites, similar amounts of variation were observed; Geobacteraceae sequences were 88 to 95% identical to each other and 87 to 93% identical to those of G. sulfurreducens.
FIG. 3.
Phylogenetic analysis of Geobacteraceae 16S rRNA and citrate synthase (gltA) mRNA transcript sequences amplified from the U(VI)-contaminated site at Rifle, Colorado, the Fe(III) reduction zone of a petroleum-contaminated aquifer in Bemidji, Minnesota, a runoff recharge pool from a highway in Plymouth, Massachusetts, that has been exposed to CMA, and a freshwater sediment fuel cell deployed in Nantucket, Massachusetts. (A) 16S rRNA nucleotide sequence comparisons; (B) citrate synthase amino acid sequence comparisons. Phylogenetic trees were constructed with the Jukes-Cantor algorithm. Desulfitobacterium hafniense was used as the outgroup for both trees, and bootstrap values were based on 100 replicates.
Citrate synthase genes that phylogenetically clustered within the Geobacter clade of the Geobacteraceae were amplified from all four sites (Fig. 3B). The phylogeny of the citrate synthase genes is consistent with the 16S rRNA gene sequence phylogeny, with the exception of the citrate synthase from Desulfuromonas acetexigens, which falls within the freshwater Geobacter clade of the family, as previously noted (11). Citrate synthase amino acid sequences from the as yet uncultured Geobacteraceae that were detected in the four environments had 63 to 90% identity and 73 to 93% similarity to each other and were 78 to 90% similar to G. sulfurreducens sequences. Like the Geobacteraceae available in pure culture, the environmental citrate synthase genes were more similar to nucleus-encoded eukaryotic citrate synthase sequences than they were to all other known prokaryotic citrate synthase genes, except the uncultivable compost organism Symbiobacterium thermophilum (46).
gltA expression patterns in the environment.
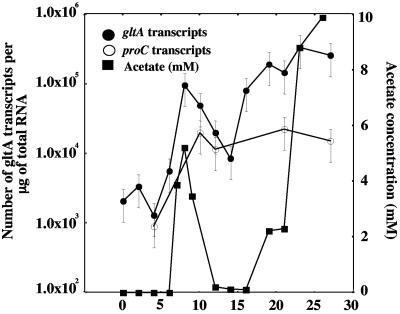
In order to evaluate gltA expression patterns in the environment, gltA primers that specifically targeted the Geobacteraceae citrate synthase genes from each site were designed for TaqMan RT-PCR analysis. There was a remarkable correspondence between acetate levels in the groundwater and levels of transcripts for gltA during the in situ uranium bioremediation field study at the Old Rifle site. As acetate concentrations rose, gltA transcript levels increased (Fig. 4). A significant increase in the number of gltA transcripts was not observed until day 8, when acetate reached the well. When acetate levels fell due to a rain event that diluted the acetate with recharge water and prevented the injection of acetate for several days, the level of gltA transcripts decreased, and then gltA transcript levels increased concurrent with a renewed increase in acetate over time. In contrast to the gltA gene, the number of transcripts for the constitutively expressed housekeeping gene proC remained relatively constant (Fig. 4).
FIG. 4.
Numbers of Geobacteraceae gltA and proC mRNA transcripts in the groundwater relative to acetate concentrations at the U(VI)-contaminated site (Rifle) after acetate was injected into the subsurface to stimulate metal reduction. Each point is the average of triplicate determinations.
Geobacteraceae gltA transcripts were also significantly higher at the CMA-exposed site in June than in October, reflecting the difference in acetate availability (Table 1). Quantitative RT-PCR analysis of mRNA extracted from the surfaces of electrodes harvesting electricity from freshwater sediments demonstrated that Geobacteraceae expressed the gltA gene during growth on the current-harvesting electrode, whereas gltA mRNA transcripts could not be detected on control electrodes emplaced in sediments but not connected to a cathode (Table 1).
Implications.
The results of this study suggest that the ability to estimate levels of expression of the citrate synthase gene from Geobacteraceae (gltA) may be a useful tool for assessing the activity of these organisms in sedimentary environments. The eukaryotic nature of the citrate synthase gene suggests that few other prokaryotes expected to be found in anoxic subsurface environments will have similar citrate synthase sequences, making it easier to design primers to specifically monitor the metabolism of Geobacteraceae. Pure culture studies suggest that levels of gltA transcripts are directly related to rates of metabolism and electron transfer to Fe(III) and electrodes. Furthermore, in situ levels of gltA transcripts in Geobacteraceae-dominated environments can be monitored, and the finding that transcript levels fluctuate in accordance with changes in acetate availability in subsurface environments suggests that gltA expression patterns are related to in situ metabolic activity. These results demonstrate that monitoring gene expression in subsurface environments can provide an indication not only of the metabolic state of Geobacteraceae (23), but also of their rates of metabolism. Such information is invaluable for making decisions about the concentrations of various nutrients and electron donors that should be added to best stimulate in situ bioremediation. This information will also aid in the development of in silico models that can predict the metabolic responses of microorganisms to various bioremediation or energy-harvesting strategies. Such studies have the potential to change the design of bioremediation and energy harvesting from an empirical practice to more of a science (28).
Acknowledgments
We thank the DOE Joint Genome Institute for providing us with preliminary sequence data for Geobacter metallireducens, Desulfuromonas acetoxidans, Pelobacter carbinolicus, and Pelobacter propionicus.
This research was supported by the Office of Science (BER), U.S. Department of Energy, with funds from the Natural and Accelerated Bioremediation Research (NABIR) program (grant DE-FG02-97ER62475), and by the Genomes to Life Program (cooperative agreement no. DE-FC02-02ER63446).
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson, R. T., and D. R. Lovley. 1998. Degradation of naphthalene and benzene under Fe(III)-reducing conditions in petroleum-contaminated aquifers. Bioremed. J. 3:121-135. [Google Scholar]
- 4.Anderson, R. T., J. Rooney-Varga, C. V. Gaw, and D. R. Lovley. 1998. Anaerobic benzene oxidation in the Fe(III)-reduction zone of petroleum-contaminated aquifers. Environ. Sci. Technol. 32:1222-1229. [Google Scholar]
- 5.Anderson, R. T., H. A. Vrionis, I. Ortiz-Bernad, C. T. Resch, A. D. Peacock, R. Dayvault, S. Marutzky, D. R. Metzler, K. Karp, M. Lowe, D. C. White, P. E. Long, and D. R. Lovley. 2003. Stimulated in situ activity of Geobacter species to remove uranium from the groundwater of a uranium-contaminated aquifer. Appl. Environ. Microbiol. 69:5884-5891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1997. Current protocols in molecular biology. John Wiley and Sons, Inc., New York, N.Y.
- 7.Baedecker, M. J., D. I. Siegel, P. Bennett, and I. M. Cozzarelli. 1989. The fate and effects of crude oil in a shallow aquifer. I. The distribution of chemical species and geochemical facies, p. 13-20. In G. E. Mallard and S. E. Ragone (ed.), U.S. Geological Survey Water Resources Division report 88-4220. U.S. Geological Survey, Reston, Va.
- 8.Balch, W. E., G. E. Fox, L. J. Magrum, C. R. Woese, and R. S. Wolfe. 1979. Methanogens: reevaluation of a unique biological group. Microbiol. Rev. 43:260-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bond, D. R., D. E. Holmes, L. M. Tender, and D. R. Lovley. 2002. Electrode-reducing microorganisms that harvest energy from marine sediments. Science 295:483-485. [DOI] [PubMed] [Google Scholar]
- 10.Bond, D. R., and D. R. Lovley. 2003. Electricity production by Geobacter sulfurreducens attached to electrodes. Appl. Environ. Microbiol. 69:1548-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bond, D. R., C. Nesbø, A. Lopez, T. Mester, F. L. Collart, and D. R. Lovley. 2005. A eukaryotic-like citrate synthase with novel biochemical properties in the Geobacteraceae. Appl. Environ Microbiol. 71:3858-3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown, N. P., C. Leroy, and C. Sandaer. 1998. A web compatible database search or multiple alignment viewer. Bioinformatics 14:380-381. [DOI] [PubMed] [Google Scholar]
- 13.Chin, K.-J., A. Esteve-Núñez, C. Leang, and D. R. Lovley. 2004. Direct correlation between rates of anaerobic respiration and levels of mRNA for key respiratory genes in Geobacter sulfurreducens. Appl. Environ. Microbiol. 70:5183-5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Church, P., D. Armstrong, G. Granato, V. Stone, K. Smith, and P. Provencher. 1996. Effectiveness of highway-drainage systems in preventing contamination of ground water by road salt, route 25, southeastern Massachusetts—description of study area, data collection programs, and methodology. U.S. Geological Survey, Reston, Va.
- 15.Coppi, M. V., C. Leang, S. J. Sandler, and D. R. Lovley. 2001. Development of a genetic system for Geobacter sulfurreducens. Appl. Environ. Microbiol. 67:3180-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cozzarelli, I. M., R. P. Eaganhouse, and M. J. Baedecker. 1990. Transformation of monoaromatic hydrocarbons to organic acids in anoxic groundwater environment. Environ. Geol. Water Sci. 16:135-141. [Google Scholar]
- 17.Eden, P. E., T. M. Schmidt, R. P. Blakemore, and N. R. Pace. 1991. Phylogenetic analysis of Aquaspirillum magnetotacticum using polymerase chain reaction-amplified 16S rRNA-specific DNA. Int. J. Syst. Bacteriol. 41:324-325. [DOI] [PubMed] [Google Scholar]
- 18.Esteve-Núñez, A., M. M. Rothermich, M. Sharma, and D. R. Lovley. 2005. Growth of Geobacter sulfurreducens under nutrient-limiting conditions in continuous culture. Environ. Microbiol. 7:641-648. [DOI] [PubMed] [Google Scholar]
- 19.Galushko, A. S., and B. Schink. 2000. Oxidation of acetate through reactions of the citric acid cycle by Geobacter sulfurreducens in pure culture and in syntrophic coculture. Arch. Microbiol. 174:314-321. [DOI] [PubMed] [Google Scholar]
- 20.Holmes, D. E., D. R. Bond, R. A. O'Neil, C. E. Reimers, L. R. Tender, and D. R. Lovley. 2004. Microbial communities associated with electrodes harvesting electricity from a variety of aquatic sediments. Microb. Ecol. 48:178-190. [DOI] [PubMed] [Google Scholar]
- 21.Holmes, D. E., K. T. Finneran, R. A. O'Neil, and D. R. Lovley. 2002. Enrichment of members of the family Geobacteraceae associated with stimulation of dissimilatory metal reduction in uranium-contaminated aquifer sediments. Appl. Environ. Microbiol. 68:2300-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holmes, D. E., K. P. Nevin, and D. R. Lovley. 2004. Comparison of 16S rRNA, nifD, recA, rpoB, and fusA genes within the family Geobacteraceae. Int. J. Syst. Evol. Microbiol. 54:1591-1599. [DOI] [PubMed] [Google Scholar]
- 23.Holmes, D. E., K. P. Nevin, and D. R. Lovley. 2004. In situ expression of Geobacteraceae nifD in subsurface sediments. Appl. Environ. Microbiol. 70:7251-7259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hult, M. F. 1984. Ground-water contamination by crude oil at Bemidji, Minnesota, research site. U.S. Geological Survey toxic waste-ground-water contamination study. U.S. Geological Survey Water Resources Investigations report 84-4188. U.S. Geological Survey, Reston, Va.
- 25.Johnson, J. P., D. E. Holmes, K. P. Nevin, and D. R. Lovley. 2005. Abstr. 105th Gen. Meet. Am. Soc. Microbiol., abstr. N-043. American Society for Microbiology, Washington, D.C.
- 26.Lane, D. L., B. Pace, G. J. Olsen, D. Stahl, M. L. Sogin, and N. R. Pace. 1985. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analysis. Proc. Natl. Acad. Sci. USA 82:6955-6959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leang, C., M. V. Coppi, and D. R. Lovley. 2003. OmcB, a c-type polyheme cytochrome, involved in Fe(III) reduction in Geobacter sulfurreducens. J. Bacteriol. 185:2096-2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lovley, D. R. 2003. Cleaning up with genomics: applying molecular biology to bioremediation. Nat. Microbiol. 1:35-44. [DOI] [PubMed] [Google Scholar]
- 29.Lovley, D. R., M. J. Baedecker, D. J. Lonergan, I. M. Cozzarelli, E. J. P. Phillips, and D. I. Siegel. 1989. Oxidation of aromatic contaminants coupled to microbial iron reduction. Nature 339:297-299. [Google Scholar]
- 30.Lovley, D. R., and F. H. Chapelle. 1995. Deep subsurface microbial processes. Rev. Geophys. 33:365-381. [Google Scholar]
- 31.Lovley, D. R., D. E. Holmes, and K. P. Nevin. 2004. Dissimilatory Fe(III) and Mn(IV) reduction. Adv. Microb. Physiol. 49:219-286. [DOI] [PubMed] [Google Scholar]
- 32.Methé, B. A., K. E. Nelson, J. A. Eisen, I. T. Paulsen, W. Nelson, J. F. Heidelberg, D. Wu, M. Wu, N. Ward, M. J. Beanan, R. J. Dodson, R. Madupu, L. M. Brinkac, S. C. Daugherty, R. T. DeBoy, A. S. Durkin, M. Gwinn, J. F. Kolonay, S. A. Sullivan, D. H. Haft, J. Selengut, T. M. Davidsen, N. Zafar, O. White, B. Tran, C. Romero, H. A. Forberger, J. Weidman, H. Khouri, T. V. Feldblyum, T. R. Utterback, S. E. Van Aken, D. R. Lovley, and C. M. Fraser. 2003. The genome of Geobacter sulfurreducens: insights into metal reduction in subsurface environments. Science 302:1967-1969. [DOI] [PubMed] [Google Scholar]
- 33.Nevin, K. P., D. E. Holmes, T. L. Woodard, E. S. Hinlein, D. W. Ostendorf, and D. R. Lovley. 2005. Geobacter bemidjiensis sp. nov. and Geobacter psychrophilus sp. nov., two novel Fe(III)-reducing subsurface isolates. Int. J. Syst. Evol. Microbiol. 55:1667-1674. [DOI] [PubMed] [Google Scholar]
- 34.Nottingham, P. M., and R. E. Hungate. 1969. Methanogenic fermentation of benzoate. J. Bacteriol. 98:1170-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ortiz-Bernad, I., R. Anderson, H. Vrionis, and D. Lovley. 2004. Vanadium respiration by Geobacter metallireducens: novel strategy for in situ removal of vanadium from groundwater. Appl. Environ. Microbiol. 70:3091-3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ostendorf, D. 1997;-2004. Series of monthly progress reports. Highway deicing agent impacts on soil and groundwater quality. Prepared under ISA 7721 and ISA 9775. Massachusetts Highway Department, Boston, Mass.
- 37.Petrie, L., N. N. North, S. L. Dollhopf, D. Balkwill, and J. E. Kostka. 2003. Enumeration and characterization of iron(III)-reducing microbial communities from acidic subsurface sediments contaminated with uranium(VI). Appl. Environ. Microbiol. 69:7467-7479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reimers, C. E., L. M. Tender, S. Fertig, and W. Wang. 2001. Harvesting energy from the marine sediment-water interface. Environ. Sci. Technol. 35:192-195. [DOI] [PubMed] [Google Scholar]
- 39.Roling, W. F. M., B. M. van Breukelen, B. L. Braster, and H. W. van Verseveld. 2001. Relationships between microbial community structure and hydrochemistry in a landfill leachate-polluted aquifer. Appl. Environ. Microbiol. 67:4619-4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rooney-Varga, J. N., R. T. Anderson, J. L. Fraga, D. Ringelberg, and D. R. Lovley. 1999. Microbial communities associated with anaerobic benzene degradation in a petroleum-contaminated aquifer. Appl. Environ. Microbiol. 65:3056-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Snoeyenbos-West, O. L., K. P. Nevin, R. T. Anderson, and D. R. Lovley. 2000. Enrichment of Geobacter species in response to stimulation of Fe(III) reduction in sandy aquifer sediments. Microb. Ecol. 39:153-167. [DOI] [PubMed] [Google Scholar]
- 42.Swofford, D. L. 1998. PAUP*. Phylogenetic analysis using parsimony (*and other methods), version 4. Sinauer Associates, Sunderland, Mass.
- 43.Tender, L. M., C. E. Reimers, H. A. Stecher, D. E. Holmes, D. R. Bond, D. A. Lowy, K. Pilobello, S. J. Fertig, and D. R. Lovley. 2002. Harnessing microbially generated power on the seafloor. Nat. Biotechnol. 20:821-825. [DOI] [PubMed] [Google Scholar]
- 44.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ueda, K., A. Yamashita, J. Ishikawa, M. Shimada, T. Watsuji, K. Morimura, H. Ikeda, M. Hattiori, and T. Beppu. 2004. Genome sequence of Symbiobacterium thermophilum, an uncultivable bacterium that depends on microbial commensalism. Nucleic Acids Res. 32:4937-4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.White, D. 2000. The physiology and biochemistry of prokaryotes, 2nd ed. Oxford University Press, New York, N.Y.