Abstract
In the past few years many waterborne outbreaks related to Cryptosporidium have been described. Current methods for detection of Cryptosporidium in water for the most part rely on viability assays which are not informative concerning the infectivity of oocysts. However, for estimation of the risk of infection with Cryptosporidium this information is required. For environmental samples the oocyst counts are often low, and the oocysts have been exposed to unfavorable conditions. Therefore, determination of the infectivity of environmental oocysts requires an assay with a high level of sensitivity. We evaluated the applicability of in vitro cell culture immunofluorescence assays with HCT-8 and Caco-2 cells for determination of oocyst infectivity in naturally contaminated water samples. Cell culture assays were compared with other viability and infectivity assays. Experiments with Cryptosporidium oocysts from different sources revealed that there was considerable variability in infectivity, which was illustrated by variable 50% infective doses, which ranged from 40 to 614 oocysts, and the results indicated that not only relatively large numbers of fresh oocysts but also aged oocysts produced infection in cell cultures. Fifteen Dutch surface water samples were tested, and the cell culture immunofluorescence assays were not capable of determining the infectivity for the low numbers of naturally occurring Cryptosporidium oocysts present in the samples. A comparison with other viability assays, such as the vital dye exclusion assay, demonstrated that surrogate methods overestimate the number of infectious oocysts and therefore the risk of infection with Cryptosporidium. For accurate risk assessment, further improvement of the method for detection of Cryptosporidium in water is needed.
Cryptosporidium is one of the important waterborne causative agents of gastrointestinal illness in humans (6, 10). More than 50 outbreaks of cryptosporidiosis associated with either contaminated drinking water or recreational water have been reported worldwide since 1983 (20). In immunocompetent individuals cryptosporidial diarrhea is usually self-limiting, lasting for about 3 weeks, but infected immunocompromised humans may develop severe and potentially fatal illness (5). At present, no consistently effective, approved therapeutic agent with anticryptosporidial activity is available.
Cryptosporidium oocysts are ubiquitous in surface water used for recreation or drinking water production (9, 21). They are extremely resistant to chlorination at the concentrations commonly used for drinking water and swimming pool water disinfection (13). The number of oocysts in finished water is usually low but relevant for public health, and the concentrations are below the detection limit of the method used. Therefore, Dutch drinking water legislation requires drinking water companies to perform a quantitative risk assessment for Cryptosporidium, based on the concentration of oocysts in the source water and the efficiency of drinking water treatment processes. The annual risk of infection for consumers of drinking water should not exceed one infection per 10,000 persons. Only infectious oocysts are a potential health risk and should be included in risk estimations.
Human volunteer studies provide the most reliable information on oocyst infectivity, but such studies are not feasible and are considered unethical for routine use. Mouse assays are the most commonly used animal models and are considered the “gold standard” for assessing Cryptosporidium infectivity. They are, however, expensive, time-consuming, and not widely applicable, and they may also be considered unethical. Moreover, data generated with mouse infectivity models vary considerably, both in different experiments (20) and between different mouse assays (26), and Cryptosporidium hominis oocysts do not efficiently infect standard mouse infectivity models (17).
The method that is widely used for detection of Cryptosporidium in water (1) includes concentration by filtration, purification by immunomagnetic separation, and immunofluorescence (IF) assay microscopy, but it cannot discriminate between infectious and noninfectious oocysts. Addition of vital dye staining (4) or in vitro excystation (19) provides information on oocyst viability, but the results do not always correlate with the outcome of in vivo (15) and in vitro (3) infectivity assays.
Cell culture assays, which have been used to study the life cycle and infection mechanism of Cryptosporidium and to test the efficacy of therapeutic agents for decades, have more recently been developed into useful tools for determination of oocyst infectivity (28). In vitro released sporozoites invade cultured cells and produce clusters of foci of infection which can be detected microscopically after labeling with specific fluorescent antibodies (23, 24). A combination of cell culturing with PCR techniques using specific primers and probes enables specific determination of Cryptosporidium parvum infectivity (7, 8).
We evaluated the applicability of cell culture infectivity assays with HCT-8 and Caco-2 cells for naturally contaminated environmental samples. The oocyst counts in such samples are often low, and the oocysts have been exposed to unfavorable environmental conditions. As a result, just a few oocysts may be capable of infecting human intestinal cells in vitro, and an assay with a high level of sensitivity is required. The sensitivity of the cell culture assays was assessed by using fresh and aged oocysts from different sources; furthermore, the relationship of these assays to other viability and infectivity methods, such as vital dye exclusion and animal infectivity, was evaluated.
MATERIALS AND METHODS
Cryptosporidium oocysts.
C. parvum oocysts were purchased from the National Institute of Veterinary Research (Brussels, Belgium), Waterborne Inc. (New Orleans, LA), and Moredun Animal Health (Moredun Scientific Limited, Penicuik, Scotland, United Kingdom) and were provided by the University of Alberta (Edmonton, Canada). We used suspensions with an excystation rate (19) of at least 70%.
Cell culture.
For routine culturing of Caco-2 (ATCC HTB-37; American Type Culture Collection, Manassas, VA) and HCT-8 (ATCC CCL-244) cells, Dulbecco modified Eagle medium containing 25 mM HEPES and 4,500 mg · liter−1 glucose and supplemented with 10% heat-inactivated fetal bovine serum, 0.1% modified Eagle medium nonessential amino acids, 0.1% l-glutamine, and 50 μg · ml−1 gentamicin (referred to below as culture medium) was used (Dulbecco modified Eagle medium and all supplements were obtained from GibcoBRL, Life Technologies, Paisley, Scotland, United Kingdom). Cells were grown in 75-cm2 culture flasks (Corning Costar, Corning, NY) in a 5% CO2 atmosphere at 37°C. Caco-2 cells were passaged once a week, and HCT-8 cells were passaged every 3 to 4 days.
For infectivity studies the cells were grown in culture medium on Lab-Tek II multichamber slides (Nunc, Life Technologies, Paisley, Scotland, United Kingdom). Prior to seeding with approximately 1 × 105 HCT-8 cells or approximately 5 × 104 Caco-2 cells, chambers were filled with 350 μl prewarmed (37°C) culture medium and slides were placed in a 5% CO2 incubator at 37°C for 30 to 60 min to enhance the attachment and growth of the cells. Cells grew to 60 to 70% confluence within 24 to 36 h for HCT-8 cells and within 7 days for Caco-2 cells.
Infection of cell monolayers.
Oocysts were pretreated by incubation in acidified Hanks' balanced salt solution (HBSS) (pH 2.7) for 1 h at 37°C and washed with HBSS (GibcoBRL). They were subsequently incubated in freshly prepared 1% (wt/vol) bovine bile (Sigma-Aldrich Chemie, Steinheim, Germany) and 0.44% (wt/vol) sodium bicarbonate (Merck, Darmstadt, Germany) for 30 min at 37°C, washed with HBSS, and finally resuspended in 100 μl HBSS (4, 19, 22).
Before inoculation, cell monolayers were carefully rinsed with Dulbecco's phosphate-buffered saline (PBS) (GibcoBRL), and then 350 μl fresh prewarmed culture medium was added. Inoculated slides were incubated at 37°C in a 5% CO2 incubator. The infection was stopped 72 h postinoculation by removal of the culture medium and fixation with 100% methanol for 10 min at room temperature.
Detection of infection.
Each infected chamber was treated with 400 μl blocking reagent (Boehringer, Mannheim, Germany) for 30 min at room temperature in the dark. Slides were subsequently stained with 75 μl SporoGlo rat polyclonal antisporozoite antibody (Waterborne Inc., New Orleans, LA) diluted 1:20 in PBS (0.01 M, pH 7.2) and 75 μl of a fluorescein isothiocyanate-labeled monoclonal antibody (Cryptosporidium and Giardia staining reagent without Evans blue; Cellabs Diagnostics, Brookvale, Australia) diluted 1:5 in PBS in each chamber. The slides were incubated at 37°C for 1 h, washed twice with PBS, dried with a medium warm hairdryer, mounted with DABCO-glycerol mounting medium, and sealed with colorless nail polish. Stained slides were screened for the presence of foci of infection at a magnification of ×250 using epifluorescence microscopy (Zeiss Axioskop; Carl Zeiss, Jena, Germany).
Comparison of different Cryptosporidium oocyst lots.
Cryptosporidium oocyst suspensions were diluted in PBS to obtain concentrations of 500, 400, 300, 200, 100, 50, 25, 10, and 5 oocysts per 100 μl. The oocysts in each dilution were enumerated in triplicate by immunofluorescence. The diluted oocyst suspensions were filtered through 1.2-μm-pore-size 13-mm-diameter membrane filters (Isopore; Millipore, Billerica, MA) and stained with 70 μl of the Cellabs monoclonal antibody at 37°C for 45 min in the dark. Subsequently, the filters were rinsed with PBS, mounted on slides with DABCO-glycerol mounting medium, covered with coverslips, and examined by epifluorescence microscopy at a magnification of ×250.
To study oocyst infectivity, four to eight replicates of each dilution were pretreated, inoculated onto HCT-8 or Caco-2 cell monolayers, incubated, stained, and examined as described above.
Fifty percent infective doses (ID50s) (the numbers of oocysts that infected 50% of exposed cell cultures or animals) were calculated by using the logit response model (11). The parameters for the logit model were calculated by regression analysis using the least-squares method.
Comparison of cell culture and mouse assays.
Five-day-old neonatal CD-1 mice were infected with C. parvum oocysts from the University of Alberta by oral administration directly into the stomach. Four cohorts of 10 mice were infected with approximately 50, 100, 200, or 400 oocysts. The exact oocyst dose was determined by IF as described above. HCT-8 or Caco-2 cell monolayers were infected with identical portions of these oocyst dilutions.
One week after infection the mice were sacrificed. The intestines were excised and homogenized in 5 ml sterile saline by using an Ultra Turrax blender (IKA Werke GmbH & Co., Staufen, Germany) at the maximum speed for 30 s. Ten microliters of each homogenate was applied to a four-well slide (Nutacon, Leimuiden, The Netherlands), dried at 37°C, fixed with 100% methanol, and stained with 12 μl of the Cellabs monoclonal antibody per well for 45 min at 37°C in the dark. Next, the slides were processed and examined as described above.
Environmental samples.
Water samples (50 to 106 liters) taken from the Rhine River at Lobith and the Meuse River at Roosteren were concentrated by using standard Envirochek filtration capsules (Pall Gelman Laboratory, Ann Arbor, MI) as described in ISO/CD 15553 (1). Concentrated samples were purified by immunomagnetic separation using the Dynal GC-Combo system (Dynal Biotech ASA, Oslo, Norway) according to the manufacturer's instructions. In brief, 1 ml of 10× SL buffer A and 1 ml of 10× SL buffer B (supplied by Dynal) were added to a 0.5-ml concentrated water sample. The final volume was adjusted to 10 ml with distilled water, and 100 μl of anti-Cryptosporidium Dynabeads and 100 μl of anti-Giardia Dynabeads were added; this was followed by incubation on a rotating mixer (25 rpm) for 1 h at room temperature. The bead-(oo)cyst complexes were collected by using a Dynal MPC-1 magnet, the supernatant was aspirated, and the complexes were resuspended in 1 ml 1× SL buffer A. The suspensions obtained were split equally into four portions; 250 μl was assayed to obtain IF counts, 250 μl was used in cell culture assays, and 500μl was sent to another laboratory for other purposes. Each portion represented 12 to 26 liters of the original sample. For cell culture infectivity assays, capturing of the bead-(oo)cyst complexes with a Dynal MPC-M magnet and careful removal of the supernatant were not followed by dissociation with 0.1 N HCl. The complexes were pretreated and used to infect Caco-2 or HCT-8 cells as described above. For each sample an infection control slide inoculated with approximately 500 oocysts of an oocyst suspension from the National Institute for Veterinary Research was assayed. For IF counting, the complexes were dissociated by incubating them in 50 μl 0.1 N HCl for 10 min at room temperature. After vigorous mixing and removal of the beads, a Dynal Spot-On slide was prepared according to the manufacturer's instructions. Slides were stained with 50 μl of the Cellabs monoclonal antibody at 37°C for 45 min in the dark, and subsequently 5 μl of propidium iodide (PI) (1 mg · ml−1 in PBS) and 5 μl DAPI (4′,6′-diamidino-2-phenylindole) (2 mg ml−1 in methanol) were added and incubated for 2 min at room temperature. The slides were then washed, processed, and examined as described above.
Ageing of Cryptosporidium oocysts in surface water.
Cryptosporidium oocysts (Moredun Animal Health) were seeded into two 250-ml Erlenmeyer flasks with 200 ml autoclaved (15 min, 121°C) river water from the Lekkanaal at a final concentration of approximately 1,000 oocysts per 100 μl and stored in the dark at 15°C with rotation at 100 rpm. Samples were taken immediately after seeding and mixing (zero time), approximately every 7 days during the next 6 weeks, and once again after storage for approximately 2 to 3 months.
Samples from flask I were tested undiluted. For the samples from flask II a dilution series (1,000, 500, 300, 100, 50, and 10 oocysts · 100 μl−1) was prepared; IF counting of the oocysts in each dilution was performed as described above for the different Cryptosporidium lots. The oocysts were also stained with 10 μl PI for 2 min at room temperature. A vital dye exclusion assay using DAPI and PI resulting in a viable count was performed as described by Campbell et al. (4). In brief, oocysts were pretreated with acidified HBSS for 1 h at 37°C, washed twice with HBSS, resuspended in 100 μl HBSS, and incubated with 10μl DAPI and 10 μl PI for 2 h at 37°C. Eight 100-μl replicates of each dilution were pretreated and used to infect Caco-2 cell monolayers as described above. The presence of foci of infection and the number of clusters of foci of infection were recorded for each well.
RESULTS
Different Cryptosporidium oocyst lots in cell culture and mouse infectivity assays.
Experiments with dilution series of Cryptosporidium oocysts from various vendors and with various lots from one vendor revealed that there was considerable variability in infectivity with HCT-8 and Caco-2 cells, which is illustrated by the variable ID50s shown in Table 1. Various experiments performed with identical lots also resulted in different ID50s. The ID50s for Moredun lot 1 oocysts were 188 with HCT-8 cells and 387 with Caco-2 cells. For Waterborne Inc. lot 1 oocysts the ID50 with HCT-8 cells was 40, whereas for lot 2 oocysts the ID50 was 107. For C. parvum oocysts from the University of Alberta no reliable ID50 could be calculated from the results of three HCT-8 experiments, whereas highly reliable, although considerably different, estimates of the ID50 with Caco-2 cells could be obtained (ID50s, 180 and 614).
TABLE 1.
ID50s for C. parvum oocysts from Moredun Animal Health, Waterborne Inc., and the University of Alberta (Canada) in cell culture and CD-1 mouse infectivity assaysa
| Oocyst source | Lot | Expt | Oocyst age (wk postproduction) | Infectivity assay | ID50 | R2 (%) |
|---|---|---|---|---|---|---|
| Moredun | 1 | 1 | 16 | HCT-8 cells | 188 | 85 |
| 2 | 2 | 15 | HCT-8 cells | 433 | 90 | |
| 1 | 3 | 17 | Caco-2 cells | 387 | 72 | |
| Waterborne | 1 | 1 | 3 | HCT-8 cells | 40 | 91 |
| 2 | 2 | 7 | HCT-8 cells | 480b | 52b | |
| 2 | 3 | 14 | HCT-8 cells | 107 | 79 | |
| 1 | 4 | 3 | Caco-2 cells | 37b | 100b | |
| Canada | 1 | 1 | 3 | HCT-8 cells | 39b | 100b |
| 1 | 2 | 8 | HCT-8 cells | 1,112b | 54b | |
| 1 | 3 | 14 | HCT-8 cells | 302b | 61b | |
| 1 | 1 | 3 | Caco-2 cells | 201b | 100b | |
| 1 | 2 | 8 | Caco-2 cells | 614 | 99 | |
| 1 | 3 | 14 | Caco-2 cells | 180 | 99 | |
| Canada | 1 | 2 | 8 | CD-1 mice | 4,195b | 19b |
| 1 | 3 | 14 | CD-1 mice | 70 | 97 |
ID50s were calculated by using the logit response model; regression analysis resulted in R2 values. ID50s with R2 values below 70% and R2 values of 100% were disregarded in comparisons.
Data disregarded in comparisons.
Replicates of dilutions appeared to produce variable levels of infection in the same experiment, which was indicated by the variable numbers of clusters of foci in a positive well (data not shown). A larger number of clusters of foci of infection was observed in positive wells with HCT-8 cells than with Caco-2 cells when the cells were infected with equal numbers of oocysts (data not shown). Regression analysis of data from some experiments resulted in low correlations (R2 values), indicating that the calculated ID50s were not reliable. For a comparison of the infectivities of oocysts having different origins with different cell lines or in different experiments, we used ID50s with correlations of 70% or more, assuming that these were reliable. Experiments that generated only two data points (R2 = 100%) were disregarded. Three experiments with neonatal CD-1 mice yielded one reliable estimate of the ID50 for C.parvum oocysts from the University of Alberta, which was 70 (Table 1).
Specificity of cell culture assays.
To test the specificity of the cell culture assays for C. parvum oocysts, an isolate of Cryptosporidium muris (Waterborne Inc.) was used. C. muris oocysts appeared to produce positive test results with both the HCT-8 and Caco-2 cell lines at doses of 50,000 to 100,000 oocysts. The supplier could, however, not guarantee 100% purity of the suspension used. Microscopic examination revealed the presence of oocysts that differed from C. muris oocysts in shape and size. These oocysts could not be typed by molecular methods due to their very low concentration.
Comparison of viability and infectivity assays.
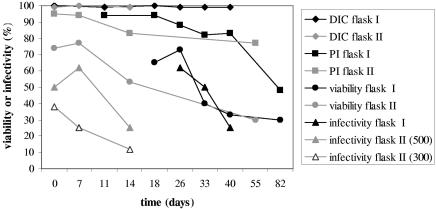
The infectivity and viability of ageing oocysts in autoclaved river water were studied for 82 days in flask I and for 55 days in flask II. At the time of inoculation, 99 to 100% of the oocysts contained sporozoites. At the end of the experiments these values had not changed. IF counting showed that the oocyst concentration in flask I remained stable for up to 82 days (approximately 1,100 oocysts · 100 μl−1). Oocyst morphology remained unchanged during this period. In flask II, which was seeded with a comparable number of oocysts, the number of intact oocysts detectable by IF rapidly declined during the first week to about 500 oocysts · 100 μl−1. The viability determined by PI exclusion after IF staining (without acid treatment) gradually declined. The PI-DAPI inclusion or exclusion after acid treatment sharply declined between days 26 and 33 in flask I and between days 7 and 14 in flask II. When applied to HCT-8 cell monolayers, the approximately 1,100 oocysts from flask I infected a gradually declining percentage of wells (Fig. 1). As a result of the initial decline, fewer oocysts from flask II were administered to the HCT-8 cells, but a trend of a gradually declining percentage of infected wells was also observed with dilutions containing both approximately 500 and approximately 300 oocysts (Fig. 1).
FIG. 1.
Ageing of C. parvum oocysts in sterile surface water at 15°C. Differential interference contrast microscopy (DIC), propidium iodide exclusion without acid treatment (PI), and the percentage of oocysts that both excluded PI after acid treatment and contained sporozoites (viability) indicate oocyst viability. Oocyst infectivity is indicated by the percentage of infected HCT-8 cell monolayers (infectivity). Flask I contained approximately 1,100 oocysts · 100 μl−1, and flask II contained approximately 500 oocysts · 100 μl−1. The results for a dilution containing approximately 300 oocysts · 100 μl−1 are also shown.
Environmental samples.
Foci of infection were observed in three of eight Meuse River samples but in none of the seven Rhine River samples. All infection control slides were positive and contained clusters of foci. The number of oocysts in the river water samples ranged from 0.1 to 1.3 oocysts per liter. Viable counting showed that 0 to 14 and 2 to 8 potentially infectious oocysts were present in the Meuse and Rhine samples, respectively (Table 2). These oocysts excluded PI and contained sporozoites, as demonstrated by DAPI staining or differential interference contrast microscopy.
TABLE 2.
Total numbers of Cryptosporidium oocysts in water samples from the Rhine River and the Meuse River as determined by immunofluorescence microscopya
| Sampling date (day/mo/yr) | River | Total no. of oocysts | No. of potentially infectious oocysts
|
Cell line | Cell culture infectivity | |
|---|---|---|---|---|---|---|
| DAPI+ PI− | DAPI− PI− | |||||
| 17/01/00 | Meuse | 4 | 0 | 4 | Caco-2 | + |
| 24/01/00 | Meuse | 6 | 6 | 0 | Caco-2 | + |
| 07/02/00 | Meuse | 2 | 2 | 0 | Caco-2 | − |
| 21/02/00 | Meuse | 10 | 0 | 5 | HCT-8 | − |
| 28/02/00 | Meuse | 16 | 1 | 6 | HCT-8 | − |
| 13/03/00 | Meuse | 25 | 2 | 12 | Caco-2 | − |
| 08/05/00 | Meuse | 6 | 0 | 2 | HCT-8 | + |
| 15/05/00 | Meuse | 0 | 0 | 0 | HCT-8 | − |
| 28/02/00 | Rhine | 13 | 1 | 7 | HCT-8 | − |
| 13/03/00 | Rhine | 9 | 1 | 2 | Caco-2 | − |
| 27/03/00 | Rhine | 2 | 0 | 2 | HCT-8 | − |
| 03/04/00 | Rhine | 4 | 0 | 2 | HCT-8 | − |
| 10/04/00 | Rhine | 5 | 1 | 3 | HCT-8 | − |
| 08/05/00 | Rhine | 18 | 0 | 6 | HCT-8 | − |
| 05/06/00 | Rhine | 7 | 0 | 3 | HCT-8 | − |
| 13/03/00 | Meuse (spiked) | 19 | 2 | 13 | Caco-2 | − |
| 15/05/00 | Meuse (spiked) | 245 | 0 | 240 | HCT-8 | + |
| 27/03/00 | Rhine (spiked) | 9 | 0 | 7 | HCT-8 | − |
| 08/05/00 | Rhine (spiked) | 159 | 9 | 126 | HCT-8 | + |
Potentially infectious oocysts excluded propidium iodide and contained sporozoites. The samples produced infection (+) or did not produce infection (−) in Caco-2 or HCT-8 cells in cell culture infectivity assays.
DISCUSSION
The variation in infectivity between C. parvum isolates from different sources, between lots from the same source, with different cell lines, and in different experiments observed in this study has also been described by Slifko et al. (26) and Rochelle et al. (20). Recently, Di Giovanni and LeChevallier (8) reported different levels of cell culture infection for the isolates that they used to evaluate their quantitative PCR-cell culture assay. Human feeding trials showed similar differences with different C. parvum isolates displaying different infectivities (16, 27). However, it must be noted that in our experiments the differences were sometimes small and the amount of data was sometimes limited. The differences may have been true differences between isolates or lots, but they may also have been due to the inaccuracy inherent in cell culture assays. ID50s were determined by using oocyst suspensions 3 to 17 weeks postproduction. As a result of the overseas source of commercial oocysts suspensions, guidelines for using oocysts within 30 days after shedding (25) were not feasible and could not be applied. Use of younger oocysts could have resulted in lower ID50s and less variability, but we have not been able to test this possibility. However, one of the drawbacks of mouse infectivity assays, the variability in the results obtained, seems to apply to cell culture infectivity assays as well.
The one reliable estimate of ID50 for C. parvum oocysts from the University of Alberta in neonatal CD-1 mice suggests that the sensitivity of the neonatal mouse assay was higher than that of the cell culture assay with Caco-2 cells. However, our data set for mouse infectivity is limited, and regression analyses showed that there was poor fit of the logit response model to data from two of three experiments. Slifko et al. (26) reported that HCT-8 cells were more sensitive to infection with C.parvum oocysts than 4-day-old BALB/c mice or neonatal CD-1 mice. They reported average logit ID50s of 8, 64, and 119 for these three assays, respectively. Rochelle et al. (20) observed cell culture infectivity equivalent to the infectivity in a standard mouse assay using CD-1 mice. They also compared cell culture infectivities with HCT-8 and Caco-2 cells and observed lower ID50s with HCT-8 cells, which indicated the higher sensitivity of HCT-8 cells. Our observation of a larger number of clusters of foci of infection in positive wells with HCT-8 cells than with Caco-2 cells suggests that HCT-8 cells support the proliferation to other life cycle stages better than Caco-2 cells support this proliferation. Upton et al. (29) showed that the HCT-8 cell line supported in vitro infection with Cryptosporidium oocysts better than 10 other cell lines.
Although cell culture assays are easier to perform than animal infectivity assays, they still require special laboratory facilities and are time-consuming. In vitro excystation and vital dye inclusion or exclusion assays are simpler and more widely applicable. However, Korich et al. (13), Finch et al. (11), and Black et al. (2) showed that there was some disparity between surrogate methods for determining oocyst infectivity and neonatal mouse assays. Bukhari et al. (3) confirmed these results and demonstrated that both in vitro excystation and the inclusion or exclusion of the vital dyes SYTO-9, SYTO-59, and DAPI-PI overestimated oocyst infectivity compared to the results obtained with CD-1 mice after treatment of fresh and environmentally aged oocysts with low concentrations of ozone. During the ageing of oocysts in autoclaved river water, cell culture infectivity appeared to decline far more rapidly than it declined with the surrogate methods that we used to demonstrate oocyst viability, including vital dyes, indicating that the surrogate methods indeed overestimated oocyst infectivity compared to the cell culture assay. We were able to detect infectivity of aged oocysts by cell culturing after 40 days in sterile river water; about 1,100 oocysts were needed for a positive result. Jenkins et al. (12) found that C. parvum oocysts remained infectious for HCT-8 cells and neonatal BALB/c mice after 7 months of storage in sterile deionized water at 15°C; however, they inoculated cells and mice with 104 oocysts.
The environmental samples that produced foci of infection in cell culture assays did not produce secondary infections resulting in large clusters of foci, indicating that the oocysts were capable of initial invasion of the cell cultures but could not proliferate to other life cycle stages. According to Slifko et al. (25) the oocysts should therefore be regarded as noninfectious for humans. However, Cryptosporidium species other than C. parvum or C. hominis may have been present in the river water samples. Our experiments with C. muris oocysts demonstrated that it is plausible that these oocysts are not capable of further development in HCT-8 and Caco-2 cells. Our dilution experiments showed that relatively large numbers of oocysts were needed for infection in cell cultures. The surface water samples examined in this study were extracted from the largest rivers in The Netherlands with the highest average Cryptosporidium loads in Dutch surface water (14). Because relatively small sample volumes (12 to 26 liters) were examined, the absence of detection of infectious oocysts in the 15samples examined does not necessarily mean that there were no infectious oocysts present in the river water. Viable counting showed that the numbers of potentially infectious oocysts present in the Meuse and Rhine samples were far less than any of the ID50s of the C. parvum suspensions used to evaluate the cell culture assays but presumably were great enough to cause infection and disease (16). Surface water samples seeded with Waterborne Inc. oocysts, which had an ID50 of 40 with HCT-8 cells, did not produce the expected infectivity with HCT-8 cells. Samples seeded with numbers of oocysts greater than this ID50 gave positive results with only one of four or five replicate monolayers. This suggests either that the sensitivity of the cell culture assay was low or that there may be inherent variation in the results obtained with this method. Quintero-Betancourt et al. (18) reported that they were able to obtain positive results with HCT-8 cells when they analyzed reclaimed effluent samples that contained at least 100 oocysts per 100 liters.
In contrast to experiments with naturally contaminated water samples, high numbers of oocysts may be used in disinfection experiments. We found that cell culture assays are useful for studying the effect of UV disinfection on oocyst infectivity. Rochelle et al. (20) found that a cell culture assay with HCT-8 cells (including reverse transcription-PCR detection of infection) and a CD-1 mice assay produced equivalent results for predicting oocyst infectivity after ozone or UV treatment. Slikfo et al. (26) reported that there were no significant differences between the results obtained by cell culture with HCT-8 cells (microscopic detection of infection) and the results obtained by mouse infectivity assays after UV or chlorine dioxide treatment.
The cell culture immunofluorescence assays described here and performed in this study are not sensitive enough to detect infectious Cryptosporidium oocysts present in Dutch surface waters used for recreation or drinking water production. In The Netherlands naturally contaminated water containing both fresh and aged oocysts generally does not contain enough infectious oocysts to produce infection in the cell culture assays used in this study. Due to this lack of sensitivity, assessment of the risk of infection with Cryptosporidium resulting from drinking water consumption or surface water recreation in The Netherlands cannot be based on oocyst infectivity at this time. However, using vital dye inclusion or exclusion assays to determine oocyst viability in environmental samples with low oocyst concentrations overestimates oocyst infectivity, which results in overestimation of the infection risk. The limit of one infection in 10,000 persons per year may be exceeded, unjustly indicating that adjustment in drinking water treatment or additional research is required. For accurate risk assessment information on oocyst infectivity is indispensable. Enhanced recovery of Cryptosporidium oocysts from water samples containing low numbers of oocysts and improved sensitivity of the cell culture assays may result in better applicability of cell culture infectivity assays to environmental water samples in the future. Molecular detection of replication intermediates in the cells instead of microscopic detection of infectivity may increase the sensitivity of cell culture assays and will be evaluated. However, the cell culture assays described here may be sensitive enough to demonstrate oocyst infectivity in sewage influents and effluents and in laboratory and pilot experiments on treatment processes.
Acknowledgments
This research was performed by the order of and for the account of the General Directorate for Environmental Protection, directorate Drinking Water, Water and Agriculture, within the framework of project 289202.
We thank Erwin Duizer for his help with the implementation of Caco-2 and HCT-8 cell culture methods, Herman Näring for performing the mouse assays, Jack Schijven for his assistance in calculating ID50s, and Willemijn Lodder, Ria de Bruin, and Harold van den Berg for their assistance in the infectivity experiments.
REFERENCES
- 1.Anonymous. 2002. Water quality—isolation and identification of Cryptosporidium oocysts and Giardia cysts form water, ISO/CD 15553, 2002-07-22. International Organisation for Standardisation, Geneva, Switzerland.
- 2.Black, E. K., G. R. Finch, R. Taghi-Kilani, and M. Belosevic. 1996. Comparison of assays for Cryptosporidium parvum oocysts viability after chemical disinfection. FEMS Microbiol. Lett. 135:187-189. [DOI] [PubMed] [Google Scholar]
- 3.Bukhari, Z., M. M. Marshall, D. G. Korich, C. R. Fricker, H. V. Smith, J. Rosen, and J. L. Clancy. 2000. Comparison of Cryptosporidium parvum viability and infectivity assays following ozone treatment of oocysts. Appl. Environ. Microbiol. 66:2972-2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campbell, A. T., L. J. Robertson, and H. V. Smith. 1992. Viability of Cryptosporidium parvum oocysts: correlation of in vitro excystation with inclusion or exclusion of fluorogenic vital dyes. Appl. Environ. Microbiol. 58:3488-3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casemore, D. P., S. E. Wright, and R. L. Coop. 1997. Cryptosporidiosis: human and animal epidemiology, p. 65-92. In R. Fayer (ed.), Cryptosporidium and cryptosporidiosis. CRC Press, Boca Raton, Fla.
- 6.de Wit, M. A. S., M. P. G. Koopmans, L. M. Kortbeek, W. J. B. Wannet, J. Vinjé, F. van Leusden, A. I. M. Bartelds, and Y. T. H. P. van Duynhoven. 2001. Sensor, a population-based cohort study on gastroenteritis in the Netherlands, incidence and etiology. Am. J. Epidemiol. 154:666-674. [DOI] [PubMed] [Google Scholar]
- 7.Di Giovanni, G. D., F. H. Hashemi, N. J. Shaw, F. A. Abrams, M. W. LeChevallier, and M. Abbaszadegan. 1999. Detection of infectious Cryptosporidium parvum oocysts in surface and filter backwash water samples by immunomagnetic separation and integrated cell culture-PCR. Appl. Environ. Microbiol. 65:3427-3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Di Giovanni, G. D., and M. W. LeChevallier. 2005. Quantitative-PCR assessment or Cryptosporidium parvum cell culture infection. Appl. Environ. Microbiol. 71:1495-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fayer, R., U. Morgan, and S. J. Upton. 2000. Epidemiology of Cryptosporidium: transmission, detection and identification. Int. J. Parasitol. 30:1305-1322. [DOI] [PubMed] [Google Scholar]
- 10.Fayer, R. 2004. Cryptosporidium: a water-borne zoonotic parasite. Vet. Parasitol. 126:37-56. [DOI] [PubMed] [Google Scholar]
- 11.Finch, G. R., C. W. Daniels, E. K. Black, F. W. Schaefer III, and M. Belosevic. 1993. Dose response of Cryptosporidium parvum in outbred neonatal CD-1 mice. Appl. Environ. Microbiol. 59:3661-3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jenkins, M., J. M. Trout, J. Higgins, M. Dorsch, D. Veal, and R. Fayer. 2003. Comparison of tests for viable and infectious Cryptosporidium parvum oocysts. Parasitol. Res. 89:1-5. [DOI] [PubMed] [Google Scholar]
- 13.Korich, D. G., J. R. Mead, M. S. Madore, N. A. Sinclair, and C. R. Sterling. 1990. Effects of ozone, chlorine dioxide, chlorine, and monochloramine on Cryptosporidium parvum oocyst viability. Appl. Environ. Microbiol. 56:1423-1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Medema, G. J., L. Heijnen, F. M. Schets, B. Wullings, W. Hoogenboezem, and H. A. M. Ketelaars. 2002. Viable and pathogenic Cryptosporidium and Giardia in source water. Application of vital dye staining, cell culture and (RT-)PCR with sequence analysis. RIWA report ISBN 90-6683-099-9. Association of River waterwork—RIWA, Nieuwegein, The Netherlands.
- 15.Neumann, N. F., L. L. Gyürék, G. R. Finch, and M. Belosevic. 2000. Intact Cryptosporidium parvum oocysts isolated after in vitro excystation are infectious to neonatal mice. FEMS Microbiol. Lett. 183:331-336. [DOI] [PubMed] [Google Scholar]
- 16.Okhuysen, P. C., C. L. Chappell, J. H. Crabb, C. R. Sterling, and H. L. Dupont. 1999. Virulence of three distinct Cryptosporidium parvum isolates for healthy adults. J. Infect. Dis. 180:1275-1281. [DOI] [PubMed] [Google Scholar]
- 17.Peng, M. M., L. Xiao, A. R. Freeman, M. J. Arrowood, A. A. Escalante, A. C. Weltman, C. S. L. Ong, W. R. MacKenzie, A. A. Lal, and C. B. Beard. 1997. Genetic polymorphism among Cryptosporidium parvum isolates: evidence of two distinct human transmission cycles. Emerg. Infect. Dis. 3:567-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quintero-Betancourt, W., A. L. Gennaccaro, T. M. Scott, and J. B. Rose. 2003. Assessment of methods for detection of infectious Cryptosporidium oocysts and Giardia cysts in reclaimed effluents. Appl. Environ. Microbiol. 69:5380-5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robertson, L. J., A. T. Campbell, and H. V. Smith. 1993. In vitro excystation of Cryptosporidium parvum. Parasitology 106:13-19. [DOI] [PubMed] [Google Scholar]
- 20.Rochelle, P. A., M. M. Marshall, J. R. Mead, A. M. Johnson, D. G. Korich, J. S. Rosen, and R. De Leon. 2002. Comparison of in vitro cell culture and a mouse assay for measuring infectivity of Cryptosporidium parvum. Appl. Environ. Microbiol. 68:3809-3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rose, J. B., J. T. Lisle, and M. LeChevallier. 1997. Waterborne cryptosporidiosis: incidence, outbreaks and treatment strategies, p. 93-110. In R. Fayer (ed.), Cryptosporidium and cryptosporidiosis. CRC Press, Boca Raton, Fla.
- 22.Schets, F. M., G. B. Engels, M. During, and A. M. de Roda Husman. 2004. Detection of infectious Cryptosporidium oocysts by cell culture: applicability to environmental samples. RIVM report 330000005/2004. National Institute for Public Health and the Environment, Bilthoven, The Netherlands. [DOI] [PMC free article] [PubMed]
- 23.Slifko, T. R., D. E. Friedman, J. B. Rose, S. J. Upton, and W. Jakubowski. 1997. Unique cultural methods used to detect viable Cryptosporidium parvum oocysts in environmental samples. Water Sci. Technol. 35:363-368. [Google Scholar]
- 24.Slifko, T. R., D. Friedman, J. B. Rose, and W. Jakubowski. 1997. An in vitro method for detecting infectious Cryptosporidium oocysts with cell culture. Appl. Environ. Microbiol. 63:3669-3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Slifko, T. R., D. E. Huffman, and J. B. Rose. 1999. A most-probable-number assay for enumeration of infectious Cryptosporidium parvum oocysts. Appl. Environ. Microbiol. 65:3936-3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Slikfo, T. R., D. E. Huffman, D. Dussert, J. H. Owens, W. Jakubowski, C. N. Haas, and J. B. Rose. 2002. Comparison of tissue culture and animal models for assessment of Cryptosporidium parvum infection. Exp. Parasitol. 101: 97-106. [DOI] [PubMed] [Google Scholar]
- 27.Teunis, P. F. M., C. L. Chappel, and P. C. Okhuysen. 2002. Dose response studies: variation between isolates. Risk Anal. 22:175-183. [DOI] [PubMed] [Google Scholar]
- 28.Upton, S. J., M. Tilley, M. V. Nesterenko, and D. B. Brillhart. 1994. A simple and reliable method of producing in vitro infections of Cryptosporidium parvum (Apicomplexa). FEMS Microbiol. Lett. 118:45-50. [DOI] [PubMed] [Google Scholar]
- 29.Upton, S. J., M. Tilley, and D. B. Brillhart. 1994. Comparative development of Cryptosporidium parvum (Apicomplexa) in 11 continuous host cell lines. FEMS Microbiol. Lett. 118:233-236. [DOI] [PubMed] [Google Scholar]