Abstract
Anaerobic or microaerophilic chemolithoautotrophic bacteria have been considered to be responsible for CO2 dark fixation in different pelagic redoxclines worldwide, but their involvement in redox processes is still not fully resolved. We investigated the impact of 17 different electron donor/acceptor combinations in water of pelagic redoxclines from the central Baltic Sea on the stimulation of bacterial CO2 dark fixation as well as on the development of chemolithoautotrophic populations. In situ, the highest CO2 dark fixation rates, ranging from 0.7 to 1.4 μmol liter−1 day−1, were measured directly below the redoxcline. In enrichment experiments, chemolithoautotrophic CO2 dark fixation was maximally stimulated by the addition of thiosulfate, reaching values of up to 9.7 μmol liter−1 CO2 day−1. Chemolithoautotrophic nitrate reduction proved to be an important process, with rates of up to 33.5 μmol liter−1 NO3− day−1. Reduction of Fe(III) or Mn(IV) was not detected; nevertheless, the presence of these potential electron acceptors influenced the development of stimulated microbial assemblages. Potential chemolithoautotrophic bacteria in the enrichment experiments were displayed on 16S ribosomal complementary DNA single-strand-conformation polymorphism fingerprints and identified by sequencing of excised bands. Sequences were closely related to chemolithoautotrophic Thiomicrospira psychrophila and Maorithyas hadalis gill symbiont (both Gammaproteobacteria) and to an uncultured nitrate-reducing Helicobacteraceae bacterium (Epsilonproteobacteria). Our data indicate that this Helicobacteraceae bacterium could be of general importance or even a key organism for autotrophic nitrate reduction in pelagic redoxclines.
Chemolithoautotrophic bacteria play an important role in biogeochemical cycles of aquatic habitats. Molecular hydrogen and reduced nitrogen (NH4+ and NO2−), sulfur (e.g., H2S and S2O32−), metals (e.g., Fe2+ and Mn2+) and carbon (e.g., CO and CH4) compounds serve as electron donors for these bacteria (37), whereas oxygen and nitrate mostly serve as electron acceptors. CO2 dark fixation has been determined in different pelagic redoxclines worldwide. For example, Taylor et al. (42) showed that bacterial chemoautotrophy, fueled by reduced sulfur species, supported an active secondary microbial food web in the redox transition zone of the Cariaco Basin. Depending on the season, dissolved inorganic carbon assimilation (27 to 159 mmol C m−2 day−1) in this zone was equivalent to 10% to 333% of phytoplankton primary production. Madrid et al. (24) hypothesized that sulfide-oxidizing epsilon symbiont relative clones were responsible for the dark CO2 fixation. Nitrate, manganese, and iron as potential electron acceptors were available for the epsilon symbiont relatives, but their reduction was not investigated. Jannasch et al. (17) isolated nine chemolithoautotrophic bacterial strains from the anoxic interface of the Black Sea. These isolates were unable to utilize nitrate, manganese, or iron oxides as electron acceptors. However, the authors postulated that this could have been due to toxic concentrations of the ions. In any case, since the oxidation of 1 mol of H2S to sulfate requires 8 mol of Fe(III), the estimated iron fluxes could have accounted for only less than 0.1% of the measured H2S oxidation rates in the suboxic zone of the Black Sea (17, 18).
The Baltic Sea itself is among the largest brackish basins of the world with periodically anoxic conditions in bottom waters. Analogously to the Black Sea, the Baltic Sea proper comprises a number of deep areas with anoxic bottom water, of which the Gotland Deep is the largest and the Landsort Deep, at 495 m, the deepest. In both deeps a stable halocline below 50 to 60 m separates the water column into the upper oxygenated layer and the underlying oxygen-deficient and anoxic/sulfidic layer (22, 28). The oxic-anoxic interfaces are generally characterized by high CO2 dark fixation rates, which may correspond to up to 30% of surface primary production (8). The simultaneous occurrence of high denitrification rates (4, 34) led to the conclusion that chemolithoautotrophic oxidation of sulfur compounds coupled to nitrate reduction should play an important role in these pelagic redoxclines. Subsequently, an epsilonproteobacterium related to Thiomicrospira denitrificans was identified as an important chemolithoautotrophic sulfide oxidizer and nitrate reducer (16, 20). Additionally, there is first evidence that this bacterium was predominantly responsible for autotrophic denitrification processes at the Gotland Deep (3). However, for this site geochemical evidence also points towards the importance of redox processes related to manganese and iron transformations (28). Additionally, sulfide and ammonium, another well known electron donator for aerobic chemolithoautotrophs, generally occur together in this pelagic redoxcline. Altogether, the importance of different potential electron donors/acceptors for different chemolithoautotrophic microorganisms is still poorly known.
Thus, our main goal was to identify bacterial phylotypes of the Gotland Deep whose activity in terms of 16S rRNA synthesis could be stimulated, along with enhanced CO2 fixation rates, upon the addition of other potential substrate combinations. Our second aim was to determine if the above-mentioned bacterium related to T. denitrificans, responsible for autotrophic nitrate reduction in the Gotland Deep, could also be a key player in autotrophic nitrate reduction in the Landsort Deep. This assumption was supported by the fact that phylogenetic relatives of T. denitrificans had been found in other marine oxic-anoxic interfaces, e.g., the Black Sea (44), the Cariaco Basin (24), or aphotic sulfidic springs (9, 10).
For this purpose, water samples of the two deeps were spiked by the addition of different substrate combinations. CO2 fixation was analyzed and stimulated bacteria identified by molecular analysis as described previously (3, 20). This approach led to the successful identification of potential key chemolithoautotrophic organisms from two pelagic redoxclines of the central Baltic Sea.
MATERIALS AND METHODS
Sampling.
Sampling was performed onboard the RV Professor Albrecht Penck during cruise 40/04/17 in the eastern Gotland Deep (Baltic Sea monitoring station 271; 57°19.2′N, 20°03′E) and in the Landsort Deep (station 284; 58°35′N, 18°14′E) (Fig. 1) in August 2004 under comparable physicochemical conditions.
FIG. 1.
Map of the central Baltic Sea and positions of sampling stations 271 and 284 (courtesy of Jan Donath, Leibniz-Institut für Ostseeforschung Warnemünde).
Physicochemical structure of the oxic-anoxic interface.
Chemical profiles of oxygen, hydrogen sulfide, ammonia, nitrite, and nitrate were determined as described elsewhere (14). Dissolved manganese and total manganese were analyzed onboard using the formaldoxime method (5). Briefly, 50-ml PE bottles were rinsed with deionized water and 20 ml 0.2-μm-filtered water sample. Subsequently, dissolved manganese was determined after filtration of 20 ml water sample into the bottle, mixed with 2 ml formaldoxime reagent, incubated for 60 min in the dark, and analyzed at 450 nm in a spectrophotometer (RF-1502; Shimadzu). Total manganese was determined accordingly from nonfiltered samples. Particulate manganese was sufficiently abundant at station 271 and could therefore be determined by subtraction of dissolved manganese from total manganese.
In situ CO2 dark fixation.
In situ CO2 fixation rates were determined throughout the redoxcline according to the method of Steemann Nielsen (40). Briefly, incubations were carried out in 120-ml Winkler bottles to which 100 μCi [14C]bicarbonate (specific activity, 53.0 mCi mmol−1; Hartmann Analytic GmbH, Braunschweig, Germany) were added. Samples were incubated at in situ temperatures for 24 h in darkness, filtered on 0.2-μm-pore-size membrane filters, and exposed to HCl fumes for 20 min, and radioactivity was counted in a scintillation counter (Packard). Parallel samples immediately fixed with formalin (1.5% final concentration) before addition of [14C]bicarbonate served as controls.
Stimulated in vitro CO2 dark fixation.
To assess the microbial diversity associated with in situ CO2 dark fixation, we performed stimulation experiments. Sulfidic water samples were collected from 227-m-deep water at station 271 or from 85-m-deep water at station 284, using 5-liter Teflon-coated Go Flo bottles (General Oceanics) attached to a polyvinyl chloride-coated stainless steel conductivity-temperature-depth rosette, and then directly transferred, air bubble free, into plastic cans. Eventually, the samples were incubated at 4°C in a container with anoxic sulfide-free seawater. Due to diffusion processes, samples were free of sulfide and contained up to 15 μmol liter−1 oxygen after 4 days of incubation. Eventually, 630 ml or 120 ml (in the latter case together with [14C]bicarbonate) was placed in transfusion bottles (Glasgerätebau Ochs, Bovenden, Germany) or Winkler bottles, respectively, and complemented by the addition of electron donor/acceptor pairs (Table 1) potentially supporting CO2 fixation activity. The electron donor/acceptor combinations were added in three replicates as described previously (20) and incubated at 10°C for 72 h in the dark. All mixtures including nitrate or nitrite were prepared with Landsort water; the others were prepared with Gotland water. Potential heterotrophic CO2 fixation was stimulated by the addition of glucose. Negative controls for all treatments were incubated at 0°C. After 0, 48, or 72 h of incubation, CO2 dark fixation was measured as described before. In parallel, biomass of the enrichments was harvested by the filtration of 600 ml enrichment water on Durapore filters (0.2-μm pore size) and stored frozen (−80°C) for later analysis. The analysis of dissolved and particulate Mn or Fe phases during stimulation experiments was based on previously described procedures (30), using a Perkin-Elmer AA spectrophotometer (models 3030 and ZL 4100 with Zeeman correction) in conjunction with an HGA 600 graphite furnace and an autosampler (model AS 60).
TABLE 1.
Electron donor/acceptor combinations used in the enrichment experimentsa
| Electron acceptor | Useb with the following electron donor:
|
||||||
|---|---|---|---|---|---|---|---|
| Na2S | Na2S2O3 | NH4Cl | MnCl2 · 4H2O | FeCl2 | CH3OH | C6H12O6c | |
| MnO2 | × | × | × | × | × | ||
| FeCl3 · 6H2O | × | × | × | × | |||
| KNO3 | × | × | × | × | × | × | × |
| KNO2 | × | ||||||
The concentration of all components was 100 μmol liter−1, except MnO2, which was at 83.5 μmol liter−1.
×, used in combination.
Stimulation of heterotrophic CO2 fixation.
Stimulation and identification of microbial phylotypes.
Nucleic acid extraction from frozen filters was performed for all thiosulfate-stimulated samples as described by Weinbauer et al. (46). Prior to reverse transcription-PCR, RNA extracts were purified from DNA by incubation with DNase I (Roche Diagnostics, Mannheim, Germany) for 60 min at 37°C, and their concentrations were determined using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies). To retrieve 16S ribosomal complementary DNA (rcDNA), 20 ng of template RNA was reverse transcribed at 42°C using the iScript cDNA synthesis kit (Bio-Rad). In addition to hexamers provided in the kit, the universal reverse primer 1492R (5′-GGTTACCTTGTTACGACTT-3′) (21) was also supplied. Bacterial Com-primers (amplifying positions 519 to 926 [Escherichia coli numbering of 16S rRNA gene]) (36) were used for 16S rcDNA amplification. Thermocycling started with an initial denaturation for 5 min at 94°C. A total of 23 cycles of 30 s at 94°C, 30 s at 55°C, and 2 min at 72°C were followed by a final elongation step of 10 min at 72°C. Generation and purification of single-stranded DNA and single-strand-conformation polymorphism (SSCP) analysis were done as described by Schwieger and Tebbe (36).
The relative fractions of 16S rcDNA bands were determined by image analysis of silver stained SSCP gels using the GelScan software (BioSciTec). The relative fraction of bands was calculated by the determination of band areas.
Reamplification of individual bands excised from the SSCP gels was performed as described by Pöhler et al. (31). PCR products were purified using the MiniElute kit (QIAGEN) as described by the manufacturer and sequenced by SEQLAB (Göttingen, Germany). Forward and reverse sequences of all samples were checked for accuracy using the SeqMan software (DNAstar). Phylogenetic affiliations of the partial sequences were initially estimated using the program BLAST (1). Sequences were aligned and analyzed by employing the ARB software package (23). Sequences with greater than 98% similarity were grouped for phylogenetic analyses. Sequences for analysis were reduced to unambiguously alignable positions by using group-specific filters. An evolutionary distance dendrogram was constructed using maximum likelihood as well as the Jukes-Cantor correction and neighbor joining. Bootstrap analyses were performed for neighbor joining with 1,000 resamplings.
Validation of SSCP-based quantification by quantitative PCR (qPCR).
An i-cycler IQ real-time system (Bio-Rad) was used to quantify the relative 16S rRNA concentration of an uncultured autotrophic nitrate-reducing Helicobacteraceae strain G138eps1 bacterium stimulated in the thiosulfate/nitrate enrichment after 72 h of incubation as described previously (16, 20). The PCR mixtures (25 μl) contained 1× iQSybrGreen Supermix (Bio-Rad), bacterial Com (900 nmol liter−1 each) or strain-specific OST 1 real-time primer system (OST 1F, 900 nmol liter−1; OST 1R, 300 nmol liter−1) (16, 20), and 3 ng of nucleic acids. An initial denaturing step at 95°C for 3 min was followed by 40 cycles of 94°C for 30 s, 59°C for 40 s, and 72°C (measuring step) for 50 s. Potential development of primer dimers was determined by a melting point analysis in a range from 40 to 94°C. Each measurement was performed in three replicates. Relative standards were prepared using serially diluted (1:10) nucleic acids again isolated from the thiosulfate/nitrate enrichment. Each sample measured was calibrated by its own respective Com and OST standard. Due to potentially lower PCR efficiencies of nondiluted cDNA extracts, each sample to be measured was always diluted 1:10. Calibration curves as well as quantifications were generated by the i-cycler IQ real-time system software. For each standard, the concentration was plotted against the cycle number at which the fluorescence signal increased above the background or cycle threshold value. Only data resulting from measurements with comparable Com- and OST-specific PCR efficiencies were used for further calculations.
Estimation of cell concentration and doubling time.
The measured carbon fixation during the stimulation experiments was assumed to represent the biomass of chemolithoautotrophic bacteria build up by exponential growth during the incubation time. Given that only new bacterial biomass was labeled by 14C, the biomass at time t = 0 h (X0) had to be calculated. Thus, the total biomass of chemolithoautotrophic bacteria at time t1 was equal to the new biomass X1 plus the unknown X0.
Normal bacterial growth was described by the equation Xt = X0 × eμ × t. The biomass built after addition of 14CO2 was measured. The unlabeled biomass of the bacterial fraction taking up 14CO2 was unknown (called X0). Thus, the following equation was used for the incubation periods: Xt + X0 = X0 × eμ × t. This equation was solved for μ, to give μ = [ln(Xt + X0) − lnX0]/t.
For the two incubation periods the following equation results when μ is assumed to be constant during the experiment: [ln(X1 + X0) − lnX0]/t1 = [ln(X2 + X0) − lnX0]/t2. Based on the new biomasses X1 and X2 determined at points t1 and t2, respectively, X0 was calculated by iteration: t1 × [ln(X2 + X0) − lnX0] − t2 × [ln(X1 + X0) − lnX0] = 0.
The initial biomass X0 was transformed to cell concentration N0 by assuming an individual cell biomass of 20 fg C (12). The mean growth rate was calculated using the equation for μ above and transformed to doubling time.
Nucleotide sequence accession numbers.
Sequences have been deposited in the EMBL nucleotide sequence database under accession numbers AJ937823 to AJ937832.
RESULTS
Physicochemical structure of the oxic-anoxic interface.
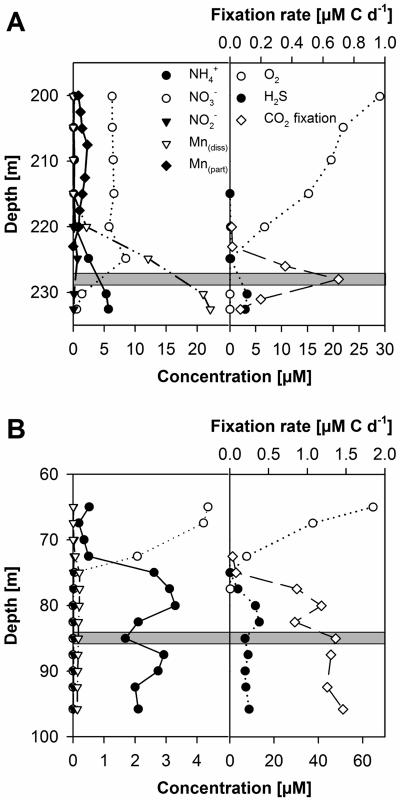
Physical and chemical parameters of stations 271 and 284 are shown in Fig. 2. Due to the invasion of oxygenated North Sea water in 2003, the pelagic redoxcline at station 271 was newly reestablished approximately 20 m above the sediment (Fig. 2A). Below the halocline at a depth of 60 m, oxygen decreased sharply to concentrations of below 50 μmol liter−1, reaching values of less than 20 μmol liter−1 at depths of around 200 m. From this depth until the redoxcline, oxygen decreased continuously. Nitrate decreased sharply from 225 to 232 m. Dissolved manganese, ammonium, and sulfide increased strongly around and below the redoxcline. Particulate manganese showed its maximum well above the redoxcline at around 210 m. The redoxcline at station 284 was established at a depth of approximately 70 to 80 m (Fig. 2B). With the exception that manganese concentrations in a range from 0.17 to 0.21 μmol liter−1 at station 284 were approximately 100-fold lower than those at station 271, the chemical profile displayed similar characteristics.
FIG. 2.
Depth profiles of NH4+, NO3−, NO2−, and particulate and dissolved Mn (left), as well as O2, H2S, and 14CO2 fixation (right), throughout the oxic-anoxic interface of Gotland Deep station 271 (A) and Landsort Deep station 284 (B) in August 2004. The shaded area indicates the CO2 fixation zone which was sampled for subsequent stimulation experiments.
CO2 fixation rates.
At station 271, the highest in situ CO2 fixation rates of 0.7 μmol liter−1 day−1 were observed in the sulfidic zone directly below the redoxcline at a depth of approximately 227 m (Fig. 2A). In the enrichment experiments, the highest CO2 dark fixation rates were detected when thiosulfate was used as an electron donor. The combinations Na2S2O3/Fe(III) and Na2S2O3/Mn(IV), and even Na2S2O3 alone, resulted in high CO2 fixation rates, but no significant reduction of the respective electron acceptors could be measured (Table 2). The combinations NH4+/Mn(IV) and methanol/Mn(IV) yielded no detectable CO2 dark fixation. Thus, the bacterial community was not further analyzed.
TABLE 2.
CO2 fixation and oxidation/reduction rates after 48 and 72 hours of incubation with different electron donator/electron acceptor combinations
| Mean (μmol liter−1)c
| |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Station and electron donor/electron acceptor | CO2 fixation
|
Oxidation of electron donor
|
Reduction of electron acceptor
|
||||||
| 0-48 h | 48-72 h | Daily rate | 0-48 h | 48-72 h | Daily rate | 0-48 h | 48-72 h | Daily rate | |
| Station 271 | |||||||||
| Methanol/Fe(III) | 0.14 (0.04) | −0.05 (0.00) | 0.03 | NDa | ND | ND | −9.1 (4.3) | 9.4 (42.3) | 0.1 |
| Fe(II)/Mn(IV) | 0.15 (0.05) | −0.10 (0.06) | 0.02 | 17.8 (4.3) | 12.4 (1.6) | 10.1 | 30.6 (0.9) | 19.7 (4.5) | 16.8 |
| NH4+/Fe(III) | 0.33 (0.33) | −0.28 (0.03) | 0.02 | −53.7 (11.8) | 68.0 (14.3) | 7.2 | ND | ND | ND |
| Na2S2O3/Fe(III) | 2.08 (0.03) | 5.65 (0.56) | 2.58 | ND | ND | ND | 1.1 (0.9) | −9.1 (1.2) | −2.7 |
| Na2S2O3/none | 2.43 (0.04) | 8.91 (0.20) | 3.78 | ND | ND | ND | NDb | ND | ND |
| Na2S2O3/Mn(IV) | 2.45 (0.01) | 1.68 (0.09) | 1.38 | ND | ND | ND | 11.3 (1.3) | −11.3 (0.0) | 0.0 |
| Glucose/Mn(IV) | 0.15 (0.11) | 0.84 (0.01) | 0.33 | ND | ND | ND | −0.7 (8.1) | −1.2 (2.2) | −0.6 |
| Glucose/Fe(III) | 0.16 (0.06) | 0.70 (0.29) | 0.29 | ND | ND | ND | −6.9 | 4.4 (2.8) | −0.8 |
| Station 284 | |||||||||
| Na2S2O3/NO3− | 4.93 (0.25) | 9.68 (0.41) | 4.87 | ND | ND | ND | 27.2 (7.3) | 33.5 (9.0) | 20.2 |
| Na2S/NO3− | 0.93 (0.64) | −0.21 (0.17) | 0.24 | 42.4 (11.2) | −19.7 (5.1) | 7.6 | ND | ND | ND |
| NH4+/NO3− | 0.04 (0.01) | 0.10 (0.09) | 0.05 | 4.9 (0.7) | −35.6 (4.8) | −10.2 | ND | ND | ND |
| Methanol/NO3− | 0.06 (0.01) | 0.02 (0.01) | 0.03 | ND | ND | ND | 21.0 (8.2) | −18.7 (7.3) | 0.8 |
| Mn(II)/NO3− | 0.07 (0.00) | 0.00 (0.00) | 0.02 | −3.4 (4.8) | 3.4 (0.0) | 0.0 | ND | ND | ND |
| Glucose/NO3− | 0.17 (0.09) | 0.22 (0.03) | 0.13 | ND | ND | ND | −10.7 (2.9) | 8.3 (2.2) | −0.8 |
ND, not determined.
0.01 ml liter−1 H2S detected.
Values in parentheses are standard deviations in micromoles per liter.
At station 284, in situ CO2 fixation rates reached approximately 1.4 μmol liter−1 day−1 in the sulfidic zone below the redoxcline beginning at a depth of approximately 85 m (Fig. 2B). No decrease of in situ CO2 fixation rates was observed down to the last investigated depth of 96 m. Highest CO2 fixation as well as nitrate reduction rates were determined for the Na2S2O3/NO3− enrichment. Other electron donators, even sulfide, yielded significantly less or, in the case of Fe(II)/NO3− and NH4+/NO2−, no detectable CO2 dark fixation (Table 2).
Heterotrophic CO2 dark fixation.
Artificially stimulated heterotrophic CO2 dark fixation reached 0.15 to 0.17 μmol liter−1 after 48 h at both stations. It increased exponentially, reaching 0.84 μmol liter−1 day−1, from 48 to 72 h. In the cases of Fe(III) and NO3−, this was accompanied by a reduction of these electron acceptors (Table 2).
Stimulated microbial communities.
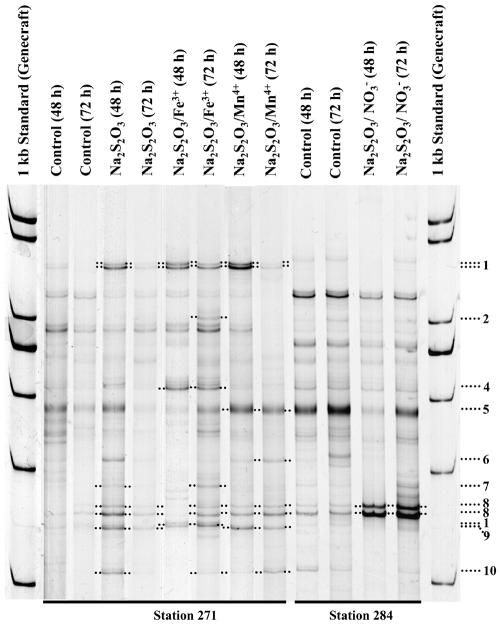
SSCP profiling revealed distinct community changes in the enrichment assays (Fig. 3). Phylogenetic analysis of SSCP band sequences revealed 16S rRNA similarities to already described species or clones in the range from 94 to 100% (Fig. 3; Table 3). With bacteria closely related or identical to the aerobically sulfide-oxidizing Thiomicrospira psychrophila and Maorithyas hadalis gill symbiont as well as the uncultured autotrophic, denitrifying Helicobacteraceae strain G138eps1 (Table 4), the highest diversity of stimulated potential chemolithoautotrophic bacteria from station 271 was observed in the Na2S2O3 enrichment after 48 h of incubation (Table 4). T. psychrophila and relatives of the uncultured Helicobacteraceae were stimulated in the Na2S2O3/Fe(III) and Na2S2O3/Mn(IV) enrichments during the whole experiment. Three organisms related to Pseudomonas stutzeri or fish-pathogenic Pseudomonas anguilliseptica and uncultured CFB group bacterium MERTZ_2CM_22 were stimulated in the Na2S2O3/Fe(III) enrichment (Fig. 3). Relatives of uncultured epsilonproteobacterium clone Nubeena319 and again the uncultured CFB group bacterium were stimulated in the Na2S2O3 and Na2S2O3/Mn(IV) enrichments (Table 4; Fig. 3 and 4).
FIG. 3.
16S rcDNA SSCP analysis of thiosulfate stimulation experiments using different electron acceptors. The original nonstimulated seawater sample served as a control. Stimulation time is given in parentheses. Numbers on the right refer to excised and sequenced bands (Table 3).
TABLE 3.
16S rRNA sequence designations, sequence origins, nucleotide sequence accession numbers, sequence lengths, and closest phylogenetic neighbors of investigated sequences
| Sequence designation (SSCP band) | Sequence origin (enrichment) | Nucleotide sequence accession number | Sequence length (bp) | Closest phylogenetic neighbor | Sequence similarity (%)a |
|---|---|---|---|---|---|
| 1 | Thiosulfate | AJ937823 | 345 | Thiomicrospira psychrophila SVAL-D | 99 |
| 2 | Thiosulfate/iron | AJ937824 | 347 | Pseudomonas stutzeri | 99 |
| 4 | Thiosulfate/iron | AJ937826 | 299 | Pseudomonas sp. strain He | 99 |
| 5 | Thiosulfate/manganese | AJ937827 | 331 | Thalassolituus oleivorans | 98 |
| 6 | Thiosulfate/manganese | AJ937828 | 300 | Maorithyas hadalis gill thioautotrophic symbiont | 99 |
| 7 | Thiosulfate/manganese | AJ937829 | 329 | Uncultured Helicobacteraceae strain G138eps1 | 96 |
| 8 | Thiosulfate | AJ937830 | 349 | Uncultured Helicobacteraceae strain G138eps1 | 100 |
| 9 | Thiosulfate | AJ937831 | 350 | Uncultured epsilonproteobacterium clone Nubeena319 | 100 |
| 10 | Thiosulfate | AJ937832 | 341 | Uncultured CFB group bacterium MERTZ_2CM_22 | 94 |
Determined by the program BLAST.
TABLE 4.
Identification of substrate-stimulated 16S rRNA phylotypes according to their relative rRNA abundances in SSCP gels
| 16S rcDNA SSCP band no. | Relative abundance (%)a with the indicated electron donor/electron acceptor combination
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Station 271
|
Station 284
|
|||||||||||
| Control
|
Na2S2O3/none
|
Na2S2O3/Fe(III)
|
Na2S2O3/Mn(IV)
|
Control
|
Na2S2O3/NO3−
|
|||||||
| 48 h | 72 h | 48 h | 72 h | 48 h | 72 h | 48 h | 72 h | 48 h | 72 h | 48 h | 72 h | |
| 1b | 7.6 | 0 | 14 | 12.8 | 28.7 | 16.6 | 31.0 | 8.2 | 0 | 0 | 0 | 0 |
| 2 | 0 | 0 | 0 | 0 | 0 | 3.5 | 0 | 0 | 0 | 3.9 | 0 | 3.1 |
| 4 | 0 | 0 | 0 | 0 | 25.0 | 11.1 | 0 | 0 | 5.2 | 3.3 | 4.7 | 3.5 |
| 5 | 20.4 | 22.5 | 18.6 | 19.2 | 13.7 | 12.2 | 32.2 | 29.8 | 26.9 | 30.2 | 11.5 | 17.6 |
| 6b | 0 | 0 | 8.7 | 0 | 0 | 0 | 0 | 9.2 | 0 | 0 | 0 | 0 |
| 7b | 5.7 | 0 | 5.5 | 0 | 0 | 3.9 | 0 | 0 | 0 | 0 | 0 | 0 |
| 8b | 3.9 | 14.0 | 17.0 | 15.8 | 6.5 | 11.1 | 13.0 | 19.9 | 7.9 | 6.3 | 50.9 | 35.0 |
| 9 | 4.4 | 0 | 11.5 | 9.2 | 0 | 0 | 12.7 | 10.3 | 0 | 0 | 0 | 0 |
| 10 | 0 | 0 | 8.4 | 0 | 0 | 3.6 | 0 | 10.2 | 4.4 | 2.6 | 0 | 1.5 |
Relative abundance in the nonstimulated control and after 48 and 72 hours of stimulation with thiosulfate and different electron acceptors. Organisms were considered stimulated if their abundance increased at least by 30% compared to the nonstimulated control (indicated in boldface).
Potentially chemolithoautotrophic bacterium.
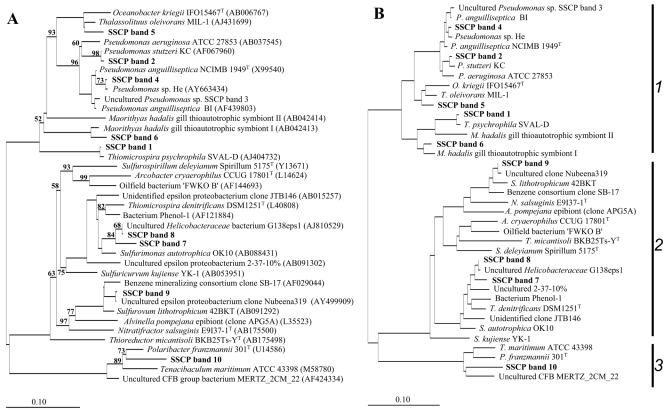
FIG. 4.
Unrooted trees showing phylogenetic relationships of the stimulated bacteria and their closest phylogenetic relatives. (A) Tree reconstructed using the neighbor-joining method and based on a comparison of approximately 300 nucleotides. Bootstrap values, expressed as a percentage of 1,000 replications, are given at branching points. Only those above 50% are shown. Database accession numbers are given in parentheses. (B) Basic tree reconstructed using the maximum-likelihood method and based on a comparison of approximately 1,400 nucleotides. Eventually, partial SSCP sequences were individually imported using ARB Parsimony. For both tree reconstruction methods, 42 members of the Verrucomicrobiaceae were used as an outgroup. Bar, 10 substitutions per 100 nucleotides. 1, Gammaproteobacteria; 2, Epsilonproteobacteria; 3, Bacteroidetes. More detailed information about the investigated sequences is given in Table 3.
With relative 16S rRNA quantities of 51% (after 48 h of incubation) and 35% (after 72 h), the Na2S2O3/NO3− enrichments were exclusively dominated by the autotrophic uncultured Helicobacteraceae strain G138eps1 (Table 4; Fig. 3). For a validation of this SSCP quantification approach, the relative 16S rRNA concentration after 72 h of incubation was additionally determined by qPCR as described previously for this specific organism (20). With a PCR efficiency of 101% and a correlation coefficient of the regression line of 0.997, its relative concentration was determined to be 32% ± 2.3%.
Estimation of doubling time and cell concentration.
Calculated bacterial cell doubling times from the two CO2 fixation measurements in all experiments using thiosulfate as an electron donor were in a range between 12 and 18 h (Table 5). Their initial cell numbers were calculated to be between 0.9 × 105 and 5.6 × 105 ml−1. Cell numbers for the thiosulfate/Mn(IV) enrichment could not be calculated, because CO2 fixation rates were not increasing exponentially with this treatment (Table 2).
TABLE 5.
Doubling time and estimated initial cell concentration of chemolithoautotrophic bacteria in different stimulation experiments with thiosulfate as the electron donator
| Cell characteristic | Value with the following electron donor/electron acceptor combination:
|
||
|---|---|---|---|
| Na2S2O3/none | Na2S2O3/Fe(III) | Na2S2O3/NO3− | |
| Doubling time (h) | 12.0 | 14.4 | 18.2 |
| Cell concn (N0) | 0.9 × 105 | 1.3 × 105 | 5.6 × 105 |
DISCUSSION
CO2 fixation rates.
High dark carbon fixation had already been observed at redoxclines of different aquatic areas (6, 24, 39, 47). Typically, ammonia oxidation and oxidation of reduced sulfur compounds were related to it using oxygen as electron acceptor. In the redoxcline of the central Baltic Sea, NH4+ was generally present in concentrations similar to those for sulfide (Fig. 2A). However, reduced sulfur compounds and not ammonium oxidation seemed to play the major role for the autotrophic CO2 fixation (Table 2). Enoksson (11) measured nitrification rates not higher than 280 nmol liter−1 per day. With a commonly used factor for conversion to carbon fixation of 0.12 (2) and also a twofold underestimation, ammonia oxidation could have been responsible for less than 0.1 μmol liter−1 carbon dioxide fixation per day. Thus, ammonia oxidation should not be the main process responsible for the observed high CO2 fixation rates.
Moreover, it is interesting to note that CO2 dark fixation was less stimulated by the addition of sulfide than by thiosulfate (Table 2). This could be due to inhibitory effects of sulfide, because as little as 300 μmol liter−1 H2S resulted in almost total inhibition of nitrous oxide reduction by denitrifying bacteria (38).
Due to the sampling procedure for in situ analyses onboard, small concentrations of oxygen possibly diffused into the samples. This potentially forced autotrophic sulfide oxidation by this artificially supplied oxygen and could have influenced the location of the in situ CO2 fixation maximum. However, highest rates of dark H14CO3− fixation in the Mariager Fjord were also observed at the lower border of the redoxcline (47).
In general, it has been suggested that 3 to 8% of the organic carbon in some heterotrophic bacteria originates from CO2 incorporated by carboxylation reactions (35). However, it can be equally important as autotrophic CO2 fixation (25) and could mask the autotrophic 14CO2 fixation signal, especially in habitats with high input of organic material (47). If heterotrophic CO2 fixation had occurred in situ, this activity should be detectable as background in all experiments. Therefore, we assume that the lowest measured CO2 fixation due to in situ anaplerotic carbon fixation accounted for less than 0.1 μmol liter−1 within 48 h. Even artificially stimulated heterotrophic CO2 dark fixation reached only 0.15 to 0.17 μM after 48 h. Higher rates (up to 0.84 μmol liter−1 day−1) were measured only after 72 h and in the case of Fe(III) and NO3− were accompanied by a reduction of these electron acceptors (Table 2). Altogether, we conclude that heterotrophic CO2 dark fixation should have been negligible for the time of our sampling for the two stations. CO2 dark fixation should have been predominantly due to the activity of sulfide-oxidizing chemolithoautotrophic bacteria. However, heterotrophic Fe(III) reduction could potentially play a role in pelagic redoxclines of the Gotland Deep.
The combinations Na2S2O3/Fe(III) and Na2S2O3/Mn(IV) and even Na2S2O3 alone resulted in high CO2 fixation rates in Gotland Deep water, but no significant reduction of the respective electron acceptors was detectable (Table 2), even assuming that soluble pools of organically complexed Fe(III) and Mn(IV) could have increased the amount of reduced substances artificially. In general, denitrification takes place already at low oxygen concentrations of approximately 10 to 15 μmol liter−1 (34). This was also demonstrated for our experiments, where autotrophic nitrate reduction was stimulated with a daily mean CO2 dark fixation of 2.5 μmol liter−1 during the first 48 h, followed by an increase up to 9.7 μmol liter−1 day−1 (Table 2). However, similar oxygen conditions did not result in analyzable Fe(III) or Mn(IV) reduction, leading to the conclusion that solely oxygen could have been reduced instead. Thus, potential autotrophic Fe(III)/Mn(IV) reduction might be restricted to strictly anaerobic areas of the pelagic redoxcline, even at high concentrations of their particulate oxides (Fig. 2A). In the Gotland Deep, respective areas could be reached with a flux of settling Mn oxides (28) and probably by Fe oxides. However, the importance of this mechanism has to be further analyzed.
The combination Fe(II)/Mn(IV) showed iron oxidation rates of 10 μmol liter−1 day−1 and manganese reduction rates of 17 μmol liter−1 day−1 (Table 2). However, it should be noted that Fe(II) readily reduces Mn(IV) in a purely chemical redox reaction, and these abiotic chemical transformations could have been inhibited in the control experiments at lower temperatures.
Stimulated microbial communities.
In Gotland Deep water, a close phylogenetic relative of the aerobically sulfide-oxidizing and chemolithoautotrophic Thiomicrospira psychrophila (19) was stimulated in all thiosulfate enrichments. However, it is also interesting to note that a member of the Pseudomonas anguilliseptica group (Fig. 4) was stimulated in the Na2S2O3/Fe(III) enrichment (Table 4). P. anguilliseptica causes red spot disease in several fishes and was originally isolated from pond-cultured Anguilla japonica eels (45). It could be speculated that iron-rich, low-oxygen marine or brackish water habitats could be natural reservoirs for these bacteria.
As already mentioned, the Na2S2O3/NO3− enrichment with Landsort Deep water was dominated solely by a potential autotrophic denitrifying bacterium with sequences 100% identical to uncultured Helicobacteraceae 16S rRNA sequences (Tables 3 and 4; Fig. 4) and related to the anaerobic chemolithoautotrophic Thiomicrospira denitrificans (43). This dominance was identified by 16S rcDNA fingerprinting, which, due to the PCR bias, can only be accepted to be “semiquantitative.” However, we were able to validate these quantifications by qPCR, because a real-time protocol for a relative quantification of the uncultured Helicobacteraceae strain G138eps1 was already established earlier (20). With nearly identical 16S rRNA abundances of 35% (analyzed by SSCP) (Table 4) and 32% (analyzed by qPCR), the SSCP approach seemed to be appropriate for quantifications in this case.
CO2 dark fixation was almost exclusively due to the activity of chemolithoautotrophic bacteria. Their calculated bacterial cell doubling in the range between 12 and 18 h (Table 5) was comparable to that of heterotrophic marine bacterial populations (15). Cell numbers reached 5.6 × 105 ml−1 for the autotrophic denitrifying Helicobacteraceae strain. Although this is approximately three times higher than earlier estimated concentrations for this bacterium determined by strain-specific quantitative PCR for station 271, it is surprisingly similar considering that two different and separated habitats, sampling times, and calculation methods were bases for our calculations (20). This demonstrated the reproducibility of our approach but also the constancy of pelagic redoxcline assemblages in the Baltic Sea. For samples of Gotland Deep water from 1998, the uncultured Helicobacteraceae strain was proposed to be a key player for autotrophic denitrification (3). With both having two bands of identical sequence, which seemed to be characteristic for this bacterium, the 16S rcDNA SSCP fingerprints from 1998 and 2004 were very comparable (Fig. 3) (see Fig. 1 in reference 16). This underlined the stability of autotrophic nitrate-reducing assemblages in pelagic redoxclines of the Baltic Sea. However, phylogenetic relatives of this epsilonproteobacterium have also been detected in other, similar habitats worldwide (9, 10, 24, 44), indicating a general importance of this epsilonproteobacterium or its phylogenetic relatives for chemolithoautotrophic CO2 fixation in pelagic redoxclines. In our experiments this phylotype was also independently stimulated in the Na2S2O3 and Na2S2O3/Mn(IV) enrichments (Table 4), possibly indicating broader physiological capacities of this organism. Physiological versatilities have already been reported for other chemolithoautotrophic bacterial groups. For example, anaerobic respiration using Fe(III) or S0 as an electron acceptor and H2 or S0 as an electron donor served as a primary energy source of the well-known chemolithoautotrophic bacterium Acidithiobacillus ferrooxidans (29). Pronk et al. (32, 33) and Das et al. (7) showed that this bacterium also grew with the oxidation of S0 by Fe(III) under oxygen-limited and acidophilic conditions. To our knowledge, this has not been found for marine environments. However, several chemolithoautotrophic and versatile Epsilonproteobacteria have been described recently. For example, oxygen could serve as an alternative electron acceptor for denitrifying Nitratiruptor tergarcus and Nitratifractor salsuginis (27). Strictly anaerobic Thioreductor micantisoli was able to oxidize hydrogen and reduce elemental sulfur or nitrate (26). Takai et al. (41) described several new strains that were able to oxidize hydrogen or reduced sulfur compounds and to reduce O2, nitrate, or elemental sulfur. All of these strains were mesophilic to thermophilic; however, sulfide-oxidizing microaerophilic Thiomicrospira sp. strain CVO was also able to grow at lower temperatures and also might utilize limiting amounts of oxygen or nitrate (13).
In conclusion, our data indicated that at pelagic oxic/anoxic interfaces of the central Baltic Sea, chemolithoautotrophic CO2 dark fixation was due to nitrate and oxygen but not to Fe(III)/Mn(IV) reduction in combination with reduced sulfur compounds. An epsilonproteobacterium appeared to be a key organism for autotrophic nitrate reduction in different pelagic redoxclines of the Baltic Sea. Ammonium did not stimulate CO2 dark fixation significantly. Thus, the relevance of a potential autotrophic Fe(III)/Mn(IV) reduction for the (bio-)geochemical cycling and fluxes of these ions has to be investigated further, especially under strictly anaerobic conditions. Moreover, our findings support the well-known fact that phylogenetic identification of microorganisms alone does not necessarily give insight into their functional role in vivo. This underlines the importance and usefulness of our approach of combining the identification of stimulated bacterial phylotypes with the analysis of CO2 fixation rates upon the addition of potential substrate combinations in order to investigate their role in biogeochemical cycles in redoxclines of the Baltic Sea.
Acknowledgments
We are grateful to the captain and crew of RV Professor Albrecht Penck. The excellent technical assistance of Ursula Hennings and Bärbel Buuk is greatly appreciated. Additionally, we thank two anonymous reviewers for their useful advice to improve the quality of this paper.
This work was funded by the Leibniz-Institut für Ostseeforschung Warnemünde.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Billen, G. 1976. Evaluation of nitrifying activity in sediments by dark 14C-bicarbonate incorporation. Water Res. 10:51-57. [Google Scholar]
- 3.Brettar, I., M. Labrenz, S. Flavier, J. Bötel, H. Kuosa, R. Christen, and M. G. Höfle. Identification of a Thiomicrospira-like Epsilonproteobacterium as catalyst for denitrification in the central Baltic Sea. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 4.Brettar, I., and G. Rheinheimer. 1991. Denitrification in the central Baltic: evidence for hydrogen sulfide oxidation as motor of denitrification at the oxic-anoxic interface. Mar. Ecol. Prog. Ser. 77:157-169. [Google Scholar]
- 5.Brewer, G. P., and D. W. Spencer. 1971. Colorimetric determination of manganese in anoxic waters. Limnol. Oceanogr. 16:107-110. [Google Scholar]
- 6.Casamayor, E. O., J. García-Cantizano, J. Mas, and C. Pedrós-Alió. 2001. Primary production in estuarine oxic/anoxic interfaces: contribution of microbial dark CO2 fixation in the Ebro River Salt Wedge Estuary. Mar. Ecol. Prog. Ser. 215:49-56. [Google Scholar]
- 7.Das, A., A. K. Mishra, and P. Roy. 1992. Anaerobic growth on elemental sulfur using dissimilar iron reduction by autotrophic Thiobacillus ferrooxidans. FEMS Microbiol. Lett. 97:167-172. [Google Scholar]
- 8.Detmer, A. E., H. C. Giesenhagen, V. M. Trenkel, H. Auf dem Venne, and F. J. Jochem. 1993. Phototrophic and heterotrophic pico- and nanoplankton in anoxic depths of the central Baltic Sea. Mar. Ecol. Prog. Ser. 99:197-203. [Google Scholar]
- 9.Engel, A. S., N. Lee, M. L. Porter, L. A. Stern, P. C. Bennett, and M. Wagner. 2003. Filamentous “Epsilonproteobacteria” dominate microbial mats from sulfidic cave springs. Appl. Environ. Microbiol. 69:5503-5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Engel, A. S., M. L. Porter, L. A. Stern, S. Quinlan, and P. C. Bennett. 2004. Bacterial diversity and ecosystem function of filamentous microbial mats from aphotic (cave) sulfidic springs dominated by chemolithoautotrophic “Epsilonproteobacteria.” FEMS Microbiol. Ecol. 51:31-53. [DOI] [PubMed] [Google Scholar]
- 11.Enoksson, V. 1986. Nitrification rates in the Baltic Sea: comparison of three isotope techniques. Appl. Environ. Microbiol. 51:244-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuhrman, J. A., T. D. Sleeter, C. A. Carlson, and L. M. Proctor. 1989. Dominance of bacterial biomass in the Sargasso Sea and its ecological implications. Mar. Ecol. Prog. Ser. 57:207-217. [Google Scholar]
- 13.Gevertz, D., A. J. Telang, G. Voordouw, and G. E. Jenneman. 2000. Isolation and characterization of strains CVO and FWKO B, two novel nitrate-reducing, sulfide-oxidizing bacteria isolated from oil field brine. Appl. Environ. Microbiol. 66:2491-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grasshoff, K., M. Erhardt, and K. Kremling. 1983. Methods of seawater analysis, 2nd ed. Verlag Chemie, Weinheim, Germany.
- 15.Hagström, Å., J. W. Ammerman, S. Henrichs, and F. Azam. 1984. Bacterioplankton growth in seawater. II. Organic matter utilization during steady-state growth in seawater cultures. Mar. Ecol. Prog. Ser. 18:41-48. [Google Scholar]
- 16.Höfle, M. G., S. Flavier, R. Christen, J. Bötel, M. Labrenz, and I. Brettar. 2005. Retrieval of nearly complete 16S rRNA gene sequences from environmental DNA following 16S rRNA-based community fingerprinting. Environ. Microbiol. 7:670-675. [DOI] [PubMed] [Google Scholar]
- 17.Jannasch, H. W., C. O. Wirsen, and S. J. Molyneaux. 1991. Chemoautotrophic sulfur-oxidizing bacteria from the Black Sea. Deep-Sea Res. 38:S1105-S1120. [Google Scholar]
- 18.Jørgensen, B. B., and F. Bak. 1991. Pathways and microbiology of thiosulfate transformations and sulfate reduction in a marine sediment (Kattegat, Denmark). Appl. Environ. Microbiol. 57:847-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knittel, K., J. Kuever, A. Meyerdierks, R. Meinke, R. Amann, and T. Brinkhoff. 2005. Thiomicrospira arctica sp. nov. and Thiomicrospira psychrophila sp. nov., psychrophilic, obligately chemolithoautotrophic, sulfur-oxidizing bacteria isolated from marine Arctic sediments. Int. J. Syst. Evol. Microbiol. 55:781-786. [DOI] [PubMed] [Google Scholar]
- 20.Labrenz, M., I. Brettar, R. Christen, S. Flavier, J. Bötel, and M. G. Höfle. 2004. Development and application of a real-time PCR approach for quantification of uncultured bacteria in the central Baltic Sea. Appl. Environ. Microbiol. 70:4971-4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Chichester, United Kingdom.
- 22.Lepland, A., and R. L. Stevens. 1998. Manganese authigenesis in the Landsort Deep, Baltic Sea. Mar. Geol. 151:1-25. [Google Scholar]
- 23.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K.-H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Madrid, V. M., G. T. Taylor, M. I. Scranton, and A. Y. Chistoserdov. 2001. Phylogenetic diversity of bacterial and archaeal communities in the anoxic zone of the Cariaco Basin. Appl. Environ. Microbiol. 67:1663-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morris, I., H. E. Glover, W. A. Kaplan, D. P. Kelly, and A. L. Weightman. 1985. Microbial activity in the Cariaco Trench. Microbios 42:133-144. [Google Scholar]
- 26.Nakagawa, S., F. Inagaki, K. Takai, K. Horikoshi, and Y. Sako. 2005. Thioreductor micantisoli gen. nov., sp. nov., a novel mesophilic, sulfur-reducing chemolithoautotroph within the ɛ-Proteobacteria isolated from hydrothermal sediments in the Mid-Okinawa Trough. Int. J. Syst. Evol. Microbiol. 55:599-605. [DOI] [PubMed] [Google Scholar]
- 27.Nakagawa, S., K. Takai, F. Inagaki, K. Horikoshi, and Y. Sako. 2005. Nitratiruptor tergarcus gen. nov., sp. nov. and Nitratifractor salsuginis gen. nov., sp. nov., nitrate-reducing chemolithoautotrophs of the ɛ-Proteobacteria isolated from a deep-sea hydrothermal system in the Mid-Okinawa Trough. Int. J. Syst. Evol. Microbiol. 55:925-933. [DOI] [PubMed] [Google Scholar]
- 28.Neretin, L. N., C. Pohl, G. Jost, T. Leipe, and F. Pollehne. 2003. Manganese cycling in the Gotland Deep, Baltic Sea. Mar. Chem. 82:125-143. [Google Scholar]
- 29.Ohmura, N., K. Sasaki, N. Matsumoto, and H. Saiki. 2002. Anaerobic respiration using Fe3+, S0, and H2 in the chemolithoautotrophic bacterium Acidithiobacillus ferrooxidans. J. Bacteriol. 184:2081-2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pohl, C., and U. Hennings. 1999. The effect of redox processes on the partitioning of Cd, Pb, Cu, and Mn between dissolved and particulate phases in the Baltic Sea. Mar. Chem. 65:41-53. [Google Scholar]
- 31.Pöhler, I., D. F. Wenderoth, K. Wendt-Potthoff, and M. G. Höfle. 2002. Bacterioplankton community structure and dynamics in enclosures during bio-remediation experiments in an acid mining lake. Water Air Soil Pollut. Focus 2:111-121. [Google Scholar]
- 32.Pronk, J. T., J. C. Debruyn, P. Bos, and J. G. Kuenen. 1992. Anaerobic growth of Thiobacillus ferrooxidans. Appl. Environ. Microbiol. 58:2227-2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pronk, J. T., K. Liem, P. Bos, and J. G. Kuenen. 1991. Energy transduction by anaerobic ferric iron respiration in Thiobacillus ferrooxidans. Appl. Environ. Microbiol. 57:2063-2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rönner, U., and F. Sörensson. 1985. Denitrification rates in the low-oxygen waters of the stratified Baltic proper. Appl. Environ. Microbiol. 50:801-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roslev, P., M. B. Larsen, D. Jørgensen, and M. Hesselsoe. 2004. Use of heterotrophic CO2 assimilation as a measure of metabolic activity in planktonic and sessile bacteria. J. Microbiol. Methods 59:381-393. [DOI] [PubMed] [Google Scholar]
- 36.Schwieger, F., and C. C. Tebbe. 1998. A new approach to utilize PCR-single-strand-conformation polymorphism for 16S rRNA gene-based microbial community analysis. Appl. Environ. Microbiol. 64:4870-4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shively, J. M., G. van Keulen, and W. G. Meijer. 1998. Something from almost nothing: carbon dioxide fixation in chemoautotrophs. Annu. Rev. Microbiol. 52:191-230. [DOI] [PubMed] [Google Scholar]
- 38.Sørensen, J., J. M. Tiedje, and R. B. Firestone. 1980. Inhibition by sulfide of nitric and nitrous oxide by denitrifying Pseudomonas fluorescens. Appl. Environ. Microbiol. 39:105-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sorokin, Y. I. 1972. The bacterial population and the processes of hydrogen sulphide oxidation in the Black Sea. J. Cons. Int. Explor. Mer. 34:423-454. [Google Scholar]
- 40.Steemann Nielsen, E. 1952. The use of radio-active carbon 14C for measuring organic production in the sea. J. Con. Perm. Int. Explor. Mer. 18:117-140. [Google Scholar]
- 41.Takai, K., F. Inagaki, S. Nakagawa, H. Hirayama, T. Nunoura, Y. Sako, K. H. Nealson, and K. Horikoshi. 2003. Isolation and phylogenetic diversity of members of previously uncultivated ɛ-Proteobacteria in deep-sea hydrothermal fields. FEMS Microbiol. Lett. 218:167-174. [DOI] [PubMed] [Google Scholar]
- 42.Taylor, G. T., M. Iabichella, T.-Y. Ho, M. I. Scranton, R. C. Thunell, F. Muller-Karger, and R. Varela. 2001. Chemoautotrophy in the redox transition zone of the Cariaco Basin: a significant midwater source of organic carbon production. Limnol. Oceanogr. 46:148-163. [Google Scholar]
- 43.Timmer-ten Hoor, A. 1975. A new type of thiosulfate oxidizing, nitrate reducing microorganism: Thiomicrospira denitrificans sp. nov. Neth. J. Sea Res. 9:344-350. [Google Scholar]
- 44.Vetriani, C., H. V. Tran, and L. J. Kerkhof. 2003. Fingerprinting microbial assemblages from the oxic/anoxic chemocline of the black sea. Appl. Environ. Microbiol. 69:6481-6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wakabayashi, H., and S. Egusa. 1972. Characteristics of a Pseudomonas sp. from an epizootic of pond-cultured eels (Anguilla japonica). Bull. Jpn. Soc. Sci. Fish. 38:577-587. [Google Scholar]
- 46.Weinbauer, M. G., I. Fritz, D. F. Wenderoth, and M. G. Höfle. 2002. Simultaneous extraction of total RNA and DNA from bacterioplankton suitable for quantitative structure and function analyses. Appl. Environ Microbiol. 68:1082-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zopfi, J., T. G. Ferdelman, B. B. Jørgensen, A. Teske, and B. Thamdrup. 2001. Influence of water column dynamics on sulfide oxidation and other major biogeochemical processes in the chemocline of Mariager Fjord (Denmark). Mar. Chem. 74:29-51. [Google Scholar]