Abstract
Yeast Saccharomyces cerevisiae cells generally cannot synthesize biotin, a vitamin required for many carboxylation reactions. Although sake yeasts, which are used for Japanese sake brewing, are classified as S. cerevisiae, they do not require biotin for their growth. In this study, we identified a novel open reading frame (ORF) in the genome of one strain of sake yeast that we speculated to be involved in biotin synthesis. Homologs of this gene are widely distributed in the genomes of sake yeasts. However, they are not found in many laboratory strains and strains used for wine making and beer brewing. This ORF was named BIO6 because it has 52% identity with BIO3, a biotin biosynthesis gene of a laboratory strain. Further research showed that yeasts without the BIO6 gene are auxotrophic for biotin, whereas yeasts holding the BIO6 gene are prototrophic for biotin. The BIO6 gene was disrupted in strain A364A, which is a laboratory strain with one copy of the BIO6 gene. Although strain A364A is prototrophic for biotin, a BIO6 disrupted mutant was found to be auxotrophic for biotin. The BIO6 disruptant was able to grow in biotin-deficient medium supplemented with 7-keto-8-amino-pelargonic acid (KAPA), while the bio3 disruptant was not able to grow in this medium. These results suggest that Bio6p acts in an unknown step of biotin synthesis before KAPA synthesis. Furthermore, we demonstrated that expression of the BIO6 gene, like that of other biotin synthesis genes, was upregulated by depletion of biotin. We conclude that the BIO6 gene is a novel biotin biosynthesis gene of S. cerevisiae.
Biotin (vitamin H) is an essential nutrient for all organisms. It is a cofactor of many enzymes involved in carboxylation reactions, such as gluconeogenesis, amino acid metabolism, fatty acid biosynthesis, and energy metabolism (21). Biotin is synthesized in many organisms from lower prokaryotes to higher plants including Escherichia coli and Arabidopsis thaliana, and its biosynthesis pathways have been well studied (2, 5, 7, 9, 11). Although there are differences in the early steps of biotin synthesis, the last four steps from pimeloyl-coenzyme A (CoA) to biotin are usually conserved (Fig. 1). However, some microbes and higher vertebrates cannot synthesize biotin by themselves and must incorporate it from the external environment. S288C (19), a typical laboratory strain of Saccharomyces cerevisiae, is an example of such organisms. It is auxotrophic for biotin and requires biotin or intermediates of biotin synthesis such as 7-keto-8-amino-pelargonic acid (KAPA) or 7,8-diamino-pelargonic acid (DAPA) in the medium (21). Only the last three steps of the biotin biosynthesis pathway (BIO3, BIO4, and BIO2) have been found in S288C, and steps before KAPA synthesis are absent or have not been elucidated (Fig. 1) (23). Some of the biotin synthesis genes are found as a cluster located on chromosome XIV. On this chromosome, the genes encoding DAPA aminotransferase (BIO3) and dethiobiotin synthetase (BIO4) are linked to a gene that codes for a transporter of KAPA (BIO5). The gene encoding biotin synthase (BIO2) (39) is found on chromosome VII.
FIG. 1.
Pathways of biotin biosynthesis.
Sake yeasts are used to make sake, which is a traditional alcoholic beverage in Japan. Although they are classified as S.cerevisiae strains (37), sake yeasts have many characteristics that differ from those of other yeast strains, such as foam formation in mash (25), tolerance of high concentrations of ethanol, strong fermentation at low temperatures, and rich flavors (12, 15, 16, 38). Another remarkable characteristic of sake yeasts is that they are prototrophic for biotin (29, 30), in contrast to other S. cerevisiae strains. However, the mechanism of their biotin prototrophy is unclear and no genes involved in biotin biosynthesis have been identified in sake yeasts. While analyzing the genome of a sake yeast strain, we identified a novel gene, BIO6, which is similar to E. coli bioA (22) and S.cerevisiae BI03 (23). Homologs of this gene were found in various sake yeast strains but not found in typical laboratory strains and other industrial strains. In this paper, we report that BIO6 is a new gene involved in a step before KAPA synthesis of the biotin synthesis pathway in S. cerevisiae.
MATERIALS AND METHODS
Strains and culture conditions.
Yeast strains used in this study are listed in Table 1. Yeast cells were cultivated at 30°C in YPD (1% yeast extract, 2% bactopeptone, and 2% glucose) medium with shaking. When needed, 20-μg/ml adenine sulfate dihydrate was added, resulting in YPAD medium. For selection of transformants, 200 μg/ml for A364A or 1 mg/ml for K7 of Geneticin (Sigma) or 0.5 μg/ml of aureobasidin A (Takara Bio) was supplemented. A synthetic minimum medium (SM) was used for a biotin auxotrophy test. One liter of this medium contains 10 g of glucose, 0.5 g of (NH4)2SO4, 0.85 g of KH2PO4, 0.15 g of K2HPO4, 0.5 g of MgSO4 · 7 H2O, 0.1 g of NaCl, 0.1 g of CaCl2 · 2H2O, 10mg of l-histidine, 20 mg of dl-methionine, 20 mg of l-tryptophan, 10 ml of 100× vitamin mixture, and 10 ml of 100× trace element mixture. One hundred milliliters of 100× vitamin mixture contains 20 mg of calcium pantothenate, 20 μg of folic acid, 100 mg of inositol, 4 mg of nicotinic acid, 2 mg of p-aminobenzoic acid, 4 mg of pyridoxine hydrochloride, 2 mg of riboflavin, and 4 mg of thiamine hydrochloride. One hundred milliliters of 100× trace element mixture contains 5 mg of H3BO3, 0.4 mg of CuSO4 · 5H2O, 1 mg of KI, 2 mg of FeCl3 · 6H2O, 4mg of MnSO4 · 4H2O, 2 mg of Na2MoO4 · 2H2O, and 4 mg of ZnSO4 · 7H2O. For growth of A364A, 3 mg of l-tyrosine, 2 mg of uracil, 3 mg of lysine monohydrochloride and 2 mg of adenine sulfate dihydrate are added to 1 liter of SM. Agarose (20 g/liter; Cambrex) was added in SM to form solid medium, because the trace amount of biotin in agar could be enough for the growth of biotin-auxotrophic yeasts. For the same reason, ultrapure water was used to prepare SM. To absolutely deplete the contaminant biotin from the medium, avidin (Sigma) was added in SM with a final concentration of 23 μg/ml to result SM-Biotin. The d-(+)-biotin was added in SM at a final concentration of 25ng/ml to result in SM+Biotin. Addition of 25-ng/ml KAPA, 5-ng/ml DAPA, or 50-ng/ml dethiobiotin (DTB) to the SM-Biotin resulted in SM+KAPA, SM+DAPA, and SM+DTB, respectively. KAPA and DAPA were kindly provided by Jack F. Kirsch of the University of California, Berkeley. For the biotin auxotrophy assay, 5 ml of cells that had been cultured overnight in YPD or YPAD liquid medium with shaking was harvested and washed three times with 20 ml of sterile ultrapure water. The cell concentrations were adjusted to an optical density at 660 nm (OD660) of 0.5 and one loopful of the 10−1 diluents was streaked on an SM+Biotin or SM-Biotin plate, followed by incubation at 30°C. For an assimilation test of intermediates of yeast biotin biosynthesis, similarly prepared cells were streaked on SM+KAPA, SM+DAPA, and SM+DTB plates, followed by incubation at 30°C for 5 days (for K7 diploid cells) or 7 to 8days (for A364A haploid cells). E. coli strains were cultured in LB (1% tryptone, 0.5% yeast extract, and 1% NaCl) medium, supplemented with 50 μg of ampicillin/ml or 25 μg of kanamycin/ml if necessary.
TABLE 1.
Yeast strains used in this study
| Straina | Remarks or genotype | Source (reference) |
|---|---|---|
| IFO10217 | Type strain of S. cerevisiae | ATCC 18824 |
| S288C | Laboratory yeast MATα | ATCC 26108 (19) |
| Σ1278B | Laboratory yeast MATa/α | ATCC 201731 |
| X2180 | Laboratory yeast MATa/α | ATCC 26109 (4) |
| A364A | Laboratory yeast; MATaade1 ade2 ura1 his7 lys2 tyr1 gal1 | ATCC 204509 |
| K6 | Sake yeast, kyokai no. 6 | Brewing Society of Japan |
| K7 | Sake yeast, kyokai no. 7 | Brewing Society of Japan |
| K8 | Sake yeast, kyokai no. 8 | Brewing Society of Japan |
| K9 | Sake yeast, kyokai no. 9 | Brewing Society of Japan |
| K10 | Sake yeast, kyokai no. 10 | Brewing Society of Japan |
| KL88 | Sake yeast | This work |
| IK5 | Sake yeast | This work |
| GR10 | Sake yeast | This work |
| TK1 | Sake yeast | This work |
| IFO 0304 | Sake yeast | IFO 0304 (17) |
| IFO 0309 | Sake yeast | IFO 0309 |
| Miyazaki | Shochu yeast | This work |
| OC-2 | Wine yeast | IAM 4274 (10) |
| EC1118 | Wine yeast | Lallemand |
| S6U | Wine yeast | Lallemand |
| IFO 2044 | Bakery yeast | IFO 2044 (20) |
| IFO 1951 | Ale yeast | IFO 1951 |
| IFO 1952 | Ale yeast | IFO 1952 |
| IFO 2106 | Distillery yeast | IFO 2106 |
| IFO 2112 | American whisky yeast | IFO 2112 |
| IFO 2114 | Japanese whisky yeast | IFO 2114 |
| YPH499 | MATaade2 ura3 his3 lys2 tyr1 leu2 | ATCC 204679 (26) |
| YPH500 | MATα ade2 ura3 his3 lys2 tyr1 leu2 | ATCC 204680 (26) |
| YHW11 | A364A bio6::kanMX4 | This work |
| YHW12 | YHW11 pAUR101-BIO6 | This work |
| YHW13 | YHW11 pAUR101 | This work |
| YHW14 | A364A bio3::kanMX4 | This work |
| YHW21 | K7 haploid, bio3::kanMX4 | This work |
All strains are S. cerevisiae except for S6U, which is a hybrid between S. cerevisiae and S. bayanus.
DNA manipulation.
Transformation of E. coli, plasmid preparation, restriction enzyme mapping, DNA ligation, and other DNA manipulations were carried out by standard techniques (24). PCR was performed with KOD DNA polymerase (Toyobo). Cosmid extraction from E. coli cells was performed using a kit (QIAprep Spin Miniprep kit; QIAGEN). Yeast genomic DNA was prepared as previously described (1). Yeast transformation was carried out as described previously (3). DNA sequences were determined with a DNA sequencer (ABI PRISM 310; Applied Biosystems).
PFGE Southern blot analysis.
Separation of chromosomes by pulse-field gel electrophoresis (PFGE) and Southern blot analysis was carried out as described previously (18). Both end fragments of the cosmid COS-55 insert were amplified by PCRs with COS-55 as a template. COS-55-F was amplified with 55-COSF1 (CAACTCCTCGGAAGTAATTATGTATGG) and 55-COSF2 (AAGAGATTG GTGATCAGATCTCCAG) as primers. COS-55-R was amplified with 55-COSR1 (ATCTGAATCTTCTGTTAAGCGGC) and 55-COSR2 (CGCAATCAAATAGTATCGAAGCTG) as primers. The resultant fragments were used as hybridization probes after being labeled with 32P.
Cloning of the BIO6 gene of K7.
The original BIO6 gene found in contig 4.1 was amplified by PCR with K7 genomic DNA as a template and ORF-F2 (CAAACATCATTGGCCACT) and ORF-R (GCTGAAGGTGTCTTGAAA) as primers. The 1.5-kb amplified fragment was cloned into the SmaI-HincII-digested pUC118 (33) to get pUC118-KBIO6. To obtain other copies of the BIO6 genes of K7 locating on different chromosomes, a cosmid library was screened by colony hybridization as described previously (18). Hybridization was performed using the COS-55-F fragment as a probe. The cosmids obtained were analyzed by shotgun sequencing to determine the locations and sequences of the BIO6 genes.
Cloning and disruption of the BIO6 gene of A364A.
The BIO6 gene of A364A was amplified by PCR with A364A genomic DNA as a template and ORF-F (TGACGAATGTGTGAACAT) and ORF-R as primers. The amplified fragment was cloned into the SmaI-HincII-digested pUC118 to get pUC118-ABIO6. To construct a BIO6-disrupted mutant of A364A, a SmaI-EcoRV fragment containing the kanMX4 gene of pFA6a-kanMX4 (34) was inserted into the SalI site of pUC118-ABIO6 to result in pUC118-ABIO6kan. The 2.2-kb SphI-KpnI fragment of this plasmid containing the kanMX4 gene flanked by the partial BIO6 sequences was used to transform A364A. Transformants were screened by Geneticin resistance. Disruption of the BIO6 gene was confirmed by PCR, and the BIO6-disrupted mutant was called YHW11. For a complementation test of YHW11, the BIO6 gene with its 1,000-bp promoter region was amplified by PCR with A364A genomic DNA as a template and ORF-F2 and ORF-R as primers. The 2.5-kb amplified fragment was cloned into the SmaI-digested pAUR101 (Takara Bio) to get pAUR101-BIO6. This plasmid was cleaved with StuI, and the resultant fragment was used to transform YHW11. Transformants were screened by aureobasidin A resistance. Introduction of the BIO6 gene with its promoter was confirmed by PCR, and the strain obtained was called YHW12.
Cloning and disruption of the BIO3 gene of A364A.
The BIO3 gene of A364A was amplified by PCR with A364A genomic DNA as a template and BIO3-F (AGGATGTCAACAGCGCAATT) and BIO3-R (AGGTCCGCTAGAATAAAT) as primers. The 1.5-kb amplified fragment was cloned into HincII-digested pUC118 to get pUC118-BIO3. To construct a BIO3-disrupted mutant of A364A, a 1.4-kb fragment containing the kanMX4 gene of pFA6a-kanMX was inserted into the BglII site of pUC118-BIO3 to get pUC118-BIO3kan. A 2.1-kb EcoRI-SphI fragment containing the kanMX4 gene flanked by the partial BIO3 sequences from pUC118-BIO3kan was used to transform A364A. Transformants were screened by Geneticin resistance. Disruption of the BIO3 gene was confirmed by PCR and the BIO3-disrupted mutant was called YHW14. The same processes were performed with K7, and the BIO3-disrupted mutant was called YHW21.
Northern blot analysis.
After being precultured overnight in 5 ml YPD or YPAD liquid medium with shaking, yeast cells were harvested and washed three times with 20 ml sterile ultrapure water. The cells were diluted in SM+Biotin or SM-Biotin liquid medium at OD660 = 0.2 (for A364A) or OD660 = 0.1 (for K7). After incubation at 30°C for 5 h with shaking, the cells were harvested and used to prepare total RNA by hot-phenol extraction (14). Northern blot analyses were performed as described previously (31). A probe for the BIO6 gene was the COS-55-F fragment. A probe for the ACT1 gene was prepared by PCR using ACT1-L (GAAGTGTGATGTCGATGTCC) and ACT1-R (CTTTCTTTCTGGAGGAGCAA) as primers.
Nucleotide sequence accession numbers.
The nucleotide sequences of COS-55, K7 BIO6, A364A BIO6, and A364A BIO3 have been submitted to the DDBJ-EMBL-GenBank database under accession numbers AB200246, AB188681, AB188515, and AB200248, respectively.
RESULTS
Identification of the BIO6 gene of K7. To characterize sake yeast, we partially sequenced the genome of sake yeast K7 by the shotgun method (our unpublished data). Almost all sequences were nearly identical to the sequences of S. cerevisiae S288C, whose genome was sequenced and registered in the databases. However, several sequences had low similarity to the registered sequences. Parts of these sequences were identical to the sequence of COS-55-F, a terminal region of a cosmid (COS-55) identified in random sequencing of a cosmid library of K7 genome (our unpublished data). COS-55 contains a 37-kb insert; its other terminal region (COS-55-R) showed high degrees of similarity to the subtelomeric region of the right arm of chromosome II of S288C, while COS-55-F did not. By assembling the sequences obtained from the shotgun method and the cosmids, we identified a contig of 6.3 kb (contig 4.1) (Fig. 2A). We discovered four ORFs on this contig. One of the ORFs (ORF2) contained COS-55-F and consisted of 1,314 nucleotides encoding 437 amino acids (Fig. 3). The amino acid sequence of this ORF was approximately 45% identical to the sequence of DAPA aminotransferase of S288C encoded by the BIO3 gene and 55% identical to the sequence of DAPA aminotransferase of E. coli encoded by the bioA gene; both of these genes are involved in the biotin synthesis pathway. We tentatively called this ORF BIO6.
FIG. 2.
Cloning of the BIO6 gene of K7. (A) Structure of contig 4.1. The arrows indicate protein coding regions and orientations. E, EcoRI; Ev, EcoRV; H, HindIII; N, NotI; P, PstI; S, SalI; Sp, SphI; X, XbaI. The COS-55-F fragment is indicated by a bar. (B) PFGE Southern blot analyses of S288C (lanes 1 and 3) and K7 (lanes 2 and 4). Chromosome DNAs prepared from approximately the same amount of cells were used for PFGE Southern blot analysis using COS-55-F (lanes 1 and 2) and COS-55-R (lanes 3 and 4) fragments as probes. The presumed chromosome numbers are indicated.
FIG. 3.
Nucleotide and deduced amino acid sequences of the BIO6 gene. The COS-55-F sequence is indicated by underlining. The SAM and PLP binding sites are indicated by double underlining.
K7 has multiple copies of the BIO6 gene.
We noticed that eight fragments in the K7 genome shotgun sequence database were identical to COS-55-F. This frequency was higher than the frequencies of the other sequences, suggesting that BIO6 exists as multiple copies in the K7 genome. Therefore, we analyzed the location of BIO6 in the K7 chromosomes by PFGE Southern blot analysis using COS-55-F and COS-55-R as probes. As shown in Fig. 2B, COS-55-R hybridized to chromosome II, but COS-55-F hybridized to chromosomes I, II, IX, and XI and possibly others. This result suggests that K7 contains at least four copies of BIO6 and COS-55 was derived from chromosome II. In a PFGE Southern blot analysis of the S288C genome, COS-55-R hybridized to chromosome II, and COS-55-F did not hybridize to any chromosomes, as expected. Screening the K7 cosmid library with COS-55-F as a probe revealed three positive cosmid clones (COS-2-1, COS-5-1, and COS-15-2). Their insert sequences were determined by the shotgun sequencing method. The sequence of COS-2-1 was identical to the sequence of contig 4.1 with partial similarity to the subtelomeric region of S288C chromosome XV left arm (data not shown). The sequence of COS-5-1 is similar to the sequence of the subtelomeric region of S288C chromosome XI left arm (data not shown). The sequence of COS-15-2 was similar to the sequence of the subtelomeric region of S288C chromosome IX left arm (data not shown). Sequence similarity and the results of PFGE Southern blot analysis suggested that COS-5-1 and COS-15-2 are located on chromosomes XI and IX of K7, respectively. Although we were not able to determine the location of COS-2-1, it might be located on the subtelomeric region of chromosome I, based on the similarity and PFGE Southern blot analysis. Taken together, these results indicate that K7 has at least four copies of the BIO6 gene on the subtelomeric regions of chromosomes I, II, IX, and XI. Perfect sequence identity among the four BIO6 ORFs, as well as highly conserved upstream regions, suggests that the BIO6 gene is functional in K7, since functional genes tend to be stable and highly preserved.
The presence of BIO6 is related to biotin prototrophy.
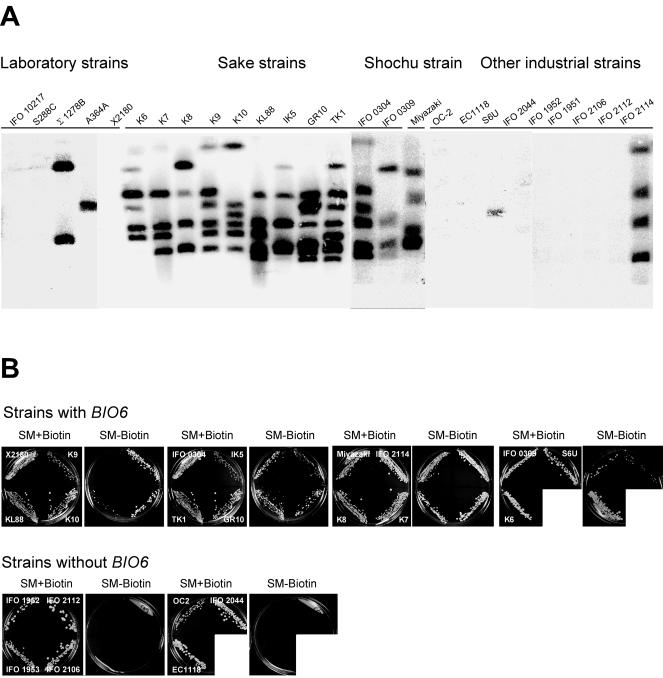
Sake yeasts, including K7, are known to be prototrophic for biotin. To determine whether BIO6 is involved in biotin prototrophy, we searched for BIO6 homologs in some laboratory and industrial strains of S. cerevisiae by PFGE Southern blot analysis. As shown in Fig. 4A, the type strain of S. cerevisiae (IFO 10217) and typical laboratory strains (S288C and X2180) did not have BIO6, while some laboratory strains (Σ1278B and A364A) had BIO6. Sake yeasts and shochu (Japanese distilled liquor) yeasts had multiple copies of BIO6, similar to K7. Other industrial yeast strains including most wine, brewery, and bakery yeasts did not have BIO6. Exceptions were a wine yeast (S6U, a hybrid of S. cerevisiae and Saccharomyces bayanus) and a distillery yeast (IFO 2114). Next, we determined the biotin requirement of these strains in a biotin-deficient medium. All strains having BIO6 grew in biotin-deficient medium and thus were prototrophic for biotin (Fig. 4B). Other strains without BIO6 were auxotrophic for biotin. These results demonstrate that BIO6 is widely distributed in S. cerevisiae and its presence is closely related to biotin prototrophy.
FIG. 4.
Relationship between the BIO6 gene and biotin prototrophy. (A) PFGE Southern blot analysis of the BIO6 gene in various yeast strains. Chromosome DNAs prepared from approximately the same amount of cells were used for PFGE Southern blot analysis using the COS-55-F fragment as a probe. (B) Biotin auxotrophic assay. Yeast strains with BIO6 (top) or without BIO6 (bottom) were streaked on SM+Biotin and SM-Biotin and incubated at 30°C for 6 days. Strain X2180 (top) was used as a negative control.
BIO6 is essential for biotin synthesis.
To confirm that BIO6 is actually involved in biotin synthesis, we attempted to disrupt the BIO6 gene. Since K7 is a diploid strain and has at least four copies of BIO6 on different chromosomes, disruption of all BIO6 genes was expected to be difficult. We chose A364A to disrupt BIO6 because it is a haploid laboratory strain and prototrophic for biotin and has only one copy of BIO6 on chromosome X or XIV (Fig. 5A). The locus of the A364A BIO6 gene does not seem to correspond to any of the K7 BIO6 genes that are located on chromosomes I, II, IX, and XI. The BIO6 gene of A364A was amplified by PCR and cloned into a plasmid vector. Sequence analysis showed that BIO6 of A364A is 98.8% identical to the BIO6 gene of K7 with the same length. The BIO6 gene of A364A was disrupted by inserting the kanMX4 gene into the BIO6 gene (Fig. 5B). The BIO6-disrupted mutant YHW11 was found to be auxotrophic for biotin (Fig. 5C). When a 2.5-kb fragment containing the complete BIO6 gene of A364A with its promoter region was reintroduced into YHW11 to get YHW12, the biotin prototrophy was recovered (Fig. 5C). These results demonstrate that the BIO6 gene is essential for biotin synthesis in A364A.
FIG. 5.
Cloning and disruption of the BIO6 gene in A364A. (A) PFGE Southern blot analyses of BIO6 of K7 and A364A. Chromosome DNAs prepared from approximately the same amount of cells were used for PFGE Southern blot analysis using the COS-55-F fragment as a probe. The presumed chromosome numbers are indicated. (B) Gene disruption of the BIO6 gene in A364A. A 1.5-kb fragment containing the BIO6 gene of A364A was amplified and cloned. The Geneticin resistance gene kanMX4, which was used as a selectable marker, was inserted in this fragment. Strain A364A was then transformed with part of this fragment. (C) Biotin auxotrophic assay. A364A, YHW11 (bio6Δ), YHW13 harboring an empty vector in YHW11 (bio6Δ-101), and YHW12 harboring a vector containing BIO6 of A364A (bio6Δ-BIO6) were tested. Cells of these strains were streaked on SM+Biotin and SM-Biotin and incubated at 30°C for 7 days.
Both BIO3 and BIO6 are needed for biotin synthesis.
Bio6p has approximately 45% identity with DAPA aminotransferase (Bio3p) of S288C. Therefore, Bio6p may act as a DAPA aminotransferase instead of a Bio3p in A364A. To preclude this possibility, we examined whether BIO3 was dispensable for biotin synthesis in A364A. First, we amplified a BIO3 homolog from A364A by PCR. Sequence analysis showed that A364A BIO3 was 99.4% identical to S288C BIO3. Then, we disrupted BIO3 of A364A by inserting the kanMX4 gene in it. The resultant strain, YHW14, was auxotrophic for biotin (Fig. 6 A). This result demonstrates that both BIO3 and BIO6 are required for biotin synthesis in A364A. When BIO3 was disrupted in K7 by the same method, the disruptant YHW21 was also auxotrophic for biotin (data not shown).
FIG. 6.
Effects of biotin synthesis intermediates on BIO6- or BIO3-disrupted strains. A364A, YHW11 (bio6Δ), and YHW14 (bio3Δ) were streaked on SM-Biotin, SM+KAPA, SM+DAPA, and SM+DTB and incubated at 30°C for 7 days.
Bio6p is involved in a different step of the biotin synthesis pathway.
Although Bio6p is similar to Bio3p, it appears to be involved in a different step of biotin synthesis because both BIO3 and BIO6 are required for biotin synthesis. Although S288C is auxotrophic for biotin, it can grow in biotin-deficient medium if an intermediate of the biotin synthesis pathway, such as KAPA, DAPA, or DTB (23), is supplemented. To identify the step of biotin synthesis in which Bio6p acts, we investigated whether the A364A bio3Δ strain (YHW14) and the bio6Δ strain (YHW11) can grow in these media. As expected from the fact that BIO3 encodes DAPA aminotransferase, YHW14 was able to grow on SM+DAPA and SM+DTB but not on SM+KAPA (Fig. 6). On the other hand, YHW11 was able to grow on each of SM+KAPA, SM+DAPA, and SM+DTB (Fig. 6). These results suggest that Bio3p and Bio6p are involved in independent steps of biotin synthesis and that Bio6p acts in a step before KAPA synthesis. YHW11 was not able to grow on the biotin-deficient medium supplemented with pimelic acid (data not shown). However, we were not able to determine whether BIO6 encodes KAPA synthase, since pimeloyl-CoA was not available.
Biotin deficiency upregulates BIO6.
The expressions of genes involved in biotin biosynthesis and biotin uptake (BIO2, BIO3, BIO4, and VHT1) were found to be inversely proportional to the extracellular biotin concentration (36). Transcription of BIO6 also appears to be regulated by biotin concentration (Fig. 7). No clear BIO6 signal was observed in K7 orA364A cells grown in medium supplemented with biotin (Fig. 7, lanes 1 and 3), but clear signals were detected in both cell types when they were grown in medium without biotin (lanes 2 and 4). The signal intensity of the K7 transcript in the induced state was much stronger than that of A364A, possibly because the K7 genome has multiple copies of BIO6.
FIG. 7.
Effect of biotin on BIO6 gene expression. Each lane was loaded with 5 μg total RNA from of K7 or A364A cells grown in the presence and absence of biotin. A COS-55-F fragment was used as a probe. Lanes 1 and 2, K7 cells grown in SM+Biotin and SM-Biotin, respectively; lanes 3 and 4, A364A cells grown in SM+Biotin and SM-Biotin, respectively. The ACT1 signal and ethidium bromide staining were determined as loading controls.
DISCUSSION
Many S. cerevisiae strains are biotin auxotrophic, which is caused by having an incomplete biotin biosynthesis pathway. Although sake yeasts were found to be prototrophic for biotin a half century ago, the mechanism of prototrophy is still unclear. In this paper, we have shown that the BIO6 gene is essential for biotin synthesis in the biotin prototrophic yeast A364A. Although we did not examine gene disruption of BIO6 of sake yeasts and other biotin prototrophic yeasts, we presume that the BIO6 genes are involved in biotin synthesis in these strains, based on the high identities of these genes and the relationship between BIO6 and biotin prototrophy (Fig. 4). Although Bio6p has similarity to DAPA aminotransferase of S288C (Bio3p) and DAPA aminotransferase of E. coli (the product of bioA), it does not act as a DAPA aminotransferase but acts in an unknown early step before KAPA. Biotin prototrophic yeasts also contain BIO3, which encodes a functional DAPA aminotransferase. However, introduction of the BIO6 gene to YPH499 or YPH500 (26), strains that are isogenic to S288C, did not rescue their biotin auxotrophy (data not shown), suggesting that some genes other than BIO6 are needed for biotin synthesis.
In E. coli, pimeloyl-CoA is converted to DAPA in two steps, one catalyzed by KAPA synthase (the product of bioF) and the other catalyzed by DAPA synthase (the product of bioA). Both KAPA synthase and DAPA synthase are vitamin B6-dependent enzymes that use pyridoxal 5′-phosphate (PLP) as a cofactor. KAPA synthase utilizes l-alanine and DAPA synthase utilizes S-adenosyl-l-methionine (SAM) as substrates (27, 28, 35). The bioA and bioF products are weakly homologous and have been suggested to be derived from a common ancestral gene (22). PLP and SAM binding-site motifs are found in similar positions in these two proteins (22). The BIO6 product is homologous to the bioA product and it also contains SAM and PLP binding-site motifs (263GLGRTG268 and 280PDIVCLGKTL289, respectively) (Fig. 8). Considering that Bio6p acts at a step before Bio3p, we hypothesize that Bio6p acts as KAPA synthase. However, further studies are needed to identify the enzymatic reaction in which Bio6p acts.
FIG. 8.
Sequence comparison of K7 Bio6p and E. coli bioA and bioF products. Conserved amino acids are indicated by white letters on black backgrounds. The SAM and PLP binding sites are indicated by asterisks and dots, respectively.
The BIO6 gene is present in a few laboratory strains of S.cerevisiae, as well as in many industrial strains of S. cerevisiae, such as sake and shochu yeasts. In most of these strains, BIO6 exists as multiple copies on different chromosomes, while it exists as a single copy in strains A364A and S6U. The BIO6 gene was not detected in the laboratory strains derived from S288C or in most wine, bakery, ale, and distillery strains. The wine strain S6U and the distillery strain IFO2114 are exceptions. Recently, the genomes of several Saccharomyces sensu stricto strains, including S. bayanus, S. paradoxus, S. mikatae, and S. kudriavzevii have been partially sequenced (13). Sequences with 81 to 90% identities to Bio6p were found in each of these four strains by BLAST searches (Fig. 9). These identities are significantly higher than the identity between Bio3p and Bio6p. This result shows that BIO6 not only exists in parts of S. cerevisiae but also exists in other strains of Saccharomyces sensu stricto yeast, which diverged approximately 5 million years ago (13). It is likely that S. cerevisiae originally had biotin synthesis genes, including BIO6. Some strains of S. cerevisiae had lost parts of the synthetic pathway including BIO6 in their evolution processes, resulting in biotin auxotrophy.
FIG. 9.
Multiple sequence alignment of Bio6p. Amino acid sequence alignment of Bio6p of K7 with those of A364A, S. bayanus (accession no. AACA0100015), S. kudriavzevii (accession no. AACI01000492), S. paradoxus (accession no. AABY01000445 and AABY01000593) and S. mikatae (accession no. AABZ01000331). Amino acids that are conserved among at least five of the six strains are indicated by white letters on black backgrounds. Positions of the SAM and PLP binding sites are shown by asterisks and dots, respectively.
Although our data demonstrate that one copy of the BIO6 gene is enough for biotin prototrophy, all Japanese sake yeasts tested hold multiple copies of the BIO6 gene. The high copy number may be related to the biotin-poor environments in which sake yeasts grow. Sake is brewed from steamed rice for >20 days at a low temperature (10 to 15°C) to produce >18% ethanol. In sake mash, biotin content is very low, which is <1% of the content in a minimal medium (8, 32). Therefore, having multiple copies of BIO6 could be advantageous to yeast cells in the mash. It is noteworthy that the original BIO6 gene of K7 is located in the subtelomeric region of chromosome II. Genes in the subtelomeric regions of chromosomes are often remodeled by chromosome rearrangements including recombination (18) and duplication (6). We hypothesize that the copy number of the BIO6 gene was increased by chromosomal rearrangements, reaching at least eight copies for K7 (diploid cells) to synthesize enough biotin in the sake mash.
The expression levels of biotin biosynthesis genes are regulated by the extracellular concentration of biotin (36). The numbers of BIO3 and BIO4 transcripts in cells grown in minimal medium (1 copy per cell) are much higher than the numbers in cells grown in rich medium (0.06 copy per cell), because the minimal medium has only a small amount of biotin. The expression level of the BIO6 gene was also increased in biotin-deficient medium (Fig. 7). Furthermore, the expression level of the BIO6 gene in K7 was higher than that in A364A under both repressed and induced conditions (Fig. 7). These data are in good agreement with the hypothesis that multiple copies of BIO6 are an advantage under biotin-deficient conditions.
Acknowledgments
We thank M. Tominaga for her help in DNA sequencing and J. F. Kirsch for providing reagents.
This work was supported by the Program for Promotion of Basic Research Activities for Innovative Biosciences (PROBRAIN).
REFERENCES
- 1.Adams, A., D. E. Gottschling, C. Kaiser, and T. Stearns. 1997. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 2.Baldet, P., and M. L. Ruffet. 1996. Biotin synthesis in higher plants: isolation of a cDNA encoding Arabidopsis thaliana bioB-gene product equivalent by functional complementation of a biotin auxotroph mutant bioB105 of Escherichia coli K12. C. R. Acad. Sci. Ser. III 319:99-106. [PubMed] [Google Scholar]
- 3.Becker, D. M., and L. Guarente. 1991. High-efficiency transformation of yeast by electroporation. Methods Enzymol. 194:182-187. [DOI] [PubMed] [Google Scholar]
- 4.Betz, H., H. Hinze, and H. Holzer. 1974. Isolation and properties of two inhibitors of proteinase B from yeast. J. Biol. Chem. 249:4515-4521. [PubMed] [Google Scholar]
- 5.Bower, S., J. B. Perkins, R. R. Yocum, C. L. Howitt, P. Rahaim, and J. Pero. 1996. Cloning, sequencing, and characterization of the Bacillus subtilis biotin biosynthetic operon. J. Bacteriol. 178:4122-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chow, T. H. C., P. Sollitti, and J. Marmur. 1989. Structure of the multigene family of MAL loci in Saccharomyces. Mol. Gen. Genet. 217:60-69. [DOI] [PubMed] [Google Scholar]
- 7.Eisenberg, M. A. 1987. Biosynthesis of biotin and lipoic acid, p. 544-550. In F. C. Neidhardt, J. L. Ingraham, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology. American Society of Microbiology, Washington, D.C.
- 8.Fukui, S., Y. Tani, and T. Kisibe. 1955. Studies on the role of B-vitamins in sake-brewing. Hakkou-kogaku 33:302-307. [Google Scholar]
- 9.Gloeckler, R., I. Ohsawa, D. Speck, C. Ledoux, S. Bernard, M. Zinsius, D. Villeval, T. Kisou, K. Kamogawa, and Y. Lemoine. 1990. Cloning and characterization of the Bacillus sphaericus genes controlling the bioconversion of pimelate into dethiobiotin. Gene 87:63-70. [DOI] [PubMed] [Google Scholar]
- 10.Hara, S., Y. Iimura, and K. Otsuka. 1980. Breeding of useful killer wine yeasts. Am. J. Enol. Vitic. 31:28-3321. [Google Scholar]
- 11.Izumi, Y., Y. Tani, and K. Ogata. 1979. Microbiological biosynthesis of biotin. Methods Enzymol. 62:326-338. [DOI] [PubMed] [Google Scholar]
- 12.Kawamura, D., and A. Toh-e. 1993. Genetic properties of low-temperature-growing yeasts. Seibutsu-kogaku 71:225-232. [Google Scholar]
- 13.Kellis, M., N. Patterson, M. Endrizzi, B. Birren, and E. S. Lander. 2003. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature 423:241-254. [DOI] [PubMed] [Google Scholar]
- 14.Köhrer, K., and H. Domdey. 1991. Preparation of high molecular weight RNA. Methods Enzymol. 194:398-405. [DOI] [PubMed] [Google Scholar]
- 15.Komoda, H., F. Mano, and Yamada. 1966. On the true flavour of various fermented beverages. I. Sake flavour. Nippon Nogeikagaku Kaishi 40:127-134. [Google Scholar]
- 16.Komoda, H., F. Mano, and Yamada. 1966. On the true flavour of various fermented beverages. II. The fragrant aroma appearing in the first period of sake mash and that of refined sake. Nippon Nogeikagaku Kaishi 40:173-177. [Google Scholar]
- 17.Kosai, J., and K. Yabe. 1897. Uber die bei der Sakebereitung beteiligten Pilze. Zentralbl. Bakteriol. Parasitenkd. Abt 2 1:619-620. [Google Scholar]
- 18.Miyashita, K., K. Sakamoto, H. Kitagaki, K. Iwashita, K. Ito, and H. Shimoi. 2004. Cloning and analysis of the AWA1 gene of a nonfoaming mutant of a sake yeast. J. Biosci. Bioeng. 97:14-18. [DOI] [PubMed] [Google Scholar]
- 19.Mortimer, R. K., and J. R. Johnston. 1986. Genealogy of principal strains of the yeast genetic stock center. Genetics 113:35-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nishimoto, F., and S. Yamashoji. 1994. Rapid assay of cell activity of yeast cell. J. Ferment. Bioeng. 77:107-108. [Google Scholar]
- 21.Ohsugi, M., and Y. Imanishi. 1985. Microbiological activity of biotin vitamers. J. Nutr. Sci. Vitaminol. 3:563-572. [DOI] [PubMed] [Google Scholar]
- 22.Otsuka, A. J., M. R. Buoncristiani, P. K. Howard, J. Flamm, C. Johnson, R. Yamamoto, K. Uchida, C. Cook, J. Ruppert, and J. Matsuzaki. 1998. The Escherichia coli biotin biosynthetic enzyme sequences predicted from the nucleotide sequence of the bio operon. J. Biol. Chem. 263:19577-19585. [PubMed] [Google Scholar]
- 23.Phalip, V., I. Kuhn, Y. Lemoine, and J. M. Jeltsch. 1999. Characterization of the biotin biosynthesis pathway in Saccharomyces cerevisiae and evidence for a cluster containing BIO5, a novel gene involved in vitamer uptake. Gene 232:43-51. [DOI] [PubMed] [Google Scholar]
- 24.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Shimoi, H., K. Sakamoto, M. Okuda, R. Atthi, K. Iwashita, and K. Ito. 2002. The AWA1 gene is required for the foam-forming phenotype and cell surface hydrophobicity of sake yeast. Appl. Environ. Microbiol. 68:2018-2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stoner, G. L., and M. A. Eisenberg. 1975. Purification and properties of 7, 8-diaminopelargonic acid aminotransferase. J. Biol. Chem. 250:4029-4036. [PubMed] [Google Scholar]
- 28.Stoner, G. L., and M. A. Eisenberg. 1975. Biosynthesis of 7, 8-diaminopelargonic acid from 7-keto-8-aminopelargonic acid and S-adenosyl-l-methionine. J. Biol. Chem. 250:4037-4043. [PubMed] [Google Scholar]
- 29.Takahashi, M. 1954. Studies on the requirements of amino acids and vitamins of yeasts. Part 1. Nippon Nogeikagaku Kaishi 28:395-398. [Google Scholar]
- 30.Takahashi, M. 1954. Studies on the requirements of amino acids and vitamins of yeasts. Part 2. Nippon Nogeikagaku Kaishi 28:398-404. [Google Scholar]
- 31.Tamura, K., Gu, Y. Q., Q. Wang, T. Yamada, K. Ito, and H. Shimoi. 2004. A hap1 mutation in a laboratory strain of Saccharomyces cerevisiae results in decreased expression of ergosterol-related genes and cellular ergosterol content compared to sake yeast. J. Biosci. Bioeng. 98:159-166. [DOI] [PubMed] [Google Scholar]
- 32.Torigata, K., and Y. Akiyama. 1968. Tests of sake brewing by yeasts after cultured with ventilation. 1. Rising and falling of vitamins contained in sake moromi and preservative tests of yeasts. J. Brew. Soc. Japan 63:60-63. [Google Scholar]
- 33.Vieira, J., and J. Messing. 1987. Production of single-stranded plasmid DNA. Methods Enzymol. 153:3-11. [DOI] [PubMed] [Google Scholar]
- 34.Wach, A. 1996. PCR-synthesis of marker cassettes with long flanking homology region for gene disruptions in Saccharomyces cerevisiae. Yeast 12:259-265. [DOI] [PubMed] [Google Scholar]
- 35.Wierenga, R. K., and W. G. J. Hol. 1983. Predicted nucleotide-binding properties of p21 protein and its cancer-associated variant. Nature 302:842-844. [DOI] [PubMed] [Google Scholar]
- 36.Woodicka, L., H. Dong, M. Mittmann, M. H. Ho, and D. J. Lockhart. 1997. Genome-wide expression minitoring in Saccharomyces cerevisiae. Nat. Biotechnol. 15:1359-1367. [DOI] [PubMed] [Google Scholar]
- 37.Yamada, Y., K. Mikata, and K. Banno. 1990. Identification of 38 brewing yeast maintained in IFO collection. Bull. JFCC 9:95-119. [Google Scholar]
- 38.Yozawa, K. 1966. Flavour components of alcoholic brewing. J. Brew. Soc. Japan 61:481-485. [Google Scholar]
- 39.Zhang, S., I. Sanyal, G. H. Bulboaca, A. Rich, and D. H. Flint. 1994. The gene for biotin synthase from Saccharomyces cerevisiae: cloning, sequencing, and complementation of Escherichia coli strains lacking biotin synthase. Arch. Biochem. Biophys. 309:29-35. [DOI] [PubMed] [Google Scholar]