Abstract
Virulence variations of Paenibacillus larvae subsp. larvae, the causative agent of American foulbrood disease of honeybees, were investigated by analysis of 16 field isolates of this pathogen, belonging to three previously characterized genotypes, as well as the type strain (ATCC 9545) of P. larvae subsp. larvae, with exposure bioassays. We demonstrated that the strain-specific 50% lethal concentrations varied within an order of magnitude and that differences in amount of time for the pathogen to kill 100% of the infected hosts (LT100) correlated with genotype. One genotype killed rather quickly, with a mean LT100 of 7.8 ± 1.7 days postinfection, while the other genotypes acted more slowly, with mean LT100s of 11.2 ± 0.8 and 11.6 ± 0.6 days postinfection.
The gram-positive, spore-forming bacterium Paenibacillus larvae subsp. larvae is the primary bacterial pathogen of honeybee brood and the causative agent of American foulbrood disease (AFB). AFB is a cosmopolitan disease and one of the major threats to beekeeping, since it is highly contagious and able to kill affected colonies. Hence, it causes considerable economic loss to beekeepers worldwide.
Spores are the only infectious form of this organism. Larvae become infected by ingestion of spore-contaminated honey. During the first 12 to 36 h after hatching, larvae are most susceptible to infection, with a dose of about 10 spores or fewer being sufficient to successfully infect and finally kill a larva (19, 20). The clinical symptoms of AFB are typical, with the brown, viscous larval remains forming a ropy thread when drawn out with a matchstick. The decaying larvae desiccate into hard scales, consisting of millions of bacterial spores.
So far, studies addressing differences in the outcomes of AFB have only focused on aspects of host tolerance (2, 3, 9, 15, 16, 20) but have neglected the possibility of variation in virulence among different strains of P. larvae subsp. larvae. Hence, although P. larvae subsp. larvae is an important pathogen, its pathogenic mechanism and virulence factors remain elusive. Molecular studies addressing these questions are hampered not only by the lack of genomic tools for this organism but also because no thorough phenotypic studies relating to the virulence of this pathogen exist.
Recently, we identified different genotypes of P. larvae subsp. larvae (7, 12) via repetitive-element PCR fingerprinting with primers ERIC, MBO REP1, and BOX A1R (18). Biochemical fingerprinting of these genotypes by a carbon source test revealed that they differ in their metabolic patterns (12). In addition, only one of the described genotypes, genotype AB, was shown to harbor plasmid DNA (12). Here we present data on the further characterization of these genotypes in terms of virulence. We demonstrate for the first time that different strains of P. larvae subsp. larvae clearly differ in virulence and that some of these differences are genotype specific. The impact of our findings for the transmission of the pathogen is discussed below.
Isolation and identification of bacterial isolates.
The P. larvae subsp. larvae type strain, ATCC 9545 (obtained from the American Type Culture Collection through U. Rdest, Biozentrum der Universität Würzburg), and 16 German field isolates of P. larvae subsp. larvae isolated from honey samples originating from clinically diseased, AFB-positive hives used in this study are listed in Table 1. Field isolates were sampled during the course of foulbrood monitoring programs between 2000 and 2004. Cultivation and identification of the isolates were performed as described previously (7, 12). Detailed biochemical and genetic analyses of the reference strain and the field strains 00-087, 01-455, 02-130, 03-125, 01-440, 02-113, 02-120, 03-159, 00-1163, 03-522, and 03-525 have been already reported (10, 12). For genetic fingerprinting of the five additional isolates used in this study, previously described techniques were employed (7, 12). All chemicals and media for microbiological work were obtained from Oxoid, Germany.
TABLE 1.
List of strains used in this study, with values for LC50 and LT100
| P. larvae subsp. larvae strain | Yr of isolation | Genotype | Estimated LC50 (CFU ml−1 larval diet) | LT100 (days p.i.)a
|
|
|---|---|---|---|---|---|
| Min/ max | Mean ± SD | ||||
| 00-1163 | 2000 | AB | 200 | 7/10 | 8.3 ± 1.1 |
| 03-194 | 2003 | AB | 260 | 7/8 | 7.3 ± 0.6 |
| 03-200 | 2003 | AB | 550 | 6/10 | 8.8 ± 1.9 |
| 03-522 | 2003 | AB | 600 | 7/10 | 8.3 ± 1.5 |
| 03-525 | 2003 | AB | 620 | 7/10 | 8.0 ± 1.0 |
| 04-309 | 2004 | AB | ≪100 | 6/7 | 6.6 ± 1.1 |
| 01-440 | 2001 | Ab | ≪100 | 10/11 | 10.7 ± 0.6 |
| 02-113 | 2002 | Ab | 200 | 11/12 | 11.8 ± 0.4 |
| 02-120 | 2002 | Ab | 110 | 10/12 | 11.0 ± 1.0 |
| 03-159 | 2003 | Ab | 270 | 11/12 | 11.7 ± 0.6 |
| 03-189 | 2003 | Ab | 800 | 10/11 | 10.5 ± 0.5 |
| 00-087 | 2000 | ab | ≪100 | 11/12 | 11.3 ± 0.6 |
| 01-455 | 2001 | ab | 550 | 12/12 | 12.0 ± 0.0 |
| 02-130 | 2002 | ab | 500 | 11/13 | 12.0 ± 1.0 |
| 03-119 | 2003 | ab | 175 | 11/12 | 11.7 ± 0.6 |
| 03-125 | 2003 | ab | 330 | 11/12 | 11.3 ± 0.6 |
| ATCC 9545b | aβ | 200 | 10/12 | 11.3 ± 0.8 | |
p.i., postinfection; min, minimum; max, maximum.
Reference strain.
Preparation of defined spore suspensions for exposure bioassays.
For the preparation of spore suspensions containing a defined concentration of CFU, around 100 P. larvae subsp. larvae colonies per strain resuspended in 300 μl brain heart infusion broth were used to inoculate the liquid part of Columbia sheep blood agar slants and incubated at 37°C for 10 days. Subsequently, the liquid part was analyzed by phase-contrast microscopy for the absence of vegetative cells. Spore concentrations were determined by cultivating serial dilutions on Columbia sheep blood agar plates as described previously (7, 12) and calculating the mean numbers of colonies grown on five plates. Suspensions were adjusted to a concentration of 1 × 107 CFU ml−1. Spore suspensions were stored at 4°C.
Exposure bioassays for the investigation of the virulence of P. larvae subsp. larvae isolates.
The virulence levels of different P. larvae subsp. larvae isolates were determined by exposure bioassays, which, in contrast to injection bioassays, require all of the steps in pathogenesis (17). For experimental infection, worker larvae collected from different colonies of Apis mellifera carnica maintained in the apiary at the Institute for Bee Research in Hohen Neuendorf, Germany, were reared in 24-well tissue culture plates according to the method of Peng and coworkers (14), with a modified larval diet consisting of 3% (wt/vol) fructose, 3% (wt/vol) glucose, and 66% (vol/vol) royal jelly (purchased from a local beekeeper) in sterile double-distilled water. Worker larvae of the first larval instar (around 12 h of age) were used throughout the experiments. Since mean weights differ significantly between different age groups (2), ages of the larvae were estimated by size. For infection, final concentrations of 100, 300, 500, 1,000, and 2,000 CFU ml−1 larval diet were adjusted by using a working solution of 1 × 105 CFU ml−1. The infectious larval diet was fed to the larvae for the first 24 h after grafting. Thereafter, normal larval diet was used for feeding. Control larvae were fed with normal larval diet throughout the entire larval stages.
Three groups of 10 larvae of the first larval instar were grafted into three wells filled with larval diet (normal or infectious) by using a special grafting tool (Graze Bienenzuchtgeräte, Germany) to avoid injuring the larvae. These three groups on one plate were treated as one replicate. One experiment consisted of four replicates: three infected groups and one noninfected control. For genotypes ab, Ab, and AB, five, five, and six strains, respectively (Table 1), with three to five concentrations each, were tested. For the reference strain, ATCC 9545, three concentrations were tested and the assays were performed three times.
Each day, the larvae were taken out of the incubator and examined under a stereo microscope. Larvae were classified as dead when they stopped respiration, lost their body elasticity, or developed marked edema and when they displayed color changes to grayish or brownish. The number of dead larvae was recorded, and surviving larvae were transferred to new wells freshly filled with food. After defecation, i.e., after clear uric acid crystals and light-yellow excretions could be observed in the remaining diet, engorged larvae were transferred into pupation plates lined with Kimwipes tissue, where they underwent the stages of pupal development. While noninfected larvae successfully underwent metamorphosis, infected larvae rarely developed beyond the stage of engorged larvae or prepupae. For the purpose of this study, mortality occurring after defecation, i.e., in the pupation plates, was referred to as “mortality after cell capping,” since the time of defecation (i.e., opening of the gut, marking the transition from larval to pupal development and the beginning of metamorphosis) of in vitro-reared larvae represents the time of capping in the colony (14). Dead animals were classified as dead from AFB only when vegetative P. larvae subsp. larvae could be cultivated from the larval remains. On no occasion was P. larvae subsp. larvae cultivated from remains of dead control animals. Experiments with a mortality exceeding 15% in the control group were excluded, as were experiments where the “natural” mortality (larval death but no growth of P. larvae subsp. larvae) in the infected groups was higher than 15%. The first three experiments were performed three times to demonstrate that the concentration-mortality relationship was reproducible.
Determination of the LC50s of different strains.
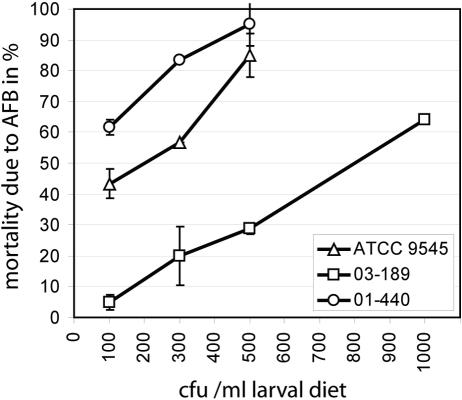
A common measure of virulence from exposure bioassays is the 50% lethal concentration (LC50), the respective concentration it takes to kill 50% of the hosts tested (17). To obtain this measure, for each strain and concentration the percentage of AFB-dead larvae was calculated and plotted against the spore concentration used for infection. Results showed a clear positive concentration-mortality relationship (Fig. 1). From the obtained graphs, an LC50 for each isolate was estimated. The LC50s varied within an order of magnitude between the isolates and revealed no correlation with genotype (Table 1). The most virulent strains (00-087, 01-440, and 04-309), in terms of spore count, killed 50% of the larvae with less than 100 CFU ml−1 larval diet, whereas it took the least virulent strain (03-189) around 800 CFU ml−1 larval diet to kill 50% of the larvae (Table 1).
FIG. 1.
Determination of LC50s. Representative results obtained for the reference strain, ATCC 9545, and strains 03-189 and 01-440 are shown. Mortality due to AFB was determined as described in the text, and the percentage of AFB-dead larvae was calculated for each concentration (100% = 30 larvae). Experiments were performed three times. The mean values ± standard deviations of mortality are plotted against the spore concentrations to determine the strain-specific LC50s.
It is well-known from field observations that some colonies show no clinical symptoms despite a high spore concentration contaminating the honey, while others exhibit clinically diseased brood although the spore concentration detectable in the honey is low (8). So far, these differences have been explained by differences in host tolerance and hygienic behavior of honeybees (2, 3, 8, 9, 15, 16, 19, 20). Indeed, a study directly comparing a susceptible bee line with a resistant bee line revealed that differences between bee strains might account for a factor of 2 in the spore dose needed for causing clinical symptoms (9).
Our results indicate another important factor involved in determining the outcome of AFB infections in honeybee colonies: variation in pathogen virulence. The LC50s of different P. larvae subsp. larvae strains, determined by exposure bioassays, varied with a factor of 10, suggesting that the impact of P. larvae subsp. larvae virulence on the outcome of an AFB infection is much greater than the influence from bee tolerance to infection, reported to vary with a factor of 2 (9).
Determination of the LT100s of different P. larvae subsp. larvae isolates.
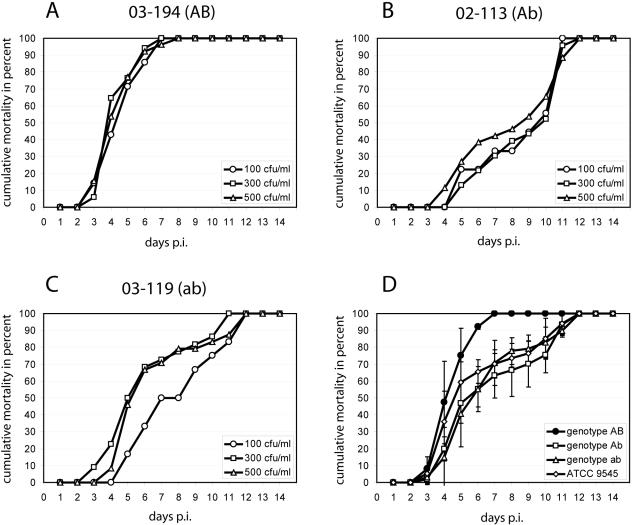
Another valid measure of virulence is the time it takes the pathogen to kill 50 or 100% of the infected hosts (LT50 or LT100, respectively) (17). To obtain the time course of infection (Fig. 2) and determine the LT100 (Table 1), the cumulative proportion of AFB-dead larvae per day was calculated for each replicate and plotted against time. Survivors were excluded from this calculation (17). Results showed that progression of the disease and time of larval death had only a minor, nonsignificant, negative correlation in some cases (Fig. 2B and C). For most of the strains tested and especially for genotype AB (Fig. 2A), the time course of infection was not even influenced by the spore concentration. For P. larvae subsp. larvae genotype AB, classical sigmoid curves were obtained, while for the other genotypes, the time course of infection revealed a biphasic curve progression in which two exponential phases of mortality were separated by a phase of reduced mortality between day 5 and day 9 postinfection. For all genotypes, the first dead larvae appeared between day 3 and day 5 postinfection. Larvae infected with genotype AB did not survive longer than 10 days postinfection (Table 1) and died rather quickly, with a mean LT100 of 7.8 ± 1.7 days postinfection (Table 2). In contrast, the other genotypes killed with mean LT100s of 11.2 ± 0.8 (Ab), 11.6 ± 0.6 (ab), and 11.3 ± 0.8 (aβ) days postinfection (Table 2), and infected larvae survived at least until day 10 postinfection (Table 1). The genotype-specific differences in disease progression became even more evident by comparing the mean cumulative mortalities of the genotypes (Fig. 2D).
FIG. 2.
Time course of infection. (A to C) Cumulative mortality was calculated per day postinfection (p.i.) as described in the text and expressed as percentage of the infected hosts (i.e., all larvae that died from AFB in this replicate). Concentrations tested are given in each panel. Results from three representative experiments are shown for each strain. (D) Cumulative mortality was calculated for each genotype from all strains as described in the text, using the concentration closest to the estimated LC50. Therefrom, the mean cumulative proportion of AFB-dead larvae per day p.i. ±standard deviation was calculated for each genotype.
TABLE 2.
P. larvae subsp. larvae genotype-specific LT100 and mortality after cell capping
| P. larvae subsp. larvae genotype | LT100 (days p.i. [mean ± SD]) | % Mortality after cell capping (mean ± SD)a |
|---|---|---|
| AB | 7.8 ± 1.7 | 5.4 ± 3.2 |
| Ab | 11.2 ± 0.8 | 26.6 ± 7.3 |
| ab | 11.6 ± 0.6 | 20.2 ± 6.3 |
| aβ | 11.3 ± 0.8 | 26.3 ± 2.8 |
One hundred percent mortality is the total number of larvae that died from AFB.
Determination of time of death with respect to cell capping.
Capping of the cells is a critical time for both the hygienic behavior of the bees and the time of death due to AFB (1). Therefore, we additionally evaluated the results by choosing this point of time as a threshold. Since the postdefecation period corresponds to the postcapping period, the time of larval defecation was used as the indicator for the beginning of metamorphosis and the time point of cell capping under normal colony conditions (14). For each strain, the mean number of AFB-dead larvae which died after defecation in the pupation plates was calculated over the entire concentration range tested and expressed as a percentage of the total number of AFB-dead larvae. Based on these values, the mean number of larvae that died from AFB after defecation was calculated for each genotype. Results showed that proportion of larvae that died after capping correlated with genotype (Table 2). Only 5.4 ± 3.2% of larvae infected with P. larvae subsp. larvae genotype AB survived until after capping, whereas 26.6 ± 7.3, 20.2 ± 6.3, and 26.3 ± 2.8% of larvae infected with genotypes Ab, ab, and aβ, respectively, died after capping. Statistical evaluation of these data by one-way analysis of variance (df = 3, F = 14.06, P = 0.0002) followed by a post hoc test (Newman-Keuls test) revealed no significant differences in time postinfection for larval mortality between genotypes ab and Ab (P = 0.29), ab and aβ (P = 0.14), or Ab and aβ (P = 0.98). In contrast, differences between the three genotypes Ab, ab, and aβ and the genotype AB for when P. larvae subsp. larvae-infected larvae died were highly significant, with P values of 0.0006, 0.002, and 0.0004, respectively.
Our data demonstrate that P. larvae subsp. larvae genotype AB killed infected larvae much more quickly and earlier than the other genotypes. Therefore, genotype AB was the most virulent genotype with respect to disease progression at the level of the individual larva. It can be hypothesized that the highly virulent (with respect to disease progression at individual larva level) AB genotype may be less virulent at colony level. The killing of most infected larvae before capping of the cells is likely to allow removal of diseased brood by nursing bees, with fewer bacterial spores produced and spread within the colony than with slower-acting strains that allow the bees to cap the cells before the host is killed. A parallel is found with Apis cerana, an Asian species of honeybee also susceptible to P. larvae subsp. larvae infections. Experiments have demonstrated that most of a P. larvae subsp. larvae-infected brood is removed before the cells are sealed for pupation, and as a consequence, colonies of A. cerana clinically diseased with AFB are less frequent than diseased colonies of A. mellifera in the same area (4). However, the hypothesis that the variations in virulence demonstrated by exposure bioassays translate to variations in virulence at colony level remains to be investigated and verified in the field.
Against the background of our results, reports on infected colonies that never developed clinical disease symptoms visible to the apiculturist (8, 13) must be reevaluated. In a colony infected by a fast-killing genotype, only sporadic cells contain the ropy stage and foulbrood scale. Since these are the visible clinical symptoms of AFB, such an infection can be overlooked or remain unrecognized for a long time. Even though the classical clinical symptoms may not be apparent in such an infected colony, the colony nevertheless should be considered clinically infected since larvae are dying from the disease already.
The existence of more- or less-virulent strains of P. larvae subsp. larvae, as demonstrated in our study, is likely to influence disease transmission and therefore to have important epidemiological consequences. P. larvae subsp. larvae is believed to be transmitted primarily through robbing of diseased colonies (horizontal transmission) (8). The mode of pathogen transmission (horizontal versus vertical) is an important factor determining virulence (11). In honeybees, which reproduce at colony level by colony fission, most pathogens are transmitted primarily vertically and, thus, are rather benign at colony level, since only rather strong colonies swarm. AFB infections are the exception to this rule, as disease transmission is actually favored by colony collapse (6). The scenario described here, with a suggested variation in virulence between genotypes at colony level (high virulence at larval level producing lower virulence at colony level), allows for selection among strains of P. larvae subsp. larvae that are more or less dependent on horizontal or vertical transmission, respectively. If all colonies that show clinical symptoms of disease are killed (as required by legislation in many countries), a selection pressure is probably imposed, selecting for less-virulent pathogen strains being primarily vertically transmitted, similarly to other honeybee pathogens (6). Recent evolutionary considerations suggest that interventions influencing disease frequency in a population, in particular, interventions that influence mode of pathogen transmission, have the potential to tip the competitive balance in favor of less-virulent pathogen strains (5). This perspective of virulence management of pathogens in honeybee pathogens needs to be evaluated in the field.
Acknowledgments
This work was supported by grants from the Ministries for Agriculture from Brandenburg and Sachsen-Anhalt, Germany.
REFERENCES
- 1.Bailey, L., and B. V. Ball (ed). 1991. Honey bee pathology, 2nd ed., p. 51. Academic Press, Ltd., New York, N.Y.
- 2.Brodsgaard, C., W. Ritter, and H. Hansen. 1998. Response of in vitro reared honeybee larvae to various doses of Paenibacillus larvae larvae spores. Apidologie 29:569-578. [Google Scholar]
- 3.Brodsgaard, C., H. Hansen, and W. Ritter. 2000. Progress of Paenibacillus larvae larvae infection in individually inoculated honeybee larvae reared singly in vitro, in micro colonies, or in full-size colonies. J. Apic. Res. 39:19-27. [Google Scholar]
- 4.Chen, Y., C. Wang, J. An, K. Ho, Y. W. Chen, C. H. Wang, and K. K. Ho. 2000. Susceptibility of the Asian honey bee, Apis cerana, to American foulbrood, Paenibacillus larvae larvae. J. Apic. Res. 39:169-175. [Google Scholar]
- 5.Ewald, P. W. 2002. Virulence management in humans, p. 399-412. In U. Dieckmann, J. A. J. Metz, M. W. Sabelis, and K. Sigmund (ed.), Adaptive dynamics of infectious diseases: in pursuit of virulence management. Cambridge University Press, Cambridge, United Kingdom.
- 6.Fries, I., and S. Camazine. 2001. Implications of horizontal and vertical pathogen transmission for honey bee epidemiology. Apidologie 32:199-214. [Google Scholar]
- 7.Genersch, E., and C. Otten. 2003. The use of repetitive element PCR fingerprinting (rep-PCR) for genetic subtyping of German field isolates of Paenibacillus larvae subsp. larvae. Apidologie 34:195-206. [Google Scholar]
- 8.Hansen, H., and C. Brodsgaard. 1999. American foulbrood: a review of its biology, diagnosis and control. Bee World 80:5-23. [Google Scholar]
- 9.Hoage, T. R., and W. C. Rothenbuhler. 1966. Larval honeybee response to various doses of Bacillus larvae spores. J. Econ. Entomol. 59:42-45. [Google Scholar]
- 10.Kilwinski, J., M. Peters, A. Ashiralieva, and E. Genersch. 2004. Proposal to reclassify Paenibacillus larvae subsp. pulvifaciens DSM 3615 (ATCC 49843) as Paenibacillus larvae subsp. larvae. Results of a comparative biochemical and genetic study. Vet. Microbiol. 104:31-42. [DOI] [PubMed] [Google Scholar]
- 11.Lipsitch, M., S. Siller, and M. A. Nowak. 1996. The evolution of virulence in pathogens with vertical and horizontal transmission. Evolution 50:1729-1741. [DOI] [PubMed] [Google Scholar]
- 12.Neuendorf, S., K. Hedtke, G. Tangen, and E. Genersch. 2004. Biochemical characterization of different genotypes of Paenibacillus larvae subsp. larvae, a honey bee bacterial pathogen. Microbiology 150:2381-2390. [DOI] [PubMed] [Google Scholar]
- 13.Nordström, S., E. Forsgren, and I. Fries. 2002. Comparative diagnosis of American foulbrood using samples of adult bees and honey. J. Apic. Sci. 46:5-12. [Google Scholar]
- 14.Peng, Y.-S. C., E. Mussen, A. Fong, M. A. Montague, and T. Tyler. 1992. Effects of chlortetracycline on honey bee worker larvae reared in vitro. J. Invertebr. Pathol. 60:127-133. [Google Scholar]
- 15.Sturtevant, A. P., and I. L. Revell. 1953. Reduction of Bacillus larvae spores in liquid food of honeybees by action of the honey stopper, and its relation to the development of American foulbrood. J. Econ. Entomol. 46:855-860. [Google Scholar]
- 16.Tarr, H. L. A. 1938. Studies on American foulbrood of bees. III. The resistance of individual larvae to inoculation with the endospores of Bacillus larvae. Ann. Appl. Biol. 25:807-814. [Google Scholar]
- 17.Thomas, S. R., and J. S. Elkinton. 2004. Pathogenicity and virulence. J. Invertebr. Pathol. 85:146-151. [DOI] [PubMed] [Google Scholar]
- 18.Versalovic, J., M. Schneider, F. J. de Bruijn, and J. R. Lupski. 1994. Genomic fingerprinting of bacteria using repetitive sequence-based polymerase chain reaction. Methods Mol. Cell. Biol. 5:25-40. [Google Scholar]
- 19.Woodrow, A. W. 1942. Susceptibility of honeybee larvae to individual inoculations with spores of Bacillus larvae. J. Econ. Entomol. 35:892-895. [Google Scholar]
- 20.Woodrow, A. W., and E. C. Holst. 1942. The mechanism of colony resistance to American foulbrood. J. Econ. Entomol. 35:327-330. [Google Scholar]