Abstract
Dynamic changes in the transcriptional expression of the tceA gene, which encodes a trichloroethene reductive dehalogenase, were characterized in a Dehalococcoides-containing microbial enrichment culture. Expression was quantified by real-time PCR as the number of tceA transcripts per tceA gene. Expression of tceA increased 40-fold after chlorinated ethene-starved cells were exposed to trichloroethene (TCE), cis-dichloroethene (DCE), or 1,1-DCE but did not increase after exposure to tetrachloroethene or vinyl chloride. Surprisingly, tceA expression also increased 30-fold after cellular exposure to the nonmetabolic substrate trans-DCE, indicating that expression of tceA is induced by both growth-supporting and non-growth-supporting chlorinated ethenes. Additional experiments revealed that the level of tceA expression was independent of the concentration of chlorinated ethenes (sum concentrations of TCE and DCEs of 2.2 to 333 μM), the concentration of the electron donor hydrogen (concentrations of 12 nM to 17 μM), and the presence of alternate bacterial electron acceptors (5 mM concentrations of fumarate, sulfate, sulfite, thiosulfate, nitrate, or nitrite) but was highly dependent on incubation temperature.
Chlorinated ethenes are common contaminants of groundwater resources. In anaerobic environments, chlorinated ethenes can be biologically degraded to less-chlorinated ethenes and, in some cases, completely dechlorinated to the nontoxic end product ethene (4, 11, 30). This process is referred to as reductive dechlorination or, when coupled with energy conservation, halorespiration (11, 12, 30).
To date, all known halorespiration processes are catalyzed by reductive dehalogenase enzymes (RDases) (11, 30). With a few exceptions, all RDases are monomeric, are tightly linked with the cytoplasmic membrane, and contain corronoid cofactors and iron-sulfur clusters (11, 30). A large number of RDases and/or their corresponding genes have been identified and linked to the dechlorination of chlorinated ethenes (16-19, 22, 25-27, 32, 35). The vinyl chloride (VC)-dechlorinating RDases are of particular interest because VC is the only proven human carcinogen of the chlorinated ethenes (15) and because the transformation of VC to ethene is often the rate-limiting step for the complete dechlorination of chlorinated ethenes to nontoxic end products (15, 33).
The first VC-dechlorinating RDase to be identified was the trichloroethene (TCE) RDase from Dehalococcoides ethenogenes strain 195 (18). This RDase reductively dechlorinates TCE, cis-dichloroethene (DCE), and 1,1-DCE at rates ranging from 5 to 12 μmol min−1 (mg of protein)−1, dechlorinates VC and trans-DCE at substantially lower rates of 0.04 to 0.45 μmol min−1 (mg of protein)−1, and cannot dechlorinate tetrachloroethene (PCE) (17). Correspondingly, D. ethenogenes strain 195 can couple growth with the dechlorination of TCE, cis-DCE, and 1,1-DCE (20, 21), cannot couple growth with the dechlorination of trans-DCE or VC (20), and exploits a separate RDase for the dechlorination of PCE (18).
The gene encoding the TCE RDase was cloned, sequenced, and designated tceA (17). A second open reading frame, designated tceB, is located downstream of tceA and encodes a hydrophobic polypeptide presumed to be involved with membrane association (17). Interestingly, while tceA is not located within close proximity to any known transcription regulators in D. ethenogenes strain 195, this strain contains 17 other putative RDase-encoding genes (36) that all have proximal transcription regulators (29). Also notable is that the tceA gene is located on a putative integrated element in D. ethenogenes strain 195 and is oriented in the opposite direction relative to all other 17 putative RDase-encoding genes (29).
Previous studies with a Dehalococcoides-containing microbial enrichment (designated ANAS) showed that quantities of tceA mRNA were 25-fold higher in TCE- and cis-DCE-exposed subcultures than in VC-exposed and chlorinated ethene-starved subcultures (14). Given that TCE and cis-DCE are metabolic substrates of tceA-containing bacteria while VC is not (20, 21), these findings suggested that tceA expression will increase only in response to growth-supporting substrates of the TCE RDase (14). These findings, however, did not reveal whether the observed differences in tceA expression resulted from gene level induction by specific chlorinated ethenes, differences in the overall metabolic and transcriptional state of the tceA-containing organism(s), or a combination of both factors.
In this study, we characterize the kinetics of the tceA expression response to cellular exposure to different chlorinated ethenes (PCE, TCE, cis-DCE, trans-DCE, 1,1-DCE, or VC) in the ANAS enrichment. Results are used to examine whether the expression of tceA is regulated at the transcription level or primarily affected by changes in the metabolic state of the tceA-containing organism(s). Further investigations examine the dependence of tceA expression on incubation conditions, including the concentration of chlorinated ethenes, the concentration of electron donor, the presence of alternate bacterial electron acceptors, and incubation temperature.
MATERIALS AND METHODS
Bacterial culture.
The anaerobic microbial enrichment investigated in this study (designated ANAS) was originally inoculated with chlorinated ethene-contaminated soil from the Alameda Naval Air Station in California and was subsequently enriched with TCE and lactate. The culture has been maintained under uniform conditions for over 6 years and can dechlorinate TCE, DCE, and VC but not PCE (28). Procedures for culture maintenance have been described previously (14, 28). Briefly, cultures were grown at 25 to 28°C in a 1.5-liter (400- ml liquid volume) continuously stirred semibatch reactor. The reactor was pressurized to 1.8 atm with N2-CO2 (90:10, vol/vol) to prevent oxygen intrusion and was periodically amended with 110 μmol of TCE (added as TCE-saturated water) and sodium lactate (final aqueous concentration of 25 mM). After complete dechlorination of TCE to ethene, the reactor was purged with N2 and 60 to 100 ml of culture was withdrawn and replaced with an equal volume of fresh anaerobic medium.
Chlorinated ethene exposure experiments.
At the completion of a dechlorination cycle, a 210-ml aliquot of stock culture was transferred to a 250-ml anaerobic bottle and starved of chlorinated ethenes for 48 h. Thereafter, 30-ml aliquots were transferred to parallel 58-ml anaerobic bottles, purged with N2-CO2 (90:10) for 1 min, and amended with sodium lactate to a final concentration of 30 mM. The bottles were further amended with 12 μmol of PCE, TCE, cis-DCE, trans-DCE, 1,1-DCE (all added as chlorinated ethene-saturated water), or VC (added as gas) and incubated at 30°C. Two 1-ml culture samples (one for mRNA analysis and one for DNA analysis) were taken immediately before and at various times after chlorinated ethene addition. Cells were immediately collected by centrifugation (12,000 × g for 5 min at 4°C), the supernatants were discarded, and the cell pellets were stored at −80°C until processing.
Incubation with trans-DCE experiment.
After completion of a dechlorination cycle, 10-ml aliquots of stock culture were transferred to parallel 160-ml anaerobic bottles that contained 90 ml of fresh anaerobic medium (10-fold dilution). The bottles were then amended with sodium lactate to an aqueous concentration of 30 mM and with 60 μmol of TCE (added as TCE-saturated water) or 20 μmol of trans-DCE (added as trans-DCE-saturated water), followed by incubation at 30°C for 10 days (two replicate bottles for each chlorinated ethene). Two additional bottles were treated similarly but starved of chlorinated ethenes to serve as controls. For DNA analyses, 1-ml culture samples were taken immediately before and 10 days after chlorinated ethene exposure as described above.
To test the hypothesis that the mean quantities of tceA genes were identical before and after 10 days of incubation, the two-tailed Student t test with unequal variances was applied. When P values were less than 0.05, mean quantities of tceA genes were considered statistically nonidentical.
Incubation condition experiments.
At the completion of a dechlorination cycle, three 100-ml aliquots of stock culture were transferred to separate 160-ml anaerobic bottles (one for each experiment) and starved of chlorinated ethenes for 48 h. Thereafter, 16-ml or 30-ml aliquots of culture were transferred to parallel 58-ml anaerobic bottles, purged with N2-CO2 (90:10) for 1 min, and amended with sodium lactate to a final concentration of 30 mM.
For the chlorinated ethene concentration experiment, the 58-ml anaerobic bottles were amended with 0.07, 0.25, 3.2, or 10.1 μmol of TCE (added as TCE-saturated water) and incubated at 30°C. The aqueous-phase sum concentrations of TCE and DCEs were maintained within a controlled range for 24 h by periodically amending bottles with additional TCE as needed. An additional subculture was starved of TCE to serve as a control.
For the alternative bacterial electron acceptor experiment, the 58-ml anaerobic bottles were amended with 6 μmol of TCE alone (added as TCE-saturated water) or with 6 μmol of TCE (added as TCE-saturated water) along with sodium fumarate, sodium sulfate, sodium sulfite, sodium thiosulfate, sodium nitrate, or sodium nitrite to aqueous concentrations of 5 mM and incubated at 30°C. An additional subculture was starved of both TCE and alternate electron acceptors to serve as a control.
For the temperature experiment, the 58-ml anaerobic bottles were amended with 6 μmol of TCE (added as TCE-saturated water) and incubated at 14, 22, or 30°C for 24 h.
For all experiments, culture samples were taken for DNA and mRNA analyses immediately before and at various times after chlorinated ethene addition as described in the previous section.
Hydrogen concentration experiment.
After completion of a dechlorination cycle, a 100-ml aliquot of stock culture was transferred to a 160-ml anaerobic bottle and amended with 8 μmol of TCE (added as TCE-saturated water). For the following 70 days, additional TCE was periodically added without supplying additional lactate until the aqueous hydrogen concentration in the bottle reached a minimum of 12 nM. Then, 23-ml culture aliquots were transferred to parallel 58-ml anaerobic bottles, purged with N2 for 30 min to remove all residual chlorinated ethenes, and starved of chlorinated ethenes for 48 h. After this starvation period, two bottles were amended with additional hydrogen to obtain aqueous hydrogen concentrations of 0.23 or 86.7 μM while a third bottle was maintained at the minimum observed aqueous hydrogen concentration of 12 nM. The bottles were then amended with 10 μmol of TCE (added as TCE-saturated water) and incubated at 30°C. During incubation, the aqueous-phase concentration of hydrogen was maintained within a controlled range by periodically adding additional hydrogen as needed. For DNA and mRNA analyses, culture samples were taken immediately before and at various times after the last addition of TCE as described above.
Isolation of nucleic acids.
Genomic DNA was isolated from frozen cell pellets of 1-ml culture samples using the UltraClean microbial DNA kit (Mo Bio Laboratories, Carlsbad, CA) according to the manufacturer's instructions.
Total RNA was isolated using the acid phenol method described previously (14). Briefly, frozen cell pellets from 1-ml culture samples were resuspended in 250 μl lysis buffer (50 mM Na acetate, 10 mM EDTA; pH 5.1), 100 μl 10% sodium dodecyl sulfate, 1.0 ml buffer-equilibrated phenol (pH 4.3) (Sigma-Aldrich, St. Louis, MO), and 2 μl of 107 transcripts μl−1 luciferase control RNA (Promega, Madison, WI). Luciferase control RNA (ref mRNA) was added as an internal reference transcript to control for mRNA losses during RNA isolation, reverse transcription, and quantification (14). Cells were lysed by bead beating, and the aqueous lysate was extracted twice with 1 volume of acid (pH 4.3) phenol-chloroform-isoamylalcohol (125:24:1) and once with 1 volume of chloroform-isoamylalcohol (24:1) (Sigma-Aldrich). RNA was precipitated by adding 0.5 volume of 7.5 M ammonium acetate and 2 volumes of 100% ethanol. The precipitate was collected by centrifugation, washed once with 80% ethanol, and resuspended in 40 μl of diethyl pyrocarbonate (DEPC)-treated water (ISC BioExpress). Contaminating DNA was removed by DNase I treatment using a DNA-free kit (Ambion, Austin, TX) according to the manufacturer's instructions. Purified RNA was stored at −80°C prior to further use.
Sequence analysis.
The following primer set was applied to ANAS genomic DNA to target a 1,792-bp section that contains the entire tceA gene of D. ethenogenes strain 195: 797F, ACGCCAAAGTGCGAAAAGC, and 2490R, TAATCTATTCCATCCTTTCTC (17). After 30 cycles of PCR, the production of a uniform-sized product was confirmed by DNA analysis on a model 2100 Bioanalyzer (Agilent, Palo Alto, CA). PCR products were cloned into the pCR 2.1-TOPO plasmid (Invitrogen, Carlsbad, CA), and 10 colonies were selected for sequencing. Sequence analysis was performed at the nucleotide and amino acid levels using the T-Coffee multiple alignment algorithm (http://www.ch.embnet.org/software/TCoffee.html) and default parameters.
Quantification of tceA genes.
To quantify tceA genes in experimental samples, quantitative PCR (qPCR) was applied to genomic DNA in conjunction with the absolute standard curve method as previously described (14). Sample DNA and 10-fold serially diluted tceA DNA standards were amplified in parallel on an ABI Prism 7000 sequence detection system (Applied Biosystems). Each 25-μl reaction volume contained 2 μl of sample or standard DNA, 1× TaqMan Universal PCR Master Mix (Applied Biosystems), 0.7 μM (each) tceA forward (ATCCAGATTATGACCCTGGTGAA) and tceA reverse (GCGGCATATATTAGGGCATCTT) primers, and 200 μM tceA probe (6-carboxyfluorescein-TGGGCTATGGCGACCGCAGG-6-carboxytetramethylrhodamine). Thermocycling conditions were as follows: 2 min at 50°C, 10 min at 95°C, and 40 cycles of 15 s at 95°C and 1 min at 60°C.
Quantification of tceA expression.
Multiplex reverse transcription (RT) followed by multiplex qPCR was used in conjunction with the absolute standard curve method to independently quantify tceA and ref mRNA as previously described (14). The tceA mRNA standard was in vitro transcribed, while the standard for ref mRNA (luciferase control RNA) was purchased from Promega. Sample RNA, serially diluted tceA mRNA standards, and serially diluted ref mRNA standards were reverse transcribed in parallel 10-μl reaction volumes using the RT core reagent kit (PE Applied Biosystems), 0.5 μM tceA reverse primer (listed above), and 0.5 μM ref reverse primer (GGAAGTTCACCGGCGTCAT). The reaction mixture was incubated for 30 min at 48°C, followed by 5 min at 95°C. To quantify contaminating genomic DNA, an additional RT reaction that did not contain reverse transcriptase was performed with each sample.
Multiplex qPCR was performed on an ABI Prism 7000 sequence detection system (Applied Biosystems). Each 25-μl reaction volume contained 2 μl of RT product, 1× TaqMan Universal PCR Master Mix (Applied Biosystems), 0.7 μM (each) tceA forward (listed above), tceA reverse (listed above), ref forward (TACAACACCCCAACATCTTCGA), and ref reverse (listed above) primers, and 200 μM (each) tceA probe (listed above) and ref probe (VIC-CGGGCGTGGCAGGTCTTCCC-6-carboxytetramethylrhodamine). Thermocycling conditions were as follows: 2 min at 50°C, 10 min at 95°C, and 40 cycles of 15 s at 95°C and 1 min at 60°C. To control for sample-specific losses of mRNA during the cell lysis, RNA isolation, and DNA removal steps, the quantity of tceA mRNA was divided by the fractional recovery of ref mRNA (14), which typically ranged between 20 and 35%. Expression of tceA was then calculated as the ref mRNA-normalized quantity of tceA mRNA per 1-ml ANAS culture sample divided by the quantity of tceA genes per 1-ml ANAS culture sample.
Chemical analyses.
Chlorinated ethenes, including isomers of DCE, were resolved and quantified by gas chromatography using a flame ionization detector and a GC-Gas Pro column (J&W Scientific, Folsom, CA). Hydrogen was quantified by gas chromatography using a reductive gas detector (Trace Analytical, Menlo Park, CA). Total mass (M) of each compound was calculated from the headspace concentration (CG) using the following equation:
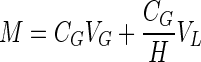 |
where VG and VL are the headspace and liquid volumes, respectively, and H is the Henry's law constant at 30°C (6, 8).
Chemicals and reagents.
All chemicals, gases, and reagents were purchased from standard commercial sources and were of reagent grade or better. All chlorinated ethenes, including specific isomers of DCE, were of high purities (>99%) (Sigma-Aldrich).
RESULTS
Identification of a tceA reductive dehalogenase gene.
Previous investigations revealed that approximately 35% of the cells in the ANAS enrichment belong to the genus Dehalococcoides (28), which is the genus affiliated with all known tceA-containing bacteria (13, 17). To determine whether the ANAS enrichment contains a tceA-like gene, a primer set targeting the entire tceA gene of D. ethenogenes strain 195 (17) was applied to ANAS genomic DNA. Sequence analysis of 10 PCR product clones revealed a single consensus sequence that was more than 97% identical at both the nucleotide and amino acid levels to the tceA genes of D. ethenogenes strain 195 (accession number AAF73916) and Dehalococcoides sp. strain FL2 (accession number AAN85588). The next closest similarities were to the VC RDase-encoding genes vcrA (accession number AAQ94119) and bvcA (accession number AAT64888) (16, 25), with 37.1 and 41.5% identities at the amino acid level, respectively.
Effect of chlorinated ethene exposure on tceA expression kinetics.
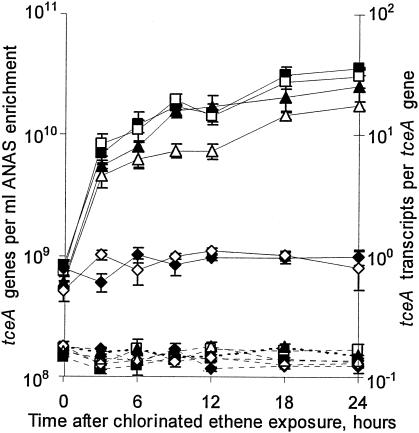
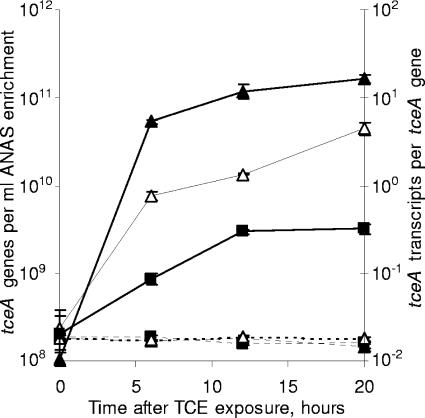
To examine whether the kinetics of tceA expression in the ANAS enrichment are affected by exposure to different chlorinated ethenes, parallel chlorinated ethene-starved subcultures were exposed to PCE, TCE, cis-DCE, trans-DCE, 1,1-DCE, or VC. Over the entire 24-h time course of the experiment, 0% of PCE, 53% of TCE, 45% of cis-DCE, 8% of trans-DCE, 50% of 1,1-DCE, and 10% of VC were dechlorinated in their respective subcultures while the number of tceA genes did not substantially change (Fig. 1). These results indicate that growth had become uncoupled from dechlorination.
FIG. 1.
The effect of cellular exposure to different chlorinated ethenes on the quantity of tceA genes (dashed lines, left y axis) and tceA transcripts per tceA gene (solid lines, right y axis). Parallel ANAS subcultures were exposed to PCE (⧫), TCE (▪), cis-DCE (□), trans-DCE (▵), 1,1-DCE (▴), or VC (⋄). All measurements are averages of triplicate qPCRs or triplicate RT-qPCRs, and error bars represent 1 standard deviation.
After exposure to TCE, cis-DCE, trans-DCE, or 1,1-DCE, tceA expression increased eightfold within 3 h and continued to increase linearly with time (r2 > 0.90) by an additional three- to fivefold over the remainder of the experiment (Fig. 1). Overall, tceA expression increased more than 40-fold after exposure to TCE, cis-DCE, or 1,1-DCE and 30-fold after exposure to trans-DCE, with final expression levels ranging from 18 to 36 transcripts per gene (Fig. 1). In contrast, exposure to PCE or VC did not substantially affect tceA expression, with expression levels ranging between 0.5 and 1.1 transcripts per gene throughout the experiment (Fig. 1). In repeated experiments, the steady and low levels of tceA expression in PCE- and VC-exposed subcultures were similar to the level of expression in subcultures starved of chlorinated ethenes (results not shown).
Incubation with trans-DCE.
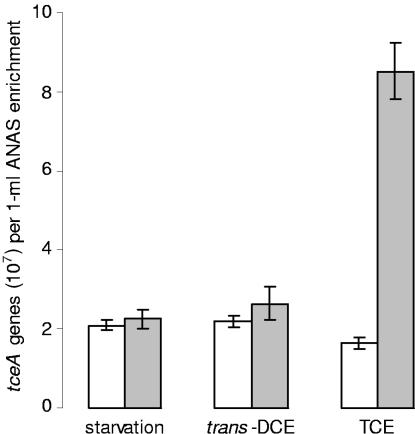
The observed increase in tceA expression after exposure to trans-DCE (Fig. 1) was surprising because the dechlorination of trans-DCE cannot support the growth of known tceA-containing bacteria (20). To determine whether trans-DCE can support the growth of the tceA-containing organism(s) in the ANAS enrichment, changes in the quantities of tceA genes were compared in parallel subcultures amended with TCE, amended with trans-DCE, or starved of chlorinated ethenes. Prior to chlorinated ethene exposure, these subcultures were diluted 10-fold to reduce the cellular density and promote the coupling of dechlorination with growth. After 10 days of incubation, the quantity of tceA genes in the subculture amended with TCE increased fivefold (P value of 1.7 × 10−4) (Fig. 2). In contrast, after 10 days of incubation with trans-DCE or 10 days of chlorinated ethene starvation, the quantities of tceA genes were statistically identical (P values of 0.12 and 0.34, respectively) (Fig. 2).
FIG. 2.
Quantity of tceA genes before and after chlorinated ethene starvation, incubation with trans-DCE, or incubation with TCE. Open bars are the initial quantities of tceA genes, and filled bars are the quantities of tceA genes after 10 days of incubation. All measurements are averages of triplicate qPCRs from duplicate biotic experiments, and error bars represent 1 standard deviation.
Effect of chlorinated ethene concentration on tceA expression.
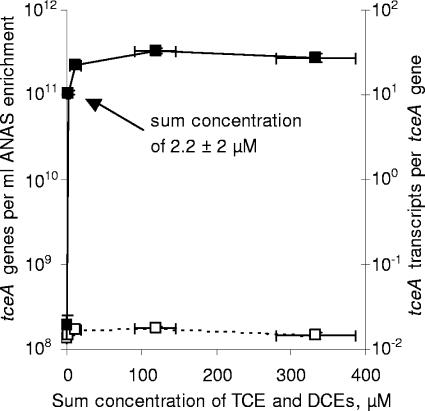
To examine whether the level of tceA expression in the ANAS enrichment depends on the concentration of chlorinated ethenes, parallel chlorinated ethene-starved ANAS subcultures were amended with different masses of TCE and incubated for 24 h. Because a significant fraction of the TCE was converted to DCE over the duration of the experiment and because the previously observed responses to TCE and the DCEs were all similar (Fig. 1), tceA expression levels are reported relative to the sum concentrations of TCE and DCEs. After 24 h of exposure, the quantities of tceA genes were not substantially different for any of the sum concentrations investigated (Fig. 3). Expression levels in the subcultures with the highest sum concentrations of TCE and DCEs (11, 119, and 333 μM) ranged from 22 to 33 transcripts per gene, while expression in the subculture with the lowest sum concentration of TCE and DCEs (2.2 μM) was only somewhat lower at 11 transcripts per gene (Fig. 3). These expression levels are fairly consistent with the levels of 18 to 36 transcripts per gene observed after 24 h of exposure to TCE, cis-DCE, trans-DCE, or 1,1-DCE (initial concentrations of 150 to 330 μM) (Fig. 1). In contrast with the subcultures exposed to chlorinated ethenes, the level of tceA expression in the subculture starved of chlorinated ethenes was only 0.03 transcript per gene (Fig. 3).
FIG. 3.
The effect of cellular exposure to different chlorinated ethene concentrations on the quantity of tceA genes (□, left y axis) and tceA transcripts per tceA gene (▪, right y axis). Parallel ANAS subcultures were amended with different masses of TCE, and the sum concentrations of TCE and DCEs were maintained within a controlled range for 24 h. All tceA measurements are averages of triplicate qPCRs or triplicate RT-qPCRs, and error bars represent 1 standard deviation (in some cases, error bars are smaller than data symbols). All sum concentrations of TCE and DCEs are median observations, and error bars are the observed ranges.
Effect of hydrogen concentration on tceA expression kinetics.
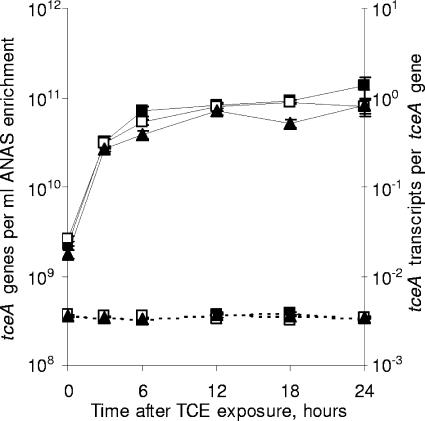
Hydrogen is the only known electron donor to support the growth of tceA-containing bacteria (21). The concentration of hydrogen, therefore, might be important for regulating a wide range of cellular functions. To examine whether the level of tceA expression in the ANAS enrichment depends on the concentration of hydrogen, parallel chlorinated ethene-starved subcultures were amended with TCE and differing masses of hydrogen. Results indicate that, for aqueous hydrogen concentrations spanning three orders of magnitude (12 nM to 17 μM), the tceA expression responses to TCE exposure were all similar (Fig. 4). Expression increased more than 10-fold within 3 h of TCE exposure and an additional 1.2- to 4-fold over the remainder of the experiment, while the number of tceA genes did not substantially change (Fig. 4).
FIG. 4.
The effect of hydrogen concentration on the quantity of tceA genes (dashed lines, left y axis) and tceA transcripts per tceA gene (solid lines, right y axis). Parallel ANAS subcultures were amended with TCE and differing masses of hydrogen to obtain aqueous hydrogen concentrations of 12 nM (▴), 220 nM (□), or 17 μM (▪). All tceA measurements are averages of triplicate qPCRs or triplicate RT-qPCRs, and error bars represent 1 standard deviation.
Effect of incubation temperature on tceA expression kinetics.
The temperature of groundwater aquifers in the United States typically ranges between 10 and 20°C (1), which is substantially lower than the standard experimental temperature of 30°C used in this study. To examine tceA expression in the ANAS enrichment at lower temperatures, parallel chlorinated ethene-starved subcultures were amended with TCE and incubated at 14, 22, or 30°C. During the 20 h of incubation, a strong correlation between temperature and the rate of increase in tceA expression was observed while the quantity of tceA genes did not substantially change (Fig. 5). After 20 h of exposure, incubation at 22°C resulted in a fourfold lower level of tceA expression than incubation at 30°C, while incubation at 14°C resulted in a 50-fold lower expression level than incubation at 30°C (Fig. 5).
FIG. 5.
The effect of incubation temperature on the quantity of tceA genes (dashed lines, left y axis) and tceA transcripts per tceA gene (solid lines, right y axis). Parallel ANAS subcultures were amended with TCE and incubated at 30°C (▴), 22°C (▵), or 14°C (▪). All tceA measurements are averages of triplicate qPCRs or triplicate RT-qPCRs, and error bars represent 1 standard deviation.
Presence of alternate bacterial electron acceptors.
The presence of alternative bacterial electron acceptors has been observed to inhibit reductive dehalogenation activity in consortia by such mechanisms as direct enzymatic inhibition, preferential use of the alternative electron acceptor, and competition with nonreductive dechlorinating bacteria that utilize the alternative electron acceptors (3, 5, 18, 24, 34). To examine whether tceA expression in the ANAS enrichment is affected by the presence of alternate bacterial electron acceptors, parallel chlorinated ethene-starved subcultures were exposed to TCE alone or to TCE along with 5 mM concentrations of fumarate, sulfate, sulfite, thiosulfate, nitrate, or nitrite. After 24 h of exposure, tceA expression levels in the TCE-exposed subcultures, including those containing alternate bacterial electron acceptors, ranged from 4 to 10 transcripts per gene. In contrast, the level of tceA expression in the subculture starved of chlorinated ethenes was only 0.05 transcript per gene.
DISCUSSION
Expression of tceA in the ANAS enrichment increased over an order of magnitude within hours after chlorinated ethene-starved cells were exposed to TCE, cis-DCE, trans-DCE, or 1,1-DCE but did not substantially change after exposure to VC or PCE (Fig. 1). Over the time course of the chlorinated ethene exposure experiment, a substantial amount of chlorinated ethenes were dechlorinated while the quantity of tceA genes did not change, suggesting that the expression responses occurred in cells that had uncoupled growth from dechlorination, similar to the behavior reported for the tceA-containing D. ethenogenes strain 195 (21).
Of the observed positive expression responses to chlorinated ethene exposure (Fig. 1), the increase in expression after exposure to trans-DCE was particularly interesting. While trans-DCE can support the growth of at least one strain of Dehalococcoides (10), it cannot support the growth of known tceA-containing bacteria (20). Similarly, trans-DCE could not support the growth of the tceA-containing organism(s) in the ANAS enrichment over the 10-day experiment, even after subcultures were diluted 10-fold to promote the coupling of growth with dechlorination (Fig. 2). The large positive expression response to trans-DCE, therefore, indicates that tceA expression can respond to non-growth-supporting substrates of the TCE RDase. This is particularly interesting, as trans-DCE can be a significant by-product of the reductive dechlorination of chlorinated ethenes (7, 23).
The positive expression response to trans-DCE was also interesting because it provides evidence of the gene level induction of tceA. Because ANAS subcultures were initially starved of chlorinated ethenes, the observed increases in expression after exposure to growth-supporting chlorinated ethenes might have resulted from broad regulatory responses to changes in metabolic activity, such as the global escalation of mRNA biosynthesis. However, the inability of trans-DCE to support growth (Fig. 2) suggests that trans-DCE exposure does not affect the metabolic state of the tceA-containing organisms. Therefore, the observed tceA expression response to trans-DCE likely results from gene level induction.
Although inducible RDase expression and reductive dehalogenation activity have been observed in a variety of dechlorinating bacteria (summarized in references 11, 24, and 30), induction of tceA expression in the ANAS enrichment was somewhat surprising given that the tceA gene of D. ethenogenes strain 195 is not located within close proximity to any known transcription regulators (29). One possible explanation for the observed induction of tceA is that its genetic organization in the ANAS enrichment differs substantially from that in D. ethenogenes strain 195 such that proximal transcription regulators exist. This explanation is unlikely, however, because a similar tceA expression response to TCE exposure has been observed in pure cultures of D. ethenogenes strain 195 (P. K. H. Lee, D. R. Johnson, V. F. Holmes, J. He, and L. Alvarez-Cohen, submitted for publication). Other possible explanations include the involvement of distally located genes or proximally located genes with as-of-yet unknown functions. Investigations at the genome-wide level should help further elucidate the regulatory system controlling tceA expression.
This study also characterized the kinetics of tceA expression (Fig. 1, 4, and 5). The expression kinetics of another RDase-encoding gene, the o-chlorophenol RDase-encoding cprA gene of Desulfitobacterium dehalogenans, has also been reported (31). In that study, cprA expression increased 15-fold after cellular exposure to the metabolic substrate 3-chloro-4-hydroxyphenylacetic acid for 30 min, which represents 0.15 × generation time (tD) of D. dehalogenans (31). Similarly, tceA expression in the ANAS enrichment increased eightfold after exposure to TCE, cis-DCE, trans-DCE, or 1,1-DCE for 3 h, which represents 0.17 × tD of the tceA-containing D. ethenogenes strain 195 (21). Although these initial responses were fairly consistent, the kinetics of tceA and cprA expression thereafter were quite different. Expression of cprA did not increase after the initial response (31), while tceA expression continued to increase linearly with time at a substantially lower but significant rate (Fig. 1).
A final objective of this work was to examine the dependence of tceA expression on incubation conditions. While tceA expression kinetics were strongly dependent on the specific types of chlorinated ethenes present in the culture (Fig. 1) and on incubation temperature (Fig. 5), tceA expression was independent of the concentration of chlorinated ethenes (Fig. 3, sum concentrations of TCE and DCEs of 2.2 to 333 μM), the concentration of the electron donor hydrogen (Fig. 4, 12 nM to 17 μM), and the presence of alternate bacterial electron acceptors (sulfate, sulfite, thiosulfate, nitrate, nitrite, and fumarate). It is possible, however, that lower concentrations of chlorinated ethenes and hydrogen would exert some effect on tceA expression. For example, the reported half-velocity coefficients for the dechlorination of TCE and cis-DCE by a Dehalococcoides-containing microbial culture are 1.4 and 3.3 μM, respectively (9), suggesting that lower concentrations in the nM range may affect expression. Additionally, the reported half-velocity coefficient for the oxidation of hydrogen by Dehalococcoides sp. strain VS is 7 nM (2), which is below the minimum hydrogen concentration of 12 nM that was achieved in the ANAS culture even after 70 days of lactate starvation. Within the experimental conditions investigated, however, the findings presented in this study suggest that the level of tceA expression in the ANAS enrichment will be dependent on the type of available chlorinated ethenes and incubation temperature but not on chlorinated ethene or hydrogen concentrations or the presence of alternate bacterial electron acceptors.
Acknowledgments
This work was supported by the Lawrence Berkeley National Laboratory through the Laboratory Directed Research and Development Program, the National Science Foundation under grant no. 0504244, and the Superfund Basic Research Project under NIEHS ES04705.
REFERENCES
- 1.Collins, W. D. 1925. Temperature of water available for individual use in the United States, p. 97-104. USGS Water Supply Papers, publication no. 520F. U.S. Geological Survey, Washington, D.C.
- 2.Cupples, A. M., A. M. Spormann, and P. L. McCarty. 2004. Vinyl chloride and cis-dichloroethene dechlorination kinetics and microorganism growth under substrate limiting conditions. Environ. Sci. Technol. 38:1102-1107. [DOI] [PubMed] [Google Scholar]
- 3.Dolfing, J., and J. E. M. Beurskens. 1995. The microbial logic and environmental significance of reductive dehalogenation. Adv. Microb. Ecol. 14:143-206. [Google Scholar]
- 4.Freedman, D. L., and J. M. Gossett. 1989. Biological reductive dechlorination of tetrachloroethylene and trichloroethylene to ethylene under methanogenic conditions. Appl. Environ. Microbiol. 55:2144-2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerritse, J., O. Drzyzga, G. Kloetstra, M. Keijmel, L. P. Wiersum, R. Hutson, M. D. Collins, and J. C. Gottschal. 1999. Influence of different electron donors and acceptors on dehalorespiration of tetrachloroethene by Desulfitobacterium frappieri TCE1. Appl. Environ. Microbiol. 65:5212-5221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gossett, J. M. 1987. Measurement of Henry's law constants for C1 and C2 chlorinated hydrocarbons. Environ. Sci. Technol. 21:202-208. [Google Scholar]
- 7.Griffin, B. M., J. M. Tiedje, and F. E. Löffler. 2004. Anaerobic microbial reductive dechlorination of tetrachloroethene to predominantly trans-1,2-dichloroethene. Environ. Sci. Technol. 38:4300-4303. [DOI] [PubMed] [Google Scholar]
- 8.Harvey, A. H. 1996. Semiempirical correlation for Henry's constants over large temperature ranges. AIChE J. 42:1491-1494. [Google Scholar]
- 9.Haston, Z. C., and P. L. McCarty. 1999. Chlorinated ethene half-velocity coefficients (KS) for reductive dehalogenation. Environ. Sci. Technol. 33:223-226. [Google Scholar]
- 10.He, J. Z., K. M. Ritalahti, K.-L. Yang, S. S. Koenigsberg, and F. E. Löffler. 2003. Detoxification of vinyl chloride to ethene coupled to growth of an anaerobic bacterium. Nature 424:62-65. [DOI] [PubMed] [Google Scholar]
- 11.Holliger, C., G. Wohlfarth, and G. Diekert. 1999. Reductive dechlorination in the energy metabolism of anaerobic bacteria. FEMS Microbiol. Rev. 22:383-398. [Google Scholar]
- 12.Holliger, C., and W. Schumacher. 1994. Reductive dehalogenation as a respiratory process. Antonie Leeuwenhoek 66:239-246. [DOI] [PubMed] [Google Scholar]
- 13.Hölscher, T., R. Krajmalnik-Brown, K. M. Ritalahti, F. von Wintzingerode, H. Görisch, F. E. Löffler, and L. Adrian. 2004. Multiple nonidentical reductive-dehalogease-homologous genes are common in Dehalococcoides. Appl. Environ. Microbiol. 70:5290-5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson, D. R., P. K. H. Lee, V. F. Holmes, and L. Alvarez-Cohen. 2005. An internal reference technique for accurately quantifying specific mRNAs by real-time PCR with application to the tceA reductive dehalogenase gene. Appl. Environ. Microbiol. 71:3866-3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kielhorn, J., C. Melber, U. Wahnschaffe, A. Aitio, and I. Mangelsdof. 2000. Vinyl chloride: still a cause for concern. Environ. Health Perspect. 108:579-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krajmalnik-Brown, R., T. Hölscher, I. N. Thomson, F. M. Saunders, K. M. Ritalahti, and F. E. Löffler. 2004. Genetic identification of a putative vinyl chloride reductase in Dehalococcoides sp. strain BAV1. Appl. Environ. Microbiol. 70:6347-6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Magnuson, J. K., M. F. Romine, D. R. Burris, and M. T. Kingsley. 2000. Trichloroethene reductive dehalogenase from Dehalococcoides ethenogenes: sequence of tceA and substrate range characterization. Appl. Environ. Microbiol. 66:5141-5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Magnuson, J. K., R. V. Stern, J. M. Gossett, S. H. Zinder, and D. R. Burris. 1998. Reductive dechlorination of tetrachloroethene to ethene by a two-component enzyme pathway. Appl. Environ. Microbiol. 64:1270-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maillard, J., W. Schumacher, F. Vazquez, C. Regeard, W. R. Hagen, and C. Holliger. 2003. Characterization of the corrinoid iron-sulfur protein tetrachloroethene reductive dehalogenase of Dehalobacter restrictus. Appl. Environ. Microbiol. 69:4628-4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maymó-Gatell, X., T. Anguish, and S. H. Zinder. 1999. Reductive dechlorination of chlorinated ethenes and 1,2-dichloroethane by “Dehalococcoides ethenogenes” 195. Appl. Environ. Microbiol. 65:3108-3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maymó-Gatell, X., Y. T. Chien, J. M. Gossett, and S. H. Zinder. 1997. Isolation of a bacterium that reductively dechlorinates tetrachloroethene to ethene. Science 276:1568-1571. [DOI] [PubMed] [Google Scholar]
- 22.Miller, E., G. Wohlfarth, and G. Diekert. 1998. Purification and characterization of the tetrachloroethene reductive dehalogenase of strain PCE-S. Arch. Microbiol. 169:497-502. [DOI] [PubMed] [Google Scholar]
- 23.Miller, G. S., C. E. Milliken, K. R. Sowers, and H. D. May. 2005. Reductive dechlorination of tetrachloroethene to trans-dichloroethene and cis-dichloroethene by PCB-dechlorinating bacterium DF-1. Environ. Sci. Technol. 39:2631-2635. [DOI] [PubMed] [Google Scholar]
- 24.Mohn, W. W., and J. M. Tiedje. 1992. Microbial reductive dehalogenation. Microbiol. Rev. 56:482-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Müller, J. A., B. M. Rosner, G. von Abendroth, G. Meshulam-Simon, P. L. McCarty, and A. M. Spormann. 2004. Molecular identification of the catabolic vinyl chloride reductase from Dehalococcoides sp. strain VS and its environmental distribution. Appl. Environ. Microbiol. 70:4880-4888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neumann, A., G. Wohlfarth, and G. Diekert. 1996. Purification and characterization of tetrachloroethene reductive dehalogenase from Dehalospirillum multivorans. J. Biol. Chem. 271:16515-16519. [DOI] [PubMed] [Google Scholar]
- 27.Okeke, B. C., Y. C. Chang, M. Hatsu, T. Suzuki, and K. Takamizawa. 2001. Purification, cloning, and sequencing of an enzyme mediating the reductive dechlorination of tetrachloroethylene (PCE) from Clostridium bifermentans DPH-1. Can. J. Microbiol. 47:448-456. [PubMed] [Google Scholar]
- 28.Richardson, R. E., V. K. Bhupathiraju, D. L. Song, T. A. Goulet, and L. Alvarez-Cohen. 2002. Phylogenetic characterization of microbial communities that reductively dechlorinate TCE based upon a combination of molecular techniques. Environ. Sci. Technol. 36:2652-2662. [DOI] [PubMed] [Google Scholar]
- 29.Seshadri, R., L. Adrian, D. E. Fouts, J. A. Eisen, A. M. Phillippy, B. A. Methe, N. L. Ward, W. C. Nelson, R. T. Deboy, H. M. Khouri, J. F. Kolonay, R. J. Dodson, S. C. Daugherty, L. M. Brinkac, S. A. Sullivan, R. Madupu, K. T. Nelson, K. H. Kang, M. Impraim, K. Tran, J. M. Robinson, H. A. Forberger, C. M. Fraser, S. H. Zinder, and J. F. Heidelberg. 2005. Genome sequence of the PCE-dechlorinating bacterium Dehalococcoides ethenogenes. Science 307:105-108. [DOI] [PubMed] [Google Scholar]
- 30.Smidt, H., and W. M. de Vos. 2004. Anaerobic microbial dehalogenation. Annu. Rev. Microbiol. 58:43-73. [DOI] [PubMed] [Google Scholar]
- 31.Smidt, H., M. van Leest, J. van der Oost, and W. M. de Vos. 2000. Transcriptional regulation of the cpr gene cluster in ortho-chlorophenol-respiring Desulfitobacterium dehalogenans. J. Bacteriol. 182:5683-5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suyama, A., M. Yamashita, S. Yoshino, and K. Furukawa. 2002. Molecular characterization of the PceA reductive dehalogenase of Desulfitobacterium sp. strain Y51. J. Bacteriol. 184:3419-3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tandoi, V., T. D. DiStefano, P. A. Bowser, J. M. Gossett, and S. H. Zinder. 1994. Reductive dehalogenation of chlorinated ethenes and halogenated ethanes by a high-rate anaerobic enrichment culture. Environ. Sci. Technol. 28:973-979. [DOI] [PubMed] [Google Scholar]
- 34.Townsend, G. T., and J. M. Suflita. 1997. Influence of sulfur oxyanions on reductive dehalogenation activities in Desulfomonile tiedjei. Appl. Environ. Microbiol. 63:3594-3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van de Pas, B. A., J. Gerritse, W. M. de Vos, G. Schraa, and A. J. M. Stams. 2001. Two distinct enzyme systems are responsible for tetrachloroethene and chlorophenol reductive dehalogenation in Desulfitobacterium strain PCE1. Arch. Microbiol. 176:165-169. [DOI] [PubMed] [Google Scholar]
- 36.Villemur, R., M. Saucier, A. Gauthier, and R. Beaudet. 2002. Occurrence of several genes encoding putative reductive dehalogenases in Desulfitobacterium hafniense/frappieri and Dehalococcoides ethenogenes. Can. J. Microbiol. 48:697-706. [DOI] [PubMed] [Google Scholar]