Abstract
Haloalkane dehalogenases are enzymes that catalyze the cleavage of the carbon-halogen bond by a hydrolytic mechanism. Genomes of Mycobacterium tuberculosis and M. bovis contain at least two open reading frames coding for the polypeptides showing a high sequence similarity with biochemically characterized haloalkane dehalogenases. We describe here the cloning of the haloalkane dehalogenase genes dmbA and dmbB from M. bovis 5033/66 and demonstrate the dehalogenase activity of their translation products. Both of these genes are widely distributed among species of the M. tuberculosis complex, including M. bovis, M. bovis BCG, M. africanum, M. caprae, M. microti, and M. pinnipedii, as shown by the PCR screening of 48 isolates from various hosts. DmbA and DmbB proteins were heterologously expressed in Escherichia coli and purified to homogeneity. The DmbB protein had to be expressed in a fusion with thioredoxin to obtain a soluble protein sample. The temperature optimum of DmbA and DmbB proteins determined with 1,2-dibromoethane is 45°C. The melting temperature assessed by circular dichroism spectroscopy of DmbA is 47°C and DmbB is 57°C. The pH optimum of DmbA depends on composition of a buffer with maximal activity at 9.0. DmbB had a single pH optimum at pH 6.5. Mycobacteria are currently the only genus known to carry more than one haloalkane dehalogenase gene, although putative haloalkane dehalogenases can be inferred in more then 20 different bacterial species by comparative genomics. The evolution and distribution of haloalkane dehalogenases among mycobacteria is discussed.
Haloalkane dehalogenases catalyze detoxification reactions and act on a broad range of halogenated aliphatic compounds (15). Haloalkane dehalogenases have primarily been isolated from bacterial strains living in soil contaminated with halogenated aliphatic compounds (16, 20, 30, 37, 38, 41, 42, 45). The enzymes are involved in biochemical pathways enabling bacteria to utilize halogenated compounds, i.e., 1,2-dichloroethane, 1-chloroalkanes, γ-hexachlorocyclohexane, 1,2-dibromoethane, and 1,3-dichloropropene as the source of carbon and energy.
Database searches revealed the presence of at least two open reading frames, rv2579 and rv2296, with sequences highly similar to haloalkane dehalogenases in the genome of the pathogenic bacterium Mycobacterium tuberculosis H37Rv (4). Dehalogenating activity was consequently confirmed in M. tuberculosis H37Rv (19). The same study demonstrated dehalogenase activity in several saprophytic mycobacterial species. The first haloalkane dehalogenase originating from a mycobacterial strain was cloned from Mycobacterium sp. strain GP1 (37). This haloalkane dehalogenase, designated DhaAf, is involved in the biochemical pathway for biodegradation of 1,2-dibromoethane. Genome sequence analysis has shown that two other representatives (fast-growing M. smegmatis MC2 155 and slow-growing M. avium subsp. paratuberculosis K10), possess open reading frames aal17946, map0345c, and map2057 coding for putative haloalkane dehalogenases. The first haloalkane dehalogenase of a bacterium isolated from animal tissue was cloned from M. avium subsp. avium N85 (18) designated DhmA. Heterologous expression of the dhmA gene in Escherichia coli resulted in a dehalogenase hydrolyzing a wide range of haloalkanes. The origin and function of the haloalkane dehalogenases in pathogenic mycobacteria is currently unclear.
In a previous study, we compared the homology model of protein Rv2579 with the crystal structure of haloalkane dehalogenase LinB from Sphingomonas paucimobilis UT26 (25) and found that 6 of 19 amino acid residues that form an active site and entrance tunnel are different between LinB and Rv2579 (32). Mutations were introduced cumulatively into the six amino acid residues of LinB resulting in a protein mutant which was designed to have an active site of Rv2579. This mutant showed haloalkane dehalogenase activity, confirming that Rv2579 is a member of haloalkane dehalogenase protein family.
We describe here the cloning and expression of two putative haloalkane dehalogenase genes (designated dmbA and dmbB, corresponding to rv2579 and rv2296 in M. tuberculosis, respectively) from the obligatory pathogen M. bovis, as well as the biochemical and kinetic characterization of proteins encoded by these genes. The distribution of mycobacterial genes dmbA, dmbB, and dhmA genes was studied in isolates of the M. tuberculosis complex, M. avium subsp. avium, M. avium subsp. hominissuis, and M. avium subsp. paratuberculosis originating from various hosts, including human beings and different geographical areas from four continents.
MATERIALS AND METHODS
Mycobacterial isolates.
Isolates of the M. tuberculosis complex (M. tuberculosis, M. bovis, M. bovis BCG, M. africanum, M. caprae, M. microti, and M. pinnipedii), M. avium subsp. avium, M. avium subsp. hominissuis, and M. avium subsp. paratuberculosis were acquired from the collection of the Veterinary Research Institute, Brno, Czech Republic, and were selected to represent a variety of hosts and countries of origin. M. avium subsp. paratuberculosis isolates were grown on Herrold Egg Youlk Medium with Mycobactin J, whereas the rest of the mycobacterial isolates were grown on solid Löwenstein-Jensen and Ogawa's medium (19).
Confirmation of mycobacterial DNA.
DNA was purified as described previously (18). The concentration of the isolated DNA was determined spectrophotometrically, diluted to a final concentration of 100 μg/ml, and used as a template for the PCR. The isolated DNA was tested for bacterial origin by PCR with 16S rRNA-targeted primers UNB51 and UNB800 (Table 1). Primers UNB51 and UNB800 were designed on the basis of the E. coli 16S rRNA gene sequence, and the resulting product of amplification (800 bp) served to reliably identify bacterial DNA. The PCR conditions used were as follows: denaturation at 94°C for 5 min, 3 cycles of denaturation at 94°C for 1 min, annealing at 58°C for 2 min, and extension at 72°C, followed by another 30 cycles of denaturation at 94°C for 20 s, annealing at 55°C for 1 min, and extension at 72°C for 1 min. The final extension was conducted at 72°C for 7 min (21). Moreover, the isolated DNA was tested for the presence of sequence motifs specific for Mycobacterium spp. (gene encoding heat-shock protein DnaJ [28]), M. tuberculosis complex (IS6110 [43]), M. avium subsp. avium (IS901 [23, 34]), and M. avium subsp. paratuberculosis (IS900 [13]). The method of Nagai et al. (28) was used to detect a specific region (230 bp) of the heat shock protein-encoding dnaJ gene using the primers YNP9 and YNP10 that target all mycobacteria (Table 1). The PCR program consisted of the following steps: denaturation at 94°C for 1 min, 30 cycles of denaturation at 94°C for 1 min, annealing at 58°C for 2 min, and extension at 72°C for 1 min, with a final extension at 72°C for 7 min. The DNA of M. avium subsp. paratuberculosis was analyzed for the presence of the insertion sequence IS900, which is specific for this subspecies. The oligonucleotides P90 and P91 (Table 1) designed by Green et al. were used in PCR to distinguish M. avium subsp. paratuberculosis from other mycobacterial species. The product of 412 bp was amplified by PCR as follows: a denaturation step at 94°C for 3 min, followed by 30 cycles of denaturation at 94°C for 1 min, annealing at 65°C for 45 s, and extension at 72°C for 2 min, with a final extension at 72°C for 2 min (13). The amplification products were separated on a 2% agarose gel, stained with ethidium bromide, and photographed under UV transillumination.
TABLE 1.
Oligonucleotides used for identification of mycobacterial DNA and cloning and screening of dehalogenase genes
| Primer | Target gene(s) | Sequence (5′-3′)a | Position | Source or reference |
|---|---|---|---|---|
| UNB51 | Bacterial 16S rRNA | GAGTTTGATCCTGGCTCA | 8-27 | 21 |
| UNB800 | Bacterial 16S rRNA | GGACTACCAGGGTATCTAAT | 787-806 | 21 |
| YNP9 | dnaJ (heat shock protein) | GGGTGACGCGGCATGGCCCA | 13-32 | 28 |
| YNP10 | dnaJ (heat shock protein) | CGGGTTTCGTCGTACTCCTT | 229-248 | 28 |
| P90 | Insertion sequence IS900 | GAAGGGTGTTCGGGGCCGTCGCTTAGG | 15-41 | 13 |
| P91 | Insertion sequence IS900 | GGCGTTGAGGTCGATCGCCCACGTGAC | 401-427 | 13 |
| DMBAf | dmbA | GCCGAATTCTAAGGAGGAATATCGATGACAGCATTCGGC | Cloning primerb | This study |
| DMBAr | dmbA | GCCAAGCTTTCAGTGATGGTGATGGTGATGGACGCCGGCGGCCGCCGACCG | Cloning primer | This study |
| DMBBf1 | dmbB | GCCGAATTCTAAGGAGGAATATCGATGGATGGATGTCCTACGC | Cloning primer | This study |
| DMBBr1 | dmbB | GCCAAGCTTTTAGTGATGGTGATGGTGATGCGTTGCCTGCTGCCAGGA | Cloning primer | This study |
| DMBBf2 | dmbB | GCCGGATCCTATGGATGTCCTACGCACC | Cloning primer | This study |
| DMBBr2 | dmbB | GCNCTGCAGTTAGTGATGGTGATGGTGATGCGTTGCCTGCTGGGAG | Cloning primer | This study |
| RV1f | dmbA | GCTATAGCTATGGCGAGCAA | 239-258 | This study |
| RV5f | dmbA | GTTGTCGTGGCCACGAAAC | 615-633 | This study |
| RV7r | dmbA | GCTGTCCTCCTGAACGAAAT | 818-837 | This study |
| RV8f | dmbA | GGGACCCGACCGCTATAGC | 228-246 | This study |
| TBC4r | dmbB | CGGGGAAAGGTGCATCGTAG | 585-604 | This study |
| TBC5f | dhmA | TTCGAAAACCTGGAGGACTAC | 31-51 | This study |
| TBC6r | dmbB, dhmA | GAAAGCCGTTGGCCACCACC | 429-448 | This study |
| TBC7f | dmbB, dhmA | GCACGGCGAGCCCACCTGGAG | 156-176 | This study |
Restriction sites are underlined.
Cloning primer, primers designed in reference to flanking regions of cloned genes. To the 5′ end of each primer specific restriction sequences were added.
Gene cloning and sequencing.
The enzymes used for DNA manipulation were obtained from New England Biolabs, Inc. (Beverly, MA), and TaKaRa Shuzo Co. (Kyoto, Japan). All other chemicals were purchased from Sigma-Aldrich (St. Louis, MO). Primer pairs flanking the rv2579 and rv2296 genes of M. tuberculosis were designed and used in a PCR with the total DNA of M. bovis 5033/66. The primers for the cloning of the dmbA gene were designated DMBAf and DMBAr, and the primers designated DMBBf1 and DMBBr1 were used for cloning of the dmbB gene (Table 1). To enable cloning into plasmid pUC18, forward and reverse primers were 5′ end modified by using the EcoRI and HindIII restriction sites, respectively. In addition, reverse primers also contained 5′ end modification, allowing the in-frame introduction of the His6 tail into C terminus of cloned DmbA and DmbB. PCR was carried out in 50-μl volumes using the Taq PCR Master Mix Kit (QIAGEN, Hilden, Germany) according to the instructions of the manufacturer. The PCR cycle for dmbA gene amplification consisted of a denaturation step for 4 min at 94°C, followed by 30 cycles of denaturation at 94°C for 40 s, annealing at 62°C for 30 s, and extension at 72°C for 90 s, with a final extension step at 72°C for 5 min. The cycling parameters for dmbB gene amplification were 94°C for 4 min, followed by 30 cycles of 94°C for 40 s, 55°C for 40 s, and 72°C for 90 s, with a final elongation step for 10 min. The same cycling parameters were used for the combinations of primers DMBBf1 and DMBBr1 and primers DMBBf2 and DMBBr2. Prior to the digestion with restriction endonucleases, the PCR products were purified with the QIAquick gel extraction kit (QIAGEN, Hilden, Germany) and ligated with the pUC18 plasmid (Invitrogen, Groningen, The Netherlands) digested and purified in the same way. Recombinant plasmids harboring inserts of the correct size were sequenced by using an ABI Prism 310 genetic analyzer (Perkin-Elmer, Norwalk, CT).
Expression and protein purification.
For the protein expression, dmbA and dmbB genes were recloned to pAQN (29). In this plasmid, His-tagged DmbA and DmbB were expressed under the control of the tac promoter. E. coli BL21(DE3) transformed with the plasmid pAQN containing dmbA or dmbB, respectively, was grown in 4 liters of Luria broth with ampicillin (100 μg/ml) at 37°C. When the culture reached an optical density of 0.6 at 600 nm, protein expression was induced by the addition of IPTG (isopropyl-β-d-thiogalactopyranoside) to a final concentration of 1 mM. Since DmbB was repeatedly purified with a low yield, in an attempt to increase its recovery, the dmbB was cloned into pTrxFus vector (Invitrogen, Amsterdam, The Netherlands) by using the primers DMBBf2 and DMBBr2. In this plasmid, His-tagged DmbB was fused to thioredoxin and expressed under the control of the PL promoter. In this case, E. coli GI724 was grown in 4 liters of medium containing 0.2% Casamino Acids, 0.5% glucose, 1 mM MgCl2, Na2HPO4 (6 g/liter), KH2PO4 (3 g/liter), NaCl (0.5 g/liter), NH4Cl (1 g/liter), and ampicillin (100 μg/ml) at 30°C. When the culture reached an optical density of 0.5 at 600 nm, protein expression was induced by the addition of l->tryptophan to a final concentration of 100 μg/ml. The cells were harvested 4 h after the induction and disrupted by sonication using a Soniprep 150 (Sanyo Gallenkamp PLC, Loughborough, United Kingdom). After centrifugation at 21,000 × g for 1 h, the supernatant was filtered through a 0.45-μm-pore-size filter and further purified on Ni-nitrilotriacetic acid Sepharose column (QIAGEN, Hilden, Germany). The His-tagged protein was bound to the resin equilibrated with a buffer containing 20 mM potassium phosphate, 0.5 M sodium chloride, and 10 mM imidazole (pH 7.5). Unbound and weakly bound proteins were washed out by a buffer with 20 mM potassium phosphate, 0.5 M sodium chloride, and 65 mM imidazole (pH 7.5), and the His-tagged haloalkane dehalogenase was eluted by using an elution buffer (20 mM potassium phosphate, 0.5 M sodium chloride, 225 mM imidazole [pH 7.5]). Protein homogeneity was verified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and the proteins were stored in a 50 mM phosphate buffer (pH 7.5) at 4°C.
CD spectra and thermal denaturation curves.
Circular dichroism (CD) spectra were recorded at room temperature with a Jasco J-810 spectrometer (Jasco, Tokyo, Japan). The data were collected from 185 to 260 nm at 100 nm/min, with a 1-s response time and a 2-nm bandwidth using a 0.1-cm quartz cuvette containing 0.15 mg of protein/ml in a 50 mM potassium phosphate buffer (pH 7.5). Each spectrum shown is the average of 10 individual scans and is corrected for absorbance caused by the buffer. For thermal denaturation, the protein solutions were heated from 22 to 72°C at 1°C/min. The changes in the ellipticity were monitored at 221, 225, and 221 nm for LinB, DmbA, and DmbB, respectively. The protein concentration of the samples was checked throughout the denaturation experiment. The recorded thermal denaturation curves of each protein were normalized to represent signal changes between approximately 1 and 0 and sequentially fitted to sigmoidal curves. The melting temperatures were evaluated from the collected data as a midpoint of the normalized thermal transition.
Biochemical characterization.
The mycobacterial putative haloalkane dehalogenases were purified to homogeneity and characterized for their pH and temperature optimum and their catalytic activity. The pH dependence of haloalkane dehalogenase activity was tested by varying the buffer components in the assay. The buffer containing 50 mM potassium acetate was used in the range from pH 4.0 to 6.0, the 50 mM phosphate buffer varied from pH 6.0 to 8.0, and the 100 mM glycine buffer varied from pH 8.0 to 10.0. The temperature dependence of the enzyme was determined by conducting the dehalogenation reaction at a temperature ranging from 20 to 60°C and the subsequent determination of products of dehalogenase activity by gas chromatography. The 1,2-dibromoethane was used as a substrate for the testing of biochemical properties of DmbA and DmbB. All activity assays were conducted in triplicates for DmbA, whereas replicated experiments could not be performed with DmbB due to the low expression yields. Steady-state kinetic constants were determined for DmbA only using two substrates: 1-chlorobutane and 1,3-dibromopropane. The kinetic data, i.e., the rate of the enzymatic reaction v against the substrate concentration [S], were fitted to the Michaelis-Menten equation as follows: v = ([E] · [S] · kcat)/(Km+[S]). The steady-state kinetic constants (kcat, Km, and Ksi) were calculated by using the computer program EZ-Fit Version 1.1 (F. W. Perrella). The turnover number, kcat, is the number of molecules converted per second by one enzyme molecule under substrate saturation conditions. The Michaelis-Menten constant, Km, is the dissociation constant and quantifies the substrate concentration at which 50% of the maximal velocity (0.5 · kcat · [E]) has been reached. The substrate inhibition constant, Ksi, is the dissociation constant for the inactive complex of enzyme with more than one substrate in the active site.
Activity assays.
The activities of purified mycobacterial dehalogenases were determined by a microtiter plate colorimetric assay using the reagents of Iwasaki (14) described previously (26). The amount of 0.2 mg of pure DmbA per ml in the glycine buffer (pH 8.5) was incubated with the halogenated substrate at a final concentration 10 mM. The progress of the reaction was monitored after 10, 20, and 30 min by measuring the activity of pure DmbA and DmbB. A more precise determination of activities was achieved by using gas chromatography (3). The reaction conditions were the same as those for the spectrophotometric assay. Steady-state kinetics were assessed by the determination of the substrate and product concentrations by using a gas chromatograph Trace GC 2000 (Finnigen, San Jose, CA) equipped with a flame ionization detector and the capillary column DB-FFAP (30 m by 0.25 mm by 0.25 μm; J&W Scientific, Folsom, CA).
Gene screening.
Primers used for the screening of putative haloalkane dehalogenase genes (Table 1) were designed according to the sequences of the dmbA (accession code Z77724), dmbB (accession code Z77163), and dhmA (accession code AJ314789). PCRs were carried out in a 20-μl volume using the Taq Master Mix Kit (QIAGEN). Three different combinations of oligonucleotides were used for screening of the dmbA gene (RV1f+RV7r, RV5f+RV7r, and RV7r+RV8f), whereas two combinations of oligonucleotides were used for screening of the dmbB (TBC4r+TBC7f and TBC6r+TBC7f) and dhmA (TBC5f+TBC6r and TBC6r+TBC7f) genes. The PCR protocol consisted of an initial denaturation step at 95°C for 3 min, followed by 35 cycles of denaturation at 94°C for 40 s and annealing for 80 s, with a final extension at 72°C for 3 min. The annealing temperatures varied for each oligonucleotide combination and were optimized by using gradient PCR. The annealing temperatures were as follows: 55°C for primer combinations RV5f-RV7r and RV7r-RV8f, 57°C for primer combinations RV1f-RV7r and TBC5f-TBC6r, 59°C for primer combination TBC4r-TBC7f, and 59°C for primer combination TBC6r-TBC7f.
Nucleotide sequence accession numbers.
The nucleotide sequences of dmbA and dmbB have been deposited in EMBL/GenBank/DDBJ databases under accession numbers AJ784272 and AJ784273, respectively.
RESULTS
Cloning and sequencing of putative haloalkane dehalogenase genes dmbA and dmbB.
The open reading frames rv2579 and rv2296 present in the genome of M. tuberculosis H37Rv have been previously identified as putative haloalkane dehalogenase genes based on the comparison of known haloalkane dehalogenases with the sequences deposited in genetic databases (19). The third open reading frame rv1833c showing a distant relationship with haloalkane dehalogenases (31.7% identity) was not considered in our study. The genome sequence of M. tuberculosis H37Rv provided data necessary for designing primers complementary to the regions flanking haloalkane dehalogenase-like genes. The DNA of M. bovis, showing a 99.9% identity of the entire genome at nucleotide level with M. tuberculosis H37Rv, was used for the cloning of genes designated dmbA and dmbB (for dehalogenases from M. bovis, paralogs A and B, respectively). Sequencing of the dmbA gene revealed 100% identity with that isolated from M. bovis MU11 (accession no. AJ243259). The translation product of the dmbA gene differs from the translation product of rv2579 of M. tuberculosis by one amino acid. The DmbA protein carries glutamine in position 293 instead of arginine present in the protein encoded by rv2579. The highest sequence identity (68%) found with biochemically characterized proteins was identified with LinB from S. paucimobilis UT26 (31). DmbB does not differ from the translation product rv2296 of M. tuberculosis. The translation product of M. tuberculosis gene dmbB shares 82% sequence identity with the haloalkane dehalogenase DhmA from M. avium subsp. avium N85 (18) and 39% sequence identity with the haloalkane dehalogenase DhlA from Xanthobacter autotrophicus GJ10 (17). The G+C content of both genes (dmbA [63.7%] and dmbB [66.5%]) does not significantly differ from an average G+C content in the genome of M. bovis and M. tuberculosis.
Expression and purification of haloalkane dehalogenases DmbA and DmbB.
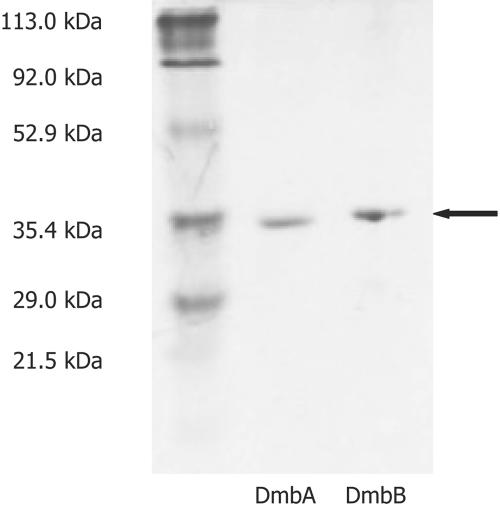
Haloalkane dehalogenase DmbA was purified with a yield of 0.1 mg per g of cell mass. Although the dmbA gene contains approximately the same number of codons rarely used in E. coli as linB gene (29), the level of expression was one magnitude lower compared to LinB using the same expression system. Haloalkane dehalogenase DmbB was purified with a yield of 0.01 mg per g of cell mass. Purified haloalkane dehalogenases DmbA and DmbB migrated as single bands on SDS-PAGE with a molecular masses of 34.5 and 34.1 kDa, respectively (Fig. 1). Proper folding of the purified DmbA and DmbB was verified by CD spectroscopy. The CD spectra of mycobacterial haloalkane dehalogenases DmbA and DmbB were compared to the spectra of haloalkane dehalogenases with known tertiary structures, i.e., LinB, DhaA, and DhlA (25, 33, 44).
FIG. 1.
SDS-PAGE of purified haloalkane dehalogenases from M. bovis. The proteins were stained in 0.25% Coomassie blue R-250.
Thermostability of haloalkane dehalogenases DmbA and DmbB.
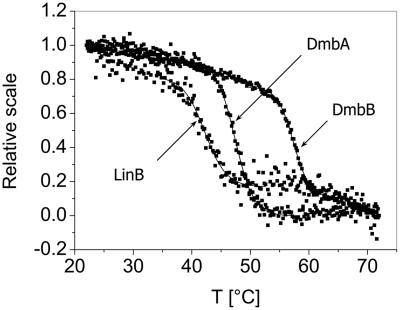
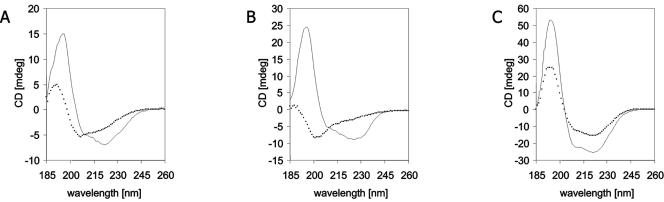
The CD spectra of mycobacterial dehalogenases were initially measured to identify suitable wavelengths for the monitoring of their thermal denaturation. Thermal denaturation was tested with LinB, DmbA, and DmbB. DmbB without fusion protein was used for thermostability measurements to prevent artifacts due to the denaturation of thioredoxin. All three proteins showed changes in ellipticity while being heated (Fig. 2). The intensity of this signal decreased with temperature for each protein following the sigmoidal curve. The melting temperatures were different for respective proteins: 42.2°C for LinB, 47.4°C for DmbA, and 57.4°C for DmbB. Comparison of the CD spectra before heating from 22 to 72°C with the heating rate 1°C/min and after heating, i.e., after cooling back in 50 min using the same heating rate and incubation at room temperature (22°C) for 10 min, is summarized in Fig. 3. All native dehalogenases showed CD spectra typical for predominantly α-helical conformation with two negative features at about 221 and 208 nm and a positive peak at about 195 nm. In comparison to the CD spectrum of native protein, the intensity of the CD spectrum of LinB after thermal denaturation was significantly lower (Fig. 3A). Moreover, these two CD spectra differ in the Θ222/Θ208 ratio, indicating some structural variation. The CD spectrum of the DmbA protein (Fig. 3B) showed the typical shape of a random coil structure with dominant-negative features at about 200 nm and a broad negative ellipticity for 230 to 215 nm after thermal denaturation. This indicates that the DmbA protein lost its α-helical conformation entirely upon heating to the denaturation temperature. Unlike LinB and DmbA, the CD spectrum of DmbB had the same shape before and after thermal denaturation (Fig. 3C). The spectrum of the DmbB protein after heating keeps characteristics of α-helical conformation with somewhat smaller intensity. A significant decrease of the DmbB concentration was not observed after thermal denaturation experiment, suggesting that the decrease of CD spectrum intensity of this protein is not caused by the loss of the protein from the solution but rather by a partial loss of the helical structure. The thermal denaturation is most likely irreversible in the case of LinB and DmbA enzymes, with no recovery of native structure after being heated to 72°C, whereas denaturation of the DmbB enzyme seems to be partially reversible.
FIG. 2.
Thermal denaturation curves of haloalkane dehalogenases LinB, DmbA, and DmbB (dotted lines). The global fit of the sigmoidal model to the data of protein denaturation curves is shown as a solid line.
FIG. 3.
Far-UV CD spectra of LinB (A), DmbA (B), and DmbB (C) dehalogenating enzymes. Solid lines represent the CD spectra of proteins before heating, and dotted lines represent the CD spectra of proteins after heating from 22°C to 72°C, cooling, and incubation at 22°C for 10 min.
Activity profiles of haloalkane dehalogenases DmbA and DmbB.
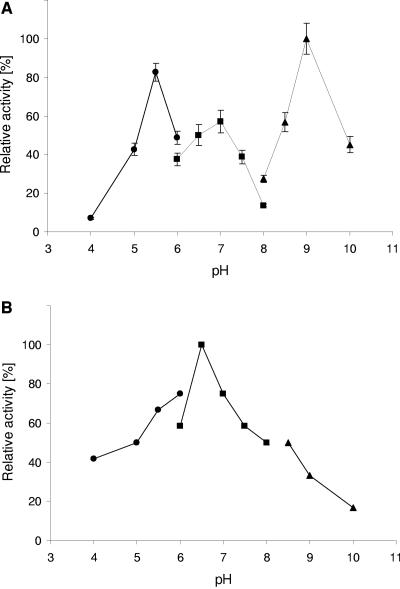
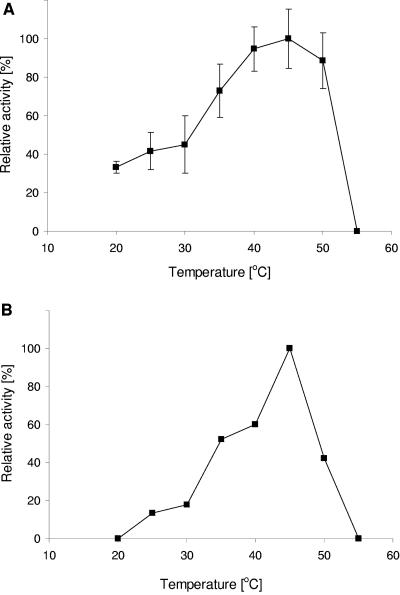
The pH optimum of DmbA and DmbB was determined over the range of pH between 4.0 and 10.0. The haloalkane dehalogenase DmbA showed more than one pH optimum (Fig. 4) with maximal activity at pH 9.0. The high activity of the enzyme at the alkaline pH was in agreement with the pH optimum of other characterized haloalkane dehalogenases. The potassium phosphate buffer had a strong inhibitory effect on DmbA. DmbB was characterized as a fusion protein with thioredoxin to overcome problems with its insolubility. The dependence of haloalkane dehalogenase DmbB activity on pH showed a different trend. The highest activity was observed at pH 6.5 in the phosphate buffer. Both haloalkane dehalogenases, DmbA and DmbB exhibited similar temperature optima at ca. 45°C (Fig. 5). Above this temperature both enzymes rapidly became inactivated. At 20°C, 32.5% of the maximum activity was observed for DmbA, whereas DmbB did not show any activity. We note that the temperature profiles of the two dehalogenases from M. bovis cannot be directly compared since DmbB was characterized in the form of a fusion protein.
FIG. 4.
Effect of pH on activity of M. bovis haloalkane dehalogenases DmbA (top panel) and DmbB (bottom panel). The data for DmbA are expressed as relative activities, and error bars represent the standard errors. Experiments with DmbB were not replicated due to insufficient protein material. Measurements were made with the substrate 1,2-dibromoethane at 37°C in 100 mM buffers: potassium acetate (•), potassium phosphate (▪), or glycine buffer (▴).
FIG. 5.
Effect of temperature on activity of haloalkane dehalogenases DmbA (top panel) and DmbB (bottom panel). The activities were determined with the substrate 1,2-dibromoethane. 2-Bromoethanol was identified as a reaction product by using gas chromatography. The data for DmbA are expressed as relative activities, and error bars represent the standard errors. Experiments with DmbB were not replicated due to insufficient protein material. The specific activity of DmbA under optimal conditions is 0.2125 μmol s−1 mg of protein−1; the specific activity of DmbB is 0.0044 μmol s−1 mg of protein−1.
Catalytic activity of DmbA.
Michaelis-Menten kinetic constants of the haloalkane dehalogenase DmbA were compared to the sixfold mutant of LinB (F143W, Q146A, D147V, L177A, I211L, and L248I), which was designed to possess an active site and an entrance tunnel of DmbA. Sixfold mutant LinB had previously been constructed by cumulative mutagenesis to mimic the active site of DmbA (32). In the present study, the pure DmbA was obtained and its dehalogenase activity was confirmed. The catalytic properties of DmbA and mutant LinB were very similar (Table 2). Both enzymes showed significant dehalogenase activity toward the same substrates. For both enzymes, the Michaelis-Menten kinetic curve for 1,3-dibromopropane indicated substrate inhibition (data not shown). The values of Km indicated a lower affinity of DmbA toward 1-chlorobutane and 1,3-dibromopropane. From the kinetic point of view, there was no biochemically significant difference between wild-type DmbA and the sixfold mutant of LinB. The observed differences in kinetic constants are within an error of independent experiments. The low yield and solubility of the haloalkane dehalogenase DmbB enabled measurements of relative activities (e.g., pH and temperature profiles) but prevented the reliable study of its kinetics.
TABLE 2.
Comparison of kinetic constants determined for wild-type LinB, sixfold LinB mutant, and wild-type DmbA
| Substrate | Mean ± SE
|
kcat/Km (mM−1 s−1) | ||
|---|---|---|---|---|
| Km (mM) | Ksi (mM) | kcat (s−1) | ||
| LinB | ||||
| 1-Chlorobutane | 0.23 ± 0.02 | NAa | 1.11 ± 0.03 | 4.83 |
| 1,3-Dibromopropane | 24.1 ± 3.23 | 0.49 ± 0.06 | 40.9 ± 5.2 | 1.70 |
| Sixfold mutant LinB | ||||
| 1-Chlorobutane | 0.14 ± 0.02 | NA | 0.58 ± 0.003 | 4.14 |
| 1,3-Dibromopropane | 10.07 ± 2.44 | 1.63 ± 0.34 | 21.50 ± 2.8 | 2.13 |
| DmbA | ||||
| 1-Chlorobutane | 0.16 ± 0.04 | NA | 0.08 ± 0.004 | 0.50 |
| 1,3-Dibromopropane | 4.52 ± 0.71 | 2.65 ± 0.49 | 9.20 ± 1.17 | 2.03 |
NA, not applicable.
Distribution of haloalkane dehalogenase genes among mycobacteria.
The identity of the all tested isolates was confirmed by PCR with the specific primers for 16S rRNA, heat shock protein DnaJ and specific insertion sequence IS900. All isolates of the M. tuberculosis complex contained both dmbB and dmbA genes, whereas all isolates of M. avium subsp. avium, M. avium subsp. hominissuis, and M. avium subsp. paratuberculosis did not encode dmbA or dmbB and instead contained the dhmA gene irrespective of the host and geographical origin (Table 3). This observation was in good agreement with available genomic data, suggesting the presence of dmbB and dmbA genes in the genome of M. bovis AF2122/97 (12) and closely related M. tuberculosis H37Rv and CDC1551 (4) and the dhmA gene in the genome M. avium 104 (18).
TABLE 3.
Results of screening for presence of haloalkane dehalogenase genes dmbA, dmbB, and dhmA in mycobacterial isolates
| Isolatea | Country | Hostd | dmbA | dmbB | dhmA |
|---|---|---|---|---|---|
| M. tuberculosis Beijing M434 | Type strain | + | + | − | |
| M. tuberculosis Beijing 02A0110 | Germany | Human | + | + | − |
| M. bovis 12/996 | Czech Republic | Cattle | + | + | − |
| M. bovis 13/2 | Czech Republic | Cattle | + | + | − |
| M. bovis CAMP2063-B | Czech Republic | Cattle | + | + | − |
| M. bovis CAMP5032/66 | Czech Republic | Cattle | + | + | − |
| M. bovis CAMP5032/69 | Czech Republic | Cattle | + | + | − |
| M. bovis CAMP5032/81 | Czech Republic | Cattle | + | + | − |
| M. bovis CAMP5033/66 | Czech Republic | Human | + | + | − |
| M. bovis CAMP5033/81 | Czech Republic | Human | + | + | − |
| M. bovis CAMP5120/74 | Czech Republic | Cattle | + | + | − |
| M. bovis CAMP5120/79 | Czech Republic | Cattle | + | + | − |
| M. bovis CAMP5120/81 | Czech Republic | Cattle | + | + | − |
| M. bovis TBC 4/58 | France | Guinea pig | + | + | − |
| M. bovis BCG25/80 | Argentina | Vaccine strain | + | + | − |
| M. bovis BCG19 | Australia | Vaccine strain | + | + | − |
| M. bovis BCG31/75 | Brazil | Vaccine strain | + | + | − |
| M. bovis BCG7/80 | Czech Republic | Vaccine strain | + | + | − |
| M. bovis BCG MB3/97* | Czech Republic | Child | + | + | − |
| M. bovis BCG MB2/97* | Czech Republic | Child | + | + | − |
| M. bovis BCG MBT/98* | Czech Republic | Child | + | + | − |
| M. bovis BCGR1/89* | Czech Republic | Child | + | + | − |
| M. bovis BCG5 | France | Vaccine strain | + | + | − |
| M. bovis BCG1/83 | Japan | Vaccine strain | + | + | − |
| M. bovis BCG28 | Norway | Vaccine strain | + | + | − |
| M. bovis B/2000 | Poland | European bison | + | + | − |
| M. bovis RUS | Slovakia | Cattle | + | + | − |
| M. bovis BCG15/80 | Sweden | Vaccine strain | + | + | − |
| M. bovis BCG | Russia | Vaccine strain | + | + | − |
| M. africanum 00A0312 | Germany | Human | + | + | − |
| M. africanum 00A0743 | Germany | Human | + | + | − |
| M. africanum 01A0228 | Germany | Human | + | + | − |
| M. africanum 01A0382 | Germany | Human | + | + | − |
| M. africanum 01A0230 | Germany | Cattle | + | + | − |
| M. caprae 14/380 | Czech Republic | Cattle | + | + | − |
| M. caprae D103/99 | Czech Republic | Human | + | + | − |
| M. caprae T158 | Czech Republic | Red deer | + | + | − |
| M. caprae 00A0317 | Germany | Cattle | + | + | − |
| M. caprae 01A0226 | Germany | Human | + | + | − |
| M. caprae M412 | Germany | Cattle | + | + | − |
| M. caprae M353 | Germany | Cattle | + | + | − |
| M. caprae HU10 | Hungary | Wild boar | + | + | − |
| M. caprae HU12 | Hungary | Cattle | + | + | − |
| M. microti 02A0953 | Germany | Fox | + | + | − |
| M. microti M432 | Germany | Coati | + | + | − |
| M. pinnipedii 01A1295 | Germany | Seal | + | + | − |
| M. pinnipedii 02A0460 | Germany | Camel | + | + | − |
| M. pinnipedii 02A0461 | Germany | Camel | + | + | − |
| M. avium subsp. avium 20/588 | Czech Republic | Pig (lymph node) | − | − | + |
| M. avium subsp. avium 20/685 | Czech Republic | Pig (mesenteric lymph node) | − | − | + |
| M. avium subsp. avium 21/320 | Czech Republic | Peregrine falcon (liver) | − | − | + |
| M. avium subsp. avium 21/345 | Czech Republic | Nutria (liver) | − | − | + |
| M. avium subsp. avium 21/383 | Czech Republic | Goldeneye (liver) | − | − | + |
| M. avium subsp. avium N32/97 | Czech Republic | Cattle (bull, mesenteric lymph node) | − | − | + |
| M. avium subsp. avium N52/97 | Czech Republic | Pig (submandibular lymph node) | − | − | + |
| M. avium subsp. avium N61/97 | Czech Republic | Pig (submandibular lymph node) | − | − | + |
| M. avium subsp. avium N65/99 | Czech Republic | Cattle (bull, mesenteric lymph node) | − | − | + |
| M. avium subsp. avium N77/96 | Czech Republic | Cattle (cow, mesenteric lymph node) | − | − | + |
| M. avium subsp. avium N90/96 | Czech Republic | Cattle (cow, mesenteric lymph node) | − | − | + |
| M. avium subsp. avium N141/97 | Czech Republic | Cattle (bull, mesenteric lymph node) | − | − | + |
| M. avium subsp. avium N216/96 | Czech Republic | Cattle (bull, mesenteric lymph node) | − | − | + |
| M. avium subsp. avium N258/96 | Czech Republic | Cattle (heifer, mesenteric lymph node) | − | − | + |
| M. avium subsp. avium N433/96 | Czech Republic | Cattle (cow, mesenteric lymph node) | − | − | + |
| M. avium subsp. avium N285/96 | Czech Republic | Cattle (bull, mesenteric lymph node) | − | − | + |
| M. avium subsp. avium 15367 | The Netherlands | Human (sputum) | − | − | + |
| M. avium subsp. avium 9501279 | The Netherlands | Pig (not known) | − | − | + |
| M. avium subsp. hominissuis N2/97 | Czech Republic | Pig (submandibular lymph node) | − | − | + |
| M. avium subsp. hominissuis N100/97 | Czech Republic | Pig (submandibular lymph node) | − | − | + |
| M. avium subsp. paratuberculosis 58 | Czech Republic | Cattle (cow, feces) | − | − | + |
| M. avium subsp. paratuberculosis 666 | Czech Republic | Cattle (cow, feces) | − | − | + |
| M. avium subsp. paratuberculosis 2108 | Denmark | Cattle (cow, feces) | − | − | + |
| M. avium subsp. paratuberculosis 4898 | Denmark | Cattle (cow, feces) | − | − | + |
| M. avium subsp. paratuberculosis 4901 | Denmark | Cattle (cow, feces) | − | − | + |
| M. avium subsp. paratuberculosis 4904 | Denmark | Cattle (cow, feces) | − | − | + |
| M. avium subsp. paratuberculosis 5388 | Denmark | Cattle (cow, feces) | − | − | + |
| M. avium subsp. paratuberculosis 5394 | Denmark | Cattle (cow, feces) | − | − | + |
| M. avium subsp. paratuberculosis 5403 | Denmark | Cattle (cow, feces) | − | − | + |
| M. avium subsp. paratuberculosis 5418 | Denmark | Cattle (cow, feces) | − | − | + |
| M. avium subsp. paratuberculosis 5429 | Denmark | Cattle (cow, feces) | − | − | + |
| M. avium subsp. paratuberculosis 5452 | Denmark | Cattle (cow, feces) | − | − | + |
| M. avium subsp. paratuberculosis 5457 | Denmark | Cattle (cow, feces) | − | − | + |
| M. avium subsp. paratuberculosis 5469 | Denmark | Cattle (cow, feces) | − | − | + |
| M. avium subsp. paratuberculosis 5474 | Denmark | Cattle (cow, feces) | − | − | + |
| M. avium subsp. paratuberculosis 5476 | Denmark | Cattle (cow, feces) | − | − | + |
| M. avium subsp. paratuberculosis 5533 | Denmark | Hamster (male, feces)b | − | − | + |
| M. avium subsp. paratuberculosis 5947 | Denmark | Hamster (male, feces)c | − | − | + |
| M. avium subsp. paratuberculosis 6371 | Denmark | Cattle (cow, feces) | − | − | + |
| M. avium subsp. paratuberculosis 6420 | Denmark | Cattle (cow, feces) | − | − | + |
| M. avium subsp. paratuberculosis 6423 | Denmark | Cattle (cow, feces) | − | − | + |
| M. avium subsp. paratuberculosis 6477 | Denmark | Cattle (cow, feces) | − | − | + |
| M. avium subsp. paratuberculosis 6564 | Denmark | Cattle (cow, feces) | − | − | + |
| M. avium subsp. paratuberculosis 6567 | Denmark | Cattle (cow, feces) | − | − | + |
| M. avium subsp. paratuberculosis 6577 | Denmark | Cattle (cow, feces) | − | − | + |
| M. avium subsp. paratuberculosis 6706 | Denmark | Cattle (cow, feces) | − | − | + |
| M. avium subsp. paratuberculosis 6732 | Denmark | Cattle (cow, feces) | − | − | + |
| M. avium subsp. paratuberculosis 6775 | Denmark | Cattle (cow, feces) | − | − | + |
| M. avium subsp. paratuberculosis 7747 | Denmark | Cattle (cow, feces) | − | − | + |
| M. avium subsp. paratuberculosis 7749 | Denmark | Cattle (cow, feces) | − | − | + |
| M. avium subsp. paratuberculosis 7840 | Denmark | Cattle (cow, feces) | − | − | + |
| M. avium subsp. paratuberculosis 1766 | Finland | Cattle (cow, feces) | − | − | + |
| M. avium subsp. paratuberculosis 1871 | Finland | Cattle (cow, feces) | − | − | + |
| M. avium subsp. paratuberculosis 1938 | Finland | Cattle (cow, feces) | − | − | + |
| M. avium subsp. paratuberculosis 2109 | Finland | Cattle (cow, feces) | − | − | + |
| M. avium subsp. paratuberculosis 2169 | Finland | Cattle (cow, feces) | − | − | + |
*, Knee joint postvaccination complications.
Hamster feces 81 days after infection. The hamster was infected with strains isolated from imported cattle from Denmark.
Hamster feces 105 days after infection. The hamster was infected with strains isolated from imported cattle from Denmark.
Host genus and species names for cattle, human (child), bison, boar, guinea pig, red deer, fox, coati, seal, camel, pig A, pig B, peregrine falcon, nutria, goldeneye, and hamster were, respectively, Bos taurus, Homo sapiens, Bison bonasus, Sus scrofa, Cavia aperea, Cervus elaphus, Vulpes vulpes, Nasua rufa, Phoca vitulina, Camelus ferus, Egretta garzetta, Sus scrofa f. domestica, Falco peregrinus, Myocastor coypus, Bucephala clangula, and Mesocricetus auratus.
DISCUSSION
M. bovis posses at least two paralogous genes, designated dmbA and dmbB, encoding for enzymes with dehalogenating activity. These two enzymes have the common fold and catalytic mechanism of known haloalkane dehalogenases, but they differ in their primary structure and biochemical properties. Mycobacterial dehalogenases DmbA and DmbB share 27% sequence identity, and each respective enzyme belongs to a separate subfamily within the haloalkane dehalogenase family (2, 8). These two subfamilies are known to differ by the topology of specificity-determining cap domain (33), substrate specificity (9), and composition and arrangement of catalytic triad (7, 22). The catalytic triad of DmbA consists of nucleophile Asp109, catalytic acid Glu133, and base His273, while the triad of DmbB consists of Asp123, Asp250, and His279. The catalytic acid is located in a very different position within an amino acid sequence in these two enzymes. It was proposed that the replacement of the catalytic acid is a result of specific adaptation to the dehalogenation of 1,2-dichloroethane in haloalkane dehalogenase DhlA from X. autotrophicus GJ10 (6, 22). Examination of the two dehalogenase enzymes with a different topological arrangement of the catalytic triad in mycobacteria suggests that the replacement of a catalytic acid must have occurred much earlier since speciation within the M. tuberculosis complex proceeded events of adaptation to anthropogenic substance such as 1,2-dichloroethane. Although the location of the catalytic acid after the β-strand 7 (in DhlA and DmbB) probably provides a functional advantage for catalysis of 1,2-dichloroethane (22) compared to the position after the β-strand 6 (in LinB and DmbA), molecular adaptation toward this substrate occurring within the last 100 years was not the major driving force for repositioning.
The activity of the protein encoded by the dehalogenase-like gene rv2579 from M. tuberculosis had been predicted by sequence analysis and computer modeling (19) long before the orthologous gene dmbA from M. bovis with 99.7% sequence identity was cloned, expressed, and functionally characterized here. To confirm the prediction and test a general concept of the functional characterization of proteins from genome projects using site-directed mutagenesis, we introduced mutations to the active site of the homologous protein LinB (32). The constructed sixfold mutant of LinB mimicking the active site of the protein encoded by rv2579, dbmA, showed clear dehalogenating activity. As the final proof of this concept, we compared the catalytic properties of DmbA with the properties of the sixfold mutant. Good agreement in the steady-state kinetic constants of both proteins confirmed that site-directed mutagenesis of orthologous proteins can serve as a useful tool for the functional annotation of proteins inferred from a genomic data.
An interesting biochemical feature of the DmbA enzyme was the lack of a single pH optimum. Each buffer used for characterization of DmbA had a different pH optimum. Evidence for the double pH optimum has been previously reported, for example, with potato invertase (40), 1,2-diacylglycerol kinase of the human erythrocyte membrane (1), UDP-glucose dehydrogenase (39), ATP-dependent DNase (11), and thermolysin-like protease (10). Haloalkane dehalogenases use three ionizable amino acids for their catalysis, but DmbA is currently the only family member showing more than one pH optimum. Observation could be related to the tendency of protein to aggregate in an acidic environment of pH between 6 and 7.5 (R. Chaloupkova et al., unpublished data). Another interesting property of studied haloalkane dehalogenases is their temperature optimum at ca. 45°C, which is significantly higher than the temperature of the typical living environment of pathogenic mycobacteria.
The presence of dehalogenase genes in mycobacteria is species specific. All of the screened isolates of the M. tuberculosis complex contained both paralogous genes dmbA and dmbB. Mycobacteria are currently the only species known to carry more than one dehalogenase gene, even though putative haloalkane dehalogenases can be inferred in more than 20 different bacterial species by sequence comparisons. We hypothesize that two mycobacterial dehalogenase genes evolved by gene duplication, which was followed by gene differentiation, and we are carrying out phylogenetic analyses to test this hypothesis. In some mycobacterial species one of the genes was lost and the second gene underwent another duplication event (M. avium subsp. paratuberculosis). Alternatively, both genes were lost during speciation (M. leprae). The acquirement of different dehalogenase genes during two independent events seems to be less likely. The presence of dehalogenase genes in every Mycobacterium strain screened in here and in species with the available genomic sequence, except for M. leprae, which undertook massive gene decay (5), confirms the standard transfer of the dehalogenase genes from ancestors to progenitor. Dehalogenase genes could have been transferred horizontally (36) from mycobacteria to species that use dehalogenating enzymes in biochemical pathways essential for growth on halogenated compounds, i.e., Rhodococcus, Xanthobacter, Pseudomonas, and Sphingomonas spp. Dehalogenase genes in these bacteria are usually part of the gene clusters located on plasmids and are regulated (35).
The function of dehalogenating enzymes in mycobacteria is currently unknown. The analysis of surrounding genes in sequenced mycobacterial genomes did not provide any clues about a possible biochemical pathway involving dehalogenation reaction. To test the hypothesis about the potential role of mycobacterial dehalogenases in pathogenesis (19), the presence of the dehalogenase genes was screened in both virulent strains M. bovis and attenuated strains M. bovis BCG. The purpose of this screening was to investigate whether the dmbA and dmbB genes are located in genomic regions of M. bovis that have been excised during serial passages of attenuated “Bacillus of Calmette and Guérin”—known as BCG (24). Both genes were found in all of the tested isolates of M. bovis and M. bovis BCG, suggesting that they are not located in excised pathogenicity regions. However, the recent study of Mattow et al. (27) suggests that the gene dmbA may be expressed in M. tuberculosis but not M. bovis BCG. These authors compared the cellular protein composition of two virulent strains of M. tuberculosis and two attenuated vaccine strains of M. bovis BCG by using high-resolution two-dimensional electrophoresis and mass spectrometry (27). The haloalkane dehalogenase DmbA was missing in two attenuated strains of M. bovis BCG (Copenhagen and Chicago) but was present in two virulent strains of M. tuberculosis (Rv2579 and Erdman). The lack of the DmbA protein in attenuated strains could be due to too low expression, repression, or lack of function.
We have shown that M. bovis carries genes coding for enzymes with dehalogenating activities in their genome and that similar genes are widely distributed among other mycobacteria irrespective of their geographical and host type origin. Mycobacterial dehalogenases DmbA and DmbB belong to a different specificity class of haloalkane dehalogenases (8). Currently, Mycobacterium is the only known genus expressing two such different enzymes within a single species. Comparison of the kinetic constants of wild-type DmbA with the previously constructed sixfold mutant LinB validated the earlier proposal of the catalytic function of this protein established from computer modeling and site-directed mutagenesis (32). Cloned genes could be used for DNA shuffling studies to reconstruct novel catalysts for degradation of important environmental pollutants, e.g., 1,2-dichloroethane and 1,2,3-trichloropropane, which was not possible previously due to the low homology of available dehalogenase genes. Characterized proteins represent a valuable material for structural and functional studies focusing on the enzymatic catalysis and protein engineering of haloalkane dehalogenases.
Acknowledgments
We thank Milan Slosarek of the Institute of Public Health (Prague, Czech Republic), Gyorgy Nagy of the Central Veterinary Institute (Budapest, Hungary), Marek Lipiec of the National Veterinary Research Institute (Pulawy, Poland), Ivan Melicharek of the State Veterinary Diagnostic Institute (Nitra, Slovakia), Jarmila Kaustova of the Institute of Health (Ostrava, Czech Republic), Dick Van Soolingen and Kristin Kremer of the National Institute of Public Health and the Environment (Bilthoven, The Netherlands) for providing some of the mycobacterial isolates. We also thank Nigel Briggs of the Brno English Centre (Brno, Czech Republic) for help with the linguistic revision of the manuscript.
This study was financially supported by grants from the Ministry of Education (MSM0021622412) and the Ministry of Agriculture (MZE0002716201) of the Czech Republic. The research work of J.D. is supported by an EMBO/HMMI grant within the Young Investigator Program.
REFERENCES
- 1.Allan, D., P. Thomas, and S. Gatt. 1980. 1,2-Diacylglycerol kinase of human erythrocyte membranes: assay with endogenously generated substrate. Biochem. J. 191:669-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barth, S., M. Fischer, R. D. Schmid, and J. Pleiss. 2004. The database of epoxide hydrolases and haloalkane dehalogenases: one structure, many functions. Bioinformatics 20:2845-2847. [DOI] [PubMed] [Google Scholar]
- 3.Chaloupkova, R., J. Sykorova, Z. Prokop, A. Jesenska, M. Monincova, M. Pavlova, M. Tsuda, Y. Nagata, and J. Damborsky. 2003. Modification of activity and specificity of haloalkane dehalogenase from Sphingomonas paucimobilis UT26 by engineering of its entrance tunnel. J. Biol. Chem. 278:52622-52628. [DOI] [PubMed] [Google Scholar]
- 4.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M.-A. Rajandream, J. Rogers, S. Rutter, K. Seeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 5.Cole, S. T., K. Eiglmeier, J. Parkhill, K. D. James, N. R. Thomson, P. R. Wheeler, N. Honore, T. Garnier, C. Churcher, D. Harris, K. Mungall, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. M. Davies, K. Devlin, S. Duthoy, T. Feltwell, A. Fraser, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, C. Lacroix, J. Maclean, S. Moule, L. Murphy, K. Oliver, M. A. Quail, M. A. Rajandream, K. M. Rutherford, S. Rutter, K. Seeger, S. Simon, M. Simmonds, J. Skelton, R. Squares, S. Squares, K. Stevens, K. Taylor, S. Whitehead, J. R. Woodward, and B. G. Barrell. 2001. Massive gene decay in the leprosy bacillus. Nature 409:1007-1011. [DOI] [PubMed] [Google Scholar]
- 6.Copley, S. D. 1998. Microbial dehalogenases: enzymes recruited to convert xenobiotic substrates. Curr. Opin. Chem. Biol. 2:613-617. [DOI] [PubMed] [Google Scholar]
- 7.Damborsky, J., and J. Koca. 1999. Analysis of the reaction mechanism and substrate specificity of haloalkane dehalogenases by sequential and structural comparisons. Protein Eng. 12:989-998. [DOI] [PubMed] [Google Scholar]
- 8.Damborsky, J., M. G. Nyandoroh, M. Nemec, I. Holoubek, A. T. Bull, and D. J. Hardman. 1997. Some biochemical properties and classification of a range of bacterial haloalkane dehalogenases. Biotechnol. Appl. Biochem. 26:19-25. [PubMed] [Google Scholar]
- 9.Damborsky, J., E. Rorije, A. Jesenska, Y. Nagata, G. Klopman, and W. J. G. M. Peijnenburg. 2001. Structure-specificity relationships for haloalkane dehalogenases. Environ. Toxicol. Chem. 20:2681-2689. [PubMed] [Google Scholar]
- 10.de Kreij, A., B. van den Burg, G. Venema, G. Vriend, V. G. Eijsink, and J. E. Nielsen. 2002. Effects of modifying the surface charge on the catalytic activity of a thermolysin-like protease. J. Biol. Chem. 277:15432-15438. [DOI] [PubMed] [Google Scholar]
- 11.Fujiyoshi, T., J. Nakayama, and M. Anai. 1982. Two pH optima of adenosine 5′-triphosphate dependent deoxyribonuclease from Bacillus laterosporus. Biochemistry 21:4159-4164. [DOI] [PubMed] [Google Scholar]
- 12.Garnier, T., K. Eiglmeier, J. C. Camus, N. Medina, H. Mansoor, M. Pryor, S. Duthoy, S. Grondin, C. Lacroix, C. Monsempe, S. Simon, B. Harris, R. Atkin, J. Doggett, R. Mayes, L. Keating, P. R. Wheeler, J. Parkhill, B. G. Barrell, S. T. Cole, S. V. Gordon, and R. G. Hewinson. 2003. The complete genome sequence of Mycobacterium bovis. Proc. Natl. Acad. Sci. USA 100:7877-7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Green, E. P., M. T. Moss, J. Hermon-Taylor, and J. J. McFadden. 1989. Insertion elements in mycobacteria. Acta Leprol. 7:239-242. [PubMed] [Google Scholar]
- 14.Iwasaki, I., S. Utsumi, and T. Ozawa. 1952. New colorimetric determination of chloride using mercuric thiocyanate and ferric ion. Bull. Chem. Soc. Japan 25:226. [Google Scholar]
- 15.Janssen, D. B. 2004. Evolving haloalkane dehalogenases. Curr. Opin. Chem. Biol. 8:150-159. [DOI] [PubMed] [Google Scholar]
- 16.Janssen, D. B., J. Gerritse, J. Brackman, C. Kalk, D. Jager, and B. Witholt. 1988. Purification and characterization of a bacterial dehalogenase with activity toward halogenated alkanes, alcohols, and ethers. Eur. J. Biochem. 171:67-92. [DOI] [PubMed] [Google Scholar]
- 17.Janssen, D. B., F. Pries, J. Ploeg, B. Kazemier, P. Terpstra, and B. Witholt. 1989. Cloning of 1,2-dichloroethane degradation genes of Xanthobacter autotrophicus GJ10 and expression and sequencing of the dhlA gene. J. Bacteriol. 171:6791-6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jesenska, A., M. Bartos, V. Czernekova, I. Rychlik, I. Pavlik, and J. Damborsky. 2002. Cloning and expression of the haloalkane dehalogenase gene dhmA from Mycobacterium avium N85 and preliminary characterization of DhmA. Appl. Environ. Microbiol. 68:3724-3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jesenska, A., I. Sedlacek, and J. Damborsky. 2000. Dehalogenation of haloalkanes by Mycobacterium tuberculosis H37Rv and other mycobacteria. Appl. Environ. Microbiol. 66:219-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keuning, S., D. B. Janssen, and B. Witholt. 1985. Purification and characterization of hydrolytic haloalkane dehalogenase from Xanthobacter autotrophicus GJ10. J. Bacteriol. 163:635-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kirschner, P., and E. C. Bottger. 1998. Species identification of mycobacteria using rDNA sequencing. Methods Mol. Biol. 101:349-361. [DOI] [PubMed] [Google Scholar]
- 22.Krooshof, G. H., E. M. Kwant, J. Damborsky, J. Koca, and D. B. Janssen. 1997. Repositioning the catalytic triad acid of haloalkane dehalogenase: effects on activity and kinetics. Biochemistry 36:9571-9580. [DOI] [PubMed] [Google Scholar]
- 23.Kunze, Z. M., F. Portaels, and J. J. McFadden. 1992. Biologically distinct subtypes of Mycobacterium avium differ in possession of insertion sequence IS901. J. Clin. Microbiol. 30:2366-2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mahairas, G. G., P. J. Sabo, M. J. Hickey, D. C. Singh, and C. K. Stover. 1996. Molecular analysis of genetic differences between Mycobacterium bovis BCG and virulent M. bovis. J. Bacteriol. 178:1274-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marek, J., J. Vevodova, I. Kuta-Smatanova, Y. Nagata, L. A. Svensson, J. Newman, M. Takagi, and J. Damborsky. 2000. Crystal structure of the haloalkane dehalogenase from Sphingomonas paucimobilis UT26. Biochemistry 39:14082-14086. [DOI] [PubMed] [Google Scholar]
- 26.Marvanova, S., Y. Nagata, M. Wimmerova, J. Sykorova, K. Hynkova, and J. Damborsky. 2001. Biochemical characterization of broad-specificity enzymes using multivariate experimental design and a colorimetric microplate assay: characterization of the haloalkane dehalogenase mutants. J. Microbiol. Methods 44:149-157. [DOI] [PubMed] [Google Scholar]
- 27.Mattow, J., P. R. Jungblut, U. E. Schaible, H. J. Mollenkopf, S. Lamer, U. Zimny-Arndt, K. Hagens, E. C. Muller, and S. H. Kaufmann. 2001. Identification of proteins from Mycobacterium tuberculosis missing in attenuated Mycobacterium bovis BCG strains. Electrophoresis 22:2936-2946. [DOI] [PubMed] [Google Scholar]
- 28.Nagai, R., S. Takewaki, A. Wada, K. Okuzumi, A. Tobita, and A. Ohkubo. 1990. Rapid detection and identification of mycobacterial DNA by PCR. Rinsho Byori 38:1247-1253. (In Japanese.) [PubMed] [Google Scholar]
- 29.Nagata, Y., K. Hynkova, J. Damborsky, and M. Takagi. 1999. Construction and characterization of histidine-tagged haloalkane dehalogenase (LinB) of a new substrate class from a γ-hexachlorocyclohexane-degrading bacterium, Sphingomonas paucimobilis UT26. Protein Expr. Purif. 17:299-304. [DOI] [PubMed] [Google Scholar]
- 30.Nagata, Y., K. Miyauchi, J. Damborsky, K. Manova, A. Ansorgova, and M. Takagi. 1997. Purification and characterization of haloalkane dehalogenase of a new substrate class from a γ-hexachlorocyclohexane-degrading bacterium, Sphingomonas paucimobilis UT26. Appl. Environ. Microbiol. 63: 3707-3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nagata, Y., T. Nariya, R. Ohtomo, M. Fukuda, K. Yano, and M. Takagi. 1993. Cloning and sequencing of a dehalogenase gene encoding an enzyme with hydrolase activity involved in the degradation of hexachlorocyclohexane in Pseudomonas paucimobilis. J. Bacteriol. 175:6403-6410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nagata, Y., Z. Prokop, S. Marvanova, J. Sykorova, M. Monincova, M. Tsuda, and J. Damborsky. 2003. Reconstruction of mycobacterial dehalogenase Rv2579 by cumulative mutagenesis of haloalkane dehalogenase LinB. Appl. Environ. Microbiol. 69:2349-2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Newman, J., T. S. Peat, R. Richard, L. Kan, P. E. Swanson, J. A. Affholter, I. H. Holmes, J. F. Schindler, C. J. Unkefer, and T. C. Terwilliger. 1999. Haloalkane dehalogenase: structure of a Rhodococcus enzyme. Biochemistry 38:16105-16114. [DOI] [PubMed] [Google Scholar]
- 34.Pavlik, I., P. Svastova, J. Bartl, L. Dvorska, and I. Rychlik. 2000. Relationship between IS901 in the Mycobacterium avium complex strains isolated from birds, animals, humans, and environment and virulence for poultry. Clin. Diagn. Lab. Immunol. 7:212-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poelarends, G., M. Zandstra, T. Bosma, L. A. Kulakov, M. J. Larkin, J. R. Marchesi, A. J. Weightman, and D. B. Janssen. 2000. Haloalkane-utilizing Rhodococcus strains isolated from geographically distinct locations possess a highly conserved gene cluster encoding haloalkane catabolism. J. Bacteriol. 182:2725-2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Poelarends, G. J., L. A. Kulakov, M. J. Larkin, J. E. T. van Hylckama Vlieg, and D. B. Janssen. 2000. Roles of horizontal transfer and gene integration in evolution of 1,3-dichloropropene- and 1,2-dibromoethane-degradative pathways. J. Bacteriol. 182:2191-2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poelarends, G. J., J. E. T. van Hylckama Vlieg, J. R. Marchesi, L. M. Freitas dos Santos, and D. B. Janssen. 1999. Degradation of 1,2-dibromoethane by Mycobacterium sp. strain GP1. J. Bacteriol. 181:2050-2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Poelarends, G. J., M. Wilkens, M. J. Larkin, J. D. vanElsas, and D. B. Janssen. 1998. Degradation of 1,3-dichloropropene by Pseudomonas cichorii 170. Appl. Environ. Microbiol. 64:2931-2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reen, R. K., D. S. Jamwal, S. C. Taneja, J. L. Koul, R. K. Dubey, F. J. Wiebel, and J. Singh. 1993. Impairment of UDP-glucose dehydrogenase and glucuronidation activities in liver and small intestine of rat and guinea pig in vitro by piperine. Biochem. Pharmacol. 46:229-238. [DOI] [PubMed] [Google Scholar]
- 40.Rorem, E. S., and S. Schwimmer. 1963. Double pH optima of potato invertase. Experientia 19:150-151. [DOI] [PubMed] [Google Scholar]
- 41.Sallis, P. J., S. J. Armfield, A. T. Bull, and D. J. Hardman. 1990. Isolation and characterization of a haloalkane halidohydrolase from Rhodococcus erythropolis Y2. J. Gen. Microbiol. 136:115-120. [DOI] [PubMed] [Google Scholar]
- 42.Scholtz, R., T. Leisinger, F. Suter, and A. M. Cook. 1987. Characterization of 1-chlorohexane halidohydrolase, a dehalogenase of wide substrate range from an Arthrobacter sp. J. Bacteriol. 169:5016-5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thierry, D., A. Brisson-Noel, V. Vincent-Levy-Frebault, S. Nguyen, J. L. Guesdon, and B. Gicquel. 1990. Characterization of a Mycobacterium tuberculosis insertion sequence, IS6110, and its application in diagnosis. J. Clin. Microbiol. 28:2668-2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Verschueren, K. H. G., F. Seljee, H. J. Rozeboom, K. H. Kalk, and B. W. Dijkstra. 1993. Crystallographic analysis of the catalytic mechanism of haloalkane dehalogenase. Nature 363:693-698. [DOI] [PubMed] [Google Scholar]
- 45.Yokota, T., T. Omori, and T. Kodama. 1987. Purification and properties of haloalkane dehalogenase from Corynebacterium sp. strain m15-3. J. Bacteriol. 169:4049-4054. [DOI] [PMC free article] [PubMed] [Google Scholar]