Abstract
Gene shuffling is a way of creating proteins with interesting new characteristics, starting from diverged sequences. We tested an alternative to gene shuffling based on plasmid recombination and found that Bacillus subtilis efficiently recombines sequences with 4% divergence, and Escherichia coli mutS is more appropriate for sequences with 22% divergence.
The technique of gene shuffling is presently essentially based on PCR (10, 12), which creates an enormous waste of useless sequences containing deletions or nonsense mutations. Much less waste is expected in vivo, since the recombination proteins are designed to promote in-register exchanges, without deletions, and since the rate of mutagenesis is lower than that of the PCR polymerases. We therefore decided to test the efficiency of creating new genes by plasmid recombination in vivo.
Experiments were conducted with two distantly related bacterial hosts, Escherichia coli and Bacillus subtilis, in which the mechanisms of homologous recombination are well known. Recombination efficiencies were measured as a function of DNA divergence (0%, 4%, and 22% divergence) and plasmid vector type (symmetrical, theta replication versus asymmetrical, rolling-circle replication). The recombination substrates were three 800-bp-long OXA genes, the OXA-7, OXA-11, and OXA-5 genes, encoding related beta-lactamases differing from the OXA-7 beta-lactamase at the nucleotide level by 0%, 4%, and 22%, respectively. They were cloned into two plasmids: pACYC184, a plasmid of E. coli that cannot replicate in B. subtilis, and pIL253, a plasmid of B. subtilis that cannot replicate in E. coli. In each bacterial host, experiments were conducted so as to maximize recombination efficiency. For E. coli electrotransformations, DNA was UV irradiated at a dose of 200 J/m2 so that the yield of recombinants was 10-fold higher than that of nonirradiated DNA. Transformation in B. subtilis is a natural process, involving the cutting of double-strand DNA and the degradation of one strand, so that DNA enters as single-strand linear DNA. In both species, recipient strains for the transformation experiments harbored a replicative plasmid sharing no significant stretch of identity with the incoming DNA, except for the presence of an OXA gene, so that the establishment of the incoming DNA is dependent on its integration into the OXA gene of the resident plasmid. The selection of clones harboring recombinant plasmids was based on a marker(s) encoded by the nonreplicative plasmid (Spcr Phlr in E. coli and Ermr in B. subtilis). An estimation of the recombination frequency was obtained by dividing the transformation efficiency of the nonreplicating plasmid by the transformation efficiency of a control, replicative plasmid. E. coli cells had a much higher competence (between 106 and 108 transformants per microgram of control DNA) than the B. subtilis cells (0.5 × 105 to 1.5 × 105 transformants per microgram of control DNA). For the B. subtilis experiments, two different recipient vectors were tested: a theta-replicating vector, pMAP176, in which replication of the two strands is simultaneous so that little single-strand DNA and no double-strand ends are present (2), and a rolling-circle-replicating vector, pMAP183, which replicates its two strands successively so that double-strand ends and single-strand DNA are present (11). The theta-replicating vectors of E. coli and B. subtilis had similar replication cycles and copy numbers (∼5). Results are shown in Table 1.
TABLE 1.
Shuffling efficiencies
| % Divergence | Recombination efficiency in indicated genetic backgrounda
|
|||||
|---|---|---|---|---|---|---|
|
E. coli theta plasmid
|
B. subtilis theta plasmid
|
B. subtilis RC plasmid
|
||||
| WT | mutS | WT | mutLS | WT | mutLS | |
| 0 | 3.6 × 10−4 | 9.3 × 10−4 | 2.0 × 10−3 | 4.2 × 10−3 | 1.5 × 10−1 | 6.4 × 10−2 |
| 4 | 3.1 × 10−8 | 2.0 × 10−5 | 2.5 × 10−4 | 9.8 × 10−3 | 3.0 × 10−1 | 7.0 × 10−2 |
| 22 | <5.5 × 10−8 | 2.6 × 10−7 | <2.5 × 10−4 | <2.5 × 10−4 | <1.4 × 10−4 | <1.4 × 10−3 |
RC, rolling circle; WT, wild type.
In wild-type E. coli, the recombination frequency between identical sequences was 3.6 × 10−4. A 4% DNA divergence reduced recombination by 10,000-fold in the wild type, but inactivation of the mutS gene had the expected effect of increasing the recombination efficiency by 1,000-fold (7) so that the net yield of transformants was 4,000 transformants per microgram of DNA. No transformants were recovered from the wild-type strain upon the selection of recombinants with 22%-divergent sequences, but in the mutS background, 40 transformants per microgram of DNA were obtained. The frequency of recombination (2 × 10−7) was lower than the rate of spontaneous Phlr Spcr mutations arising in E. coli mutS (5 × 10−6), so that the plasmid content of ∼100 clones was analyzed to detect these recombinations.
In the wild-type B. subtilis host, the recombination frequency between identical sequences was 2 × 10−3 on theta plasmids and 100-fold more efficient on the rolling-circle vector. This confirms earlier reports showing that rolling-circle plasmids are hyperrecombinogenic (4). Moderate divergence (4%) decreased recombination in wild-type B. subtilis by 10-fold only (in the theta plasmid) or not at all (in the rolling-circle plasmid). Indeed, moderate divergence is much more “tolerated” in wild-type B. subtilis than in wild-type E. coli (5, 7). The ΔmutLS mutation increased recombination efficiency in the B. subtilis theta plasmid by a factor of 40. No recombinants were detected for sequences with a divergence of 22% (frequency below 10−4). It has been shown previously that in B. subtilis, recombination frequencies remained high, with sequences having up to 7% divergence, but then decreased sharply irrespective of the presence or absence of MutL and MutS (5). We speculate, therefore, that 22%-divergent sequences are unlikely to recombine in B. subtilis.
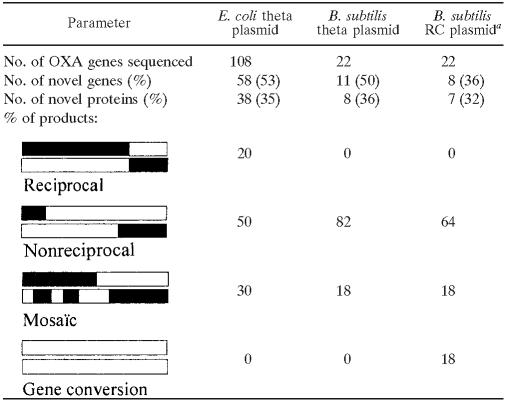
Chimeric genes resulting from recombination between the 4%-divergent OXA genes were sequenced in 54 plasmids of E. coli and 22 plasmids of B. subtilis (11 for each plasmid type). Results are reported in Table 2. According to classical crossing-over models, the two DNA strands of a recombined plasmid are expected to be slightly different. Depending on which strand will give a progeny, different products may thus be recovered. “Reciprocal products” are expected if the progeny derives from the strand cut by the RuvABC resolvase (for E. coli) or by RuvAB and probably RecU (for B. subtilis) (1). However, if the other strand is kept, and depending on its processing, “nonreciprocal products” could be observed. More-complex situations can be encountered, such as multiple crossovers giving “mosaic” products and “gene conversion” products, where a gene conversion process precedes the crossover step. Under all conditions tested, the main category was “nonreciprocal” products (50 to 82% of the recombinants). Mosaic products were also found in all cases (18 to 30%). Interestingly, reciprocal products were recovered only in E. coli. Conversely, gene conversion products were found only in B. subtilis, with the rolling-circle plasmid vector.
TABLE 2.
Recombinant-product analysis
RC, rolling circle.
All strains and plasmids used in this study are described in Table 3.
TABLE 3.
Strains and plasmids
| Name | Genotype | Source/construction |
|---|---|---|
| E. coli strains | ||
| AB1157 Nal | Like AB1157 but Nalr | M. Radman strain collection |
| MIXP1 | mutS::Tn5 (Kanr) | P1 transduction of mutS::Tn5 into AB1157 Nal |
| Plasmids | ||
| pHV1210 | pE194-pBR322 hybrid | Noirot et al. (6) |
| pIC156 | pUC1318-Spcr | Steinmetz and Richter (9) |
| pUC-Phleo | pUC19-Phlr | E. Dervyn collection |
| pMIX92 | pUC19-Spcr-Phlr | Integration of the Spcr cassette of pIC156 (BamHI EcoRI) into pUC19-Phlr |
| pMIX93 | pACYC184-OXA-7 | Integration of OXA-7 or E. coli into pACYC184 (ScaI-PpuMI) |
| pMIX95 | pACYC184-OXA-11 | Integration of OXA-11 of P. aeruginosa into pACYC184 (BamHI-EcoRI) |
| pMIX96 | pACYC184-OXA-5 | Integration of OXA-5 of P. aeruginosa plasmid pMON812 into pACYC184 BamHI-EcoO109I |
| pMAP177 | pACYC184-Ermr | Integration of the Ermr cassette of pHV1210 (ClaI SacI) into pACYC184 (ClaI-FspI) |
| pMAP178 | pACYC184-Ermr-OXA-7 | Integration of the Ermr cassette of pHV1210 (ClaI SacI) into pMIX93 (ClaI SacI) |
| pMAP179 | pACYC184-Ermr-OXA-11 | Integration of the Ermr cassette of pHV1210 (ClaI SacI) into pMIX95 (ClaI-FspI) |
| pMAP180 | pACYC184-Ermr-OXA-5 | Integration of the Ermr cassette of pHV1210 (ClaI SacI) into pMIX96 (ClaI-FspI) |
| B. subtilis strains | ||
| PB1856 | ΔmutLS | Ginetti et al. (3) |
| SB202 | WTa | |
| MAS 831 | ΔmutLS | Transformation of the mutLS::Cmr allele from strain PB1856 into SB202 |
| Plasmids | ||
| pIL253 | Simon and Chopin (8) | |
| pIL252 | Simon and Chopin (8) | |
| pMIX90 | pIL253-Spcr | Integration of the Spcr cassette of pIC156 (SacI) into pIL253 |
| pMIX91 | pIL253-Spcr-Phlr | Integration of the PhIr cassette of pUC19-Phlco (EcoRI SalI) into pMIX90 |
| pMIX94 | pMIX91-OXA-7 | Integration of OXA-7 from pMIX93 into pMIX91 |
| pMIX98 | pMIX91-OXA-11 | Integration of OXA-11 from pMIX95 into pMIX91 |
| pMAP176 | pIL252-Spcr-OXA-7 | Integration of the Spcr-OXA-7 cassette of pTG2 into pIL252 (EcoRI-Eco1091 sites) |
| pMAP183 | pUB110-OXA-7 | Integration of the OXA-7 cassette of pMAP178 (PstI-NruI) into the PvuII site of pUB110 (orientation such that OXA-7 and the Kanr gene are convergent) |
WT, wild type.
We conclude that plasmid vectors are appropriate tools to create new genes by recombination, with B. subtilis being a slightly better host for a low level of divergence (4%) and the E. coli mutS strain being more appropriate for a high level of divergence (up to 22%). This in vivo technique should therefore apply to attempts at creating new gene combinations starting from ortholog genes of related species, or paralogs within a species, so that the divergence at the DNA level is less than 20%. It does not apply, however, to the creation of hybrid genes composed of a combination of two unrelated sequences. Nevertheless, we predict that in vivo recombination should also be possible when the starting sequences are composed of highly conserved regions interspersed with regions of high divergence.
REFERENCES
- 1.Ayora, S., B. Carrasco, E. Doncel, R. Lurz, and J. C. Alonso. 2004. Bacillus subtilis RecU protein cleaves Holliday junctions and anneals single-stranded DNA. Proc. Natl. Acad. Sci. USA 101:452-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bruand, C., E. Le Chatelier, S. D. Ehrlich, and L. Janniere. 1993. A fourth class of theta-replicating plasmids: the pAM beta 1 family from gram-positive bacteria. Proc. Natl. Acad. Sci. USA 90:11668-11672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ginetti, F., M. Perego, A. M. Albertini, and A. Galizzi. 1996. Bacillus subtilis mutS mutL operon: identification, nucleotide sequence and mutagenesis. Microbiology 142:2021-2029. [DOI] [PubMed] [Google Scholar]
- 4.Janniere, L., C. Bruand, and S. D. Ehrlich. 1990. Structurally stable Bacillus subtilis cloning vectors. Gene 87:53-61. [DOI] [PubMed] [Google Scholar]
- 5.Majewski, J., and F. M. Cohan. 1999. DNA sequence similarity requirements for interspecific recombination in Bacillus. Genetics 153:1525-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Noirot, P., M.-A. Petit, and S. D. Ehrlich. 1987. Plasmid replication stimulates DNA recombination in Bacillus subtilis. J. Mol. Biol. 196:39-48. [DOI] [PubMed] [Google Scholar]
- 7.Rayssiguier, C., D. S. Thaler, and M. Radman. 1989. The barrier to recombination between Escherichia coli and Salmonella typhimurium is disrupted in mismatch-repair mutants. Nature 342:396-401. [DOI] [PubMed] [Google Scholar]
- 8.Simon, D., and A. Chopin. 1988. Construction of a vector plasmid family and its use for molecular cloning in Streptococcus lactis. Biochimie 70:559-566. [DOI] [PubMed] [Google Scholar]
- 9.Steinmetz, M., and R. Richter. 1994. Plasmids designed to alter the antibiotic resistance expressed by insertion mutations in Bacillus subtilis, through in vivo recombination. Gene 142:79-83. [DOI] [PubMed] [Google Scholar]
- 10.Stemmer, W. P. 1994. Rapid evolution of a protein in vitro by DNA shuffling. Nature 370:389-391. [DOI] [PubMed] [Google Scholar]
- 11.te Riele, H., B. Michel, and S. D. Ehrlich. 1986. Single-stranded plasmid DNA in Bacillus subtilis and Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 83:2541-2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao, H., L. Giver, Z. Shao, J. A. Affholter, and F. H. Arnold. 1998. Molecular evolution by staggered extension process (StEP) in vitro recombination. Nat. Biotechnol. 16:258-261. [DOI] [PubMed] [Google Scholar]