Abstract
A series of new expression vectors (pPro) have been constructed for the regulated expression of genes in Escherichia coli. The pPro vectors contain the prpBCDE promoter (PprpB) responsible for expression of the propionate catabolic genes (prpBCDE) and prpR encoding the positive regulator of this promoter. The efficiency and regulatory properties of the prpR-PprpB system were measured by placing the gene encoding the green fluorescent protein (gfp) under the control of the inducible PprpB of E. coli. This system provides homogenous expression in individual cells, highly regulatable expression over a wide range of propionate concentrations, and strong expression (maximal 1,500-fold induction) at high propionate concentrations. Since the prpBCDE promoter has CAP-dependent activation, the prpR-PprpB system exhibited negligible basal expression by addition of glucose to the medium.
Inducible expression systems are essential molecular tools for production of recombinant proteins in cells, for synthesis and degradation of small molecules catalyzed by the enzymes expressed from the expression system, and for testing the function of unknown genes or proteins in cells. Although there are very good expression systems for high-level production of recombinant proteins, the more subtle approach of metabolic optimization has been hampered by the lack of appropriate systems for fine-tuning gene expression, and in particular expression systems that have homogeneous control in all cells of the culture.
It has been reported that the all-or-none phenomenon occurs because the expression of the gene encoding the transporter for the inducer is controlled by the inducer itself (8, 28, 35). To alleviate the all-or-none phenomenon, control of expression of the gene encoding the transporter for the inducer itself was decoupled by placing the transporter gene under control of an inducer-independent promoter or using a synthetic inducer analogue that does not require active transport across the cell membrane (21, 22). Since arabinose does not have a synthetic analogue that can freely diffuse across the cell membrane, the arabinose transporter gene was placed under control of a promoter that was not regulated by arabinose and homogenous expression from PBAD promoter was achieved (22). However, the araC-PBAD system requires genetically modified Escherichia coli strain as a host and is somewhat weaker than the Ptac promoter (1, 2). On the other hand, the Plac promoter system has an effective gratuitous inducer (IPTG [isopropyl-β-d-thiogalactopyranoside]) that can diffuse across the cell membrane without the aid of the lactose transporter (4); it has been shown that use of the gratuitous inducer IPTG eliminates the all-or-none response for the Ptac and Ptrc promoters (20). However, IPTG is expensive (38), not digestible, and toxic (11). As such, IPTG contamination of recombinant protein products is undesirable (11).
Other expression systems available include the tetracycline-inducible promoter Ptet, the alkane-inducible promoter PalkB, and the phosphate-regulated promoters Pugp. Pugp appears to be 80% as efficient in promoting expression as Ptac and about seven times stronger Ptet (38). Ptet expression level is independent of the host strain background and is approximately the same as the lac promoter (36). The alkS gene and the alkB promoter (PaklB) of Pseudomonas oleovorans GPo1 were assembled into a convenient alkane response genetic expression cassette, which can produce the membrane component of alkane hydroxylase, AlkB, up to 10% of total cell protein (27, 31).
In Salmonella enterica serovar Typhimurium LT2, the prpRBCDE genes encode a transcriptional activator and four enzymes of the 2-methylcitrate (2-MC) cycle that permit growth on propionate as a sole carbon and energy source by catalyzing its conversion to pyruvate (14-17). Propionate, a membrane-permeable weak acid (6), is metabolized into 2-MC by enzymes expressed from chromosomal prpEC. 2-MC-activated PrpR binds to an enhancer-like element located at a distance 5′ to the prpBCDE promoter, contacts the σ54-dependent RNA polymerase by means of DNA loop formation, and activates transcription of the prpBCDE operon. A closely related gene cluster was found in the E. coli genome (7). In a previous study, we showed that E. coli prpBCDE expression was propionate inducible (23).
In the present study, the prpBCDE promoter (PprpB) has been incorporated into a plasmid vector for expression of foreign genes. We compared the strength, basal expression, and induced expression from PprpB with PBAD and Ptrc using gfp to monitor gene expression.
MATERIALS AND METHODS
Bacterial strains and media.
The bacterial strains used in the present study are listed in Table 1. All DNA manipulations were performed in E. coli DH10B by using established protocols (33). Strain JSB, a sbm-ygfD-ygfG-ygfH-ygfI deletion mutant of E. coli BL21(DE3), was constructed by a PCR-mediated gene disruption method (9). Cultures were grown in Luria-Bertani (LB) broth at 37°C. Cell growth was monitored as the optical density at a wavelength of 600 nm (OD600). Media were amended with arabinose, IPTG, or sodium propionate (pH 8.0) as indicated. The following antibiotics were used at the concentrations indicated: ampicillin, 100 μg/ml; chloramphenicol, 30 μg/ml; and kanamycin, 50 μg/ml.
TABLE 1.
E. coli strains and plasmids used in this study
| E. coli strain or plasmid | Descriptiona | Reference or source |
|---|---|---|
| E. coli | ||
| DH10B | F′ mcrA Δ(mrr-hsdRMS mcrBC) φ80dlacZΔM15 ΔlacX74 deoR recA1 araΔ139 Δ(ara leu)7697 galU galK λ−rpsL endA1 nupG Strr | Life Technologies |
| BL21(DE3) | F′ ompT[lon] hsdSB(rB− mB−) gal dcm λDE3 | Novagen |
| JSB | BL21(DE3)/Δ(sbm-ygfDGHI) | This work |
| Plasmids | ||
| P70GL | pBAD24 carrying gfpuv and lacZ; Apr | 37 |
| pTrc99A-gfp | pTrc99A carrying gfpuv; Apr | This study |
| pTrc99A | trc promoter, lacIq, pBR322 ori; Apr | Amersham Pharmacia Biotech |
| pPro18 | 2-MC inducible, pBR322 ori; Apr | This study |
| pPro18-Cm | 2-MC inducible, pBR322 ori; Cmr | This study |
| pPro18-Kan | 2-MC inducible, pBR322 ori; Kmr | This study |
| pPro24 | 2-MC inducible, pBR322 ori; Apr | This study |
| pPro30 | 2-MC inducible, p15A ori; Apr | This study |
| pPro33 | 2-MC inducible, p15A ori; Cmr | This study |
| pPro24-gfp | pPro24 carrying gfpuv; Apr | This study |
| pPro33-gfp | pPro33 carrying gfpuv; Cmr | This study |
| pBAD18 | Arabinose inducible, pBR322 ori; Apr | 12 |
| pBAD24 | Arabinose inducible, pBR322 ori; Apr | 12 |
| pBAD18-Cm | Arabinose inducible, pBR322 ori; Cmr | 12 |
| pBAD18-Kan | Arabinose inducible, pBR322 ori; Kmr | 12 |
| pBAD30 | Arabinose inducible, p15A ori; Apr | 12 |
| pBAD33 | Arabinose inducible, p15A ori; Cmr | 12 |
| pBAD24-gfp | pBAD24 carrying gfpuv; Apr | This study |
Apr, ampicillin resistance; Cmr, chloramphenicol resistance; Kmr, kanamycin resistance; ori, replication origin.
Construction of pPro vectors.
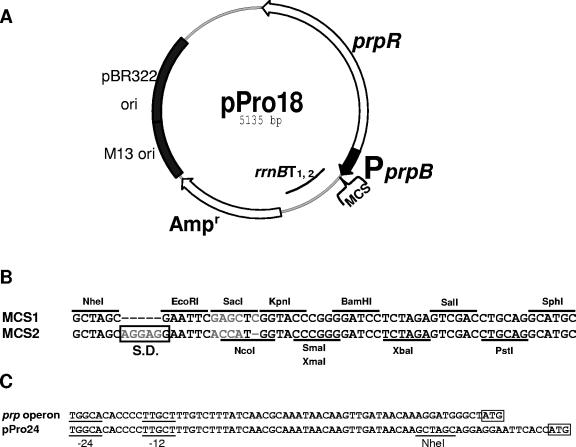
Constructed vectors differ in multicloning sites, antibiotic resistant gene, and copy number. The DNA containing PprpB and prpR was amplified from E. coli BL21(DE3) genomic DNA. To construct pPro vectors, araC and PBAD on the pBAD series of expression vectors were replaced with prpR and PprpB (Fig. 1).
FIG. 1.
Map of pPro18 as a representative of the pPro vectors. All pPro vectors have MCS1 (A) with the exception of pPro24, which has MCS2 (B). All restriction endonuclease sites are unique except for PstI in pPro18, pPro18-Cm, and pPro30; EcoRI in pPro18-Cm and pPro33; and SmaI and XmaI in pPro18-Kan. (C) The −12/−24 regions of the prpBCDE promoter and integrated NheI site are indicated. The ATG start sites for PrpB in prpBCDE or for cloned genes on pPro24 are boxed. Abbreviations: rrnBT1, 2, part of the strong ribosomal rrnB terminators; ori, origin of replication; S.D., Shine-Dalgarno box.
(i) pPro18.
The PCR product containing prpR and PprpB was digested with ClaI and NheI and ligated to the large fragments of pBAD18 resulting from digestion with the same enzymes, creating pPro18 (Fig. 1A).
(ii) pPro18-Cm.
The ClaI-NheI prpR-PprpB region from pPro18 was ligated to the large fragments of pBAD18-Cm resulting from digestion with the same enzymes, creating pPro18-Cm.
(iii) pPro18-Kan.
The Bst1107I-NheI prpR-PprpB region from pPro18 was ligated to the large fragments of pBAD18-Kan resulting from digestion with the same enzymes, creating pPro18-Kan.
(iv) pPro24.
The ClaI-NheI prpR-PprpB region from pPro18 was ligated to the large fragments of pBAD24 resulting from digestion with the same enzymes, creating pPro24 (Fig. 1C).
(v) pPro30.
After deletion of the NheI site in the front of pACYC origin region of pBAD30 by partial digestion with NheI, T4 DNA polymerase treatment, and self-ligation, the ClaI-NheI prpR-PprpB region from pPro18 was ligated to the large fragments of pBAD30 resulting from digestion with the same enzymes, creating pPro30.
(vi) pPro33.
After deletion of the NheI site in the front of pACYC origin region of pBAD33 by partial digestion with NheI, T4 DNA polymerase treatment, and self-ligation, the ClaI-NheI prpR-PprpB region from pPro18 was ligated to the large fragments of pBAD33 resulting from digestion with the same enzymes, creating pPro33.
Plasmid construction.
The reporter plasmids were made to test the utility of the pPro vectors for the regulated expression of genes. The promoter-reporter plasmids pBAD24-gfp, pPro24-gfp, and pTrc99A-gfp were constructed by subcloning the PCR-amplified gfpuv gene encoding the UV-excitable green fluorescent protein (GFP) from p70GL into the multiple cloning site (MCS) of pBAD24, pPro24, and pTrc99A, respectively. GFP was used to provide an indirect, quantitative measurement of the transcriptional properties of the cloned gene (10, 40).
Transcriptional fusion studies of PBAD, PprpB, or Ptrc promoters linked to the gfp reporter gene.
A seed culture was made by inoculating cells into LB medium containing ampicillin (100 μg/ml) and growing the cells overnight at 37°C. A total of 50 μl each of the seed cultures were inoculated into 5 ml of fresh LB medium supplemented with ampicillin (100 μg/ml). The cells were grown at 37°C and when the OD600 reached 0.5, the cells harboring pBAD24-gfp, pPro24-gfp, and pTrc99A-gfp were induced with arabinose, propionate, or IPTG, respectively.
GFP fluorescence in batch cultures of E. coli containing the reporter plasmids expressing gfp was measured in a Tecan SpectraFluor Plus plate reader (Tecan-US, Durham, NC) by using an excitation wavelength of 405 nm and an emission wavelength of 535 nm. GFP fluorescence was normalized for cell density (GFP fluorescence per OD600 unit). The GFP content of individual cells was determined as described previously (21) with a Beckman-Coulter EPICS XL flow cytometer (Beckman Instruments) equipped with an argon laser (emission at 488 nm/15 mV) and a 525-nm band-pass filter.
RESULTS
Characteristics and construction of vectors.
We have developed a prpBCDE promoter expression system (prpR-PprpB) that can be induced by using 2-MC. The system is composed of the prpBCDE promoter, PprpB, and a transcriptional activator gene, prpR, that activates the expression of cloned genes under the control of PprpB in the presence of 2-MC. The 2-MC is made from propionate via propionyl-coenzyme A (CoA) by using the chromosomally encoded prpCE gene products (14, 29, 30). Thus, the promoter was inactive in E. coli DH10B, which lacks the prp operon (23). In addition to 2-MC and PrpR, expression of the prpBCDE operon of enteric bacteria—which codes for 2-methylcitrate synthase, 2-methylcitrate dehydratase, 2-methylcitrate lyase (three key enzymes in the methylcitrate cycle), and propionyl-CoA synthetase—is dependent on the cyclic AMP-CRP complex, IHF, NtrA, and sigma-54-dependent RNA polymerase (5, 16, 23, 29, 30, 39). Although seemingly complex, these regulatory arrangements allow bacteria to produce enzymes for propionate catabolism only when they are needed, suggesting that PprpB must be inducible.
The pPro vectors contain prpR and PprpB, followed by an MCS and rrnB transcription terminators derived from the arabinose-inducible pBAD expression vectors (Fig. 1A). They also carry a pBR322 origin of replication (pPro18/18-Cm/ 18-Kan/24) or a p15A origin replication (pPro30/33), an M13 intragenic region for phage packaging and production of single-stranded DNA, and an antibiotic resistance gene (Fig. 1A). Only pPro24, which carries MCS2, contains an optimized Shine-Dalgarno site (SD) (34) and a translational start codon (ATG) at the NcoI site in order to easily clone genes that lack sequences for initiation of translation (Fig. 1B and C). The pPro30/33 vectors, which harbor the p15A origin of replication from pACYC184, can be used to reduce gene expression (due to lower copy number) and are compatible with pBR322-derived plasmids (pPro18/18-Cm/18-Kan/24) to stably coexpress different genes on separate plasmids in a single host. Since the pBAD30/33 vectors have two NheI sites, the NheI site located in front of the p15A origin of pPro30/33 was removed by partial digestion, T4 polymerase treatment, and self-ligation so that the NheI site in MCS1 would be available for cloning purposes.
Regulated expression of the PprpB-gfp gene.
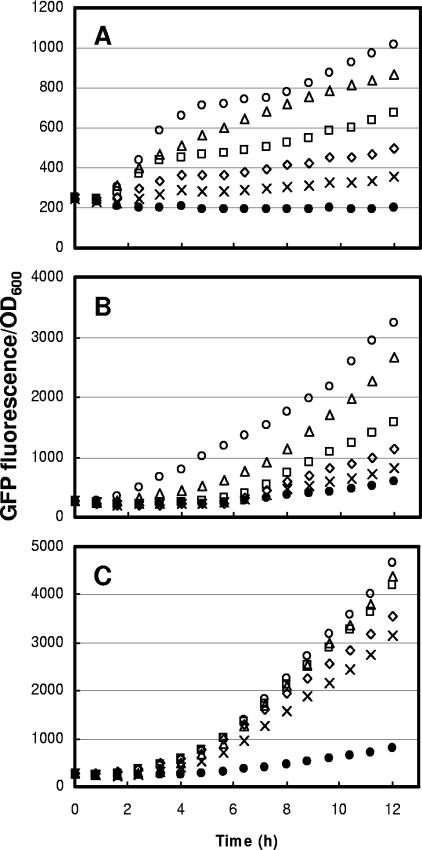
A useful property of a controllable expression system is that the expression level of a recombinant product is proportional to the amount of inducer added to the cell culture. To determine the range of inducibility, the gene encoding GFP (gfp) was placed under the control of PprpB and used as a reporter for promoter activity. We compared the expression levels of PprpB-gfp to those of PBAD-gfp and Ptrc-gfp in E. coli BL21(DE3) (Fig. 2). The prpR-PprpB system was regulated over a wide range of inducer concentrations, and production of GFP varied directly and linearly with the extracellular propionate concentration (Fig. 2B). In a similar manner, the expression of gfp under the control of PBAD was regulatable with arabinose concentration in the medium (Fig. 2A). However, as described above, it has been reported that the variation in expression level with inducer concentration is due a variation in the percentage of induced cells in the population rather than a variation in the expression level in individual cells (see reference 22 and discussion below). Although it does not suffer from all-or-none gene expression, the Ptrc system did not show high dose-dependent inducibility with the addition of IPTG (Fig. 2C). The reason for this highly cooperative induction is that IPTG induces its own transporter (24). It has been reported that this problem is alleviated by deletion of the lac permease (19).
FIG. 2.
Comparison of culture-average fluorescence (fluorescence per OD600 unit) of E. coli harboring the pBAD24-gfp (A), pPro24-gfp (B), or pTrc99A-gfp (C). Overnight-grown cells carrying the plasmid in LB at 37°C were subcultured (1:100) into fresh LB medium (5 ml) with ampicillin (100 μg/ml), grown at 37°C in a shaking incubator until the OD600 reached ca. 0.5, and then exposed to different concentrations of inducer in 96-well plates at 37°C with shaking in a Tecan SpectraFluor Plus plate reader. The data are raw: e.g., the background fluorescence intensity was not removed by using background subtraction. Symbols indicate inducer concentrations. (A) Open circles, 20 mM arabinose; open triangles, 5 mM; open rectangles, 1.25 mM; open diamonds, 0.31 mM; crosses, 0.08 mM; solid circles, 0 mM. (B) Open circles, 50 mM propionate; open triangles, 12.6 mM; open rectangles, 3.2 mM; open diamonds, 0.8 mM; crosses, 0.2 mM; solid circles, 0 mM. (C) Open circles, 1 mM IPTG; open triangles, 0.25 mM; open rectangles, 0.063 mM; open diamonds, 0.016 mM; crosses, 0.004 mM; solid circles, 0 mM.
In terms of the dynamic response to inducer addition, the PBAD system has been reported to have a very fast rate of induction (12). In contrast, the Ptrc system exhibited a time delay before expression of GFP. Like the PBAD system the prpR-PprpB expression system showed significant GFP fluorescence after about 90 min of induction upon addition of 50 mM propionate.
In the absence of inducer, PBAD showed much lower basal expression throughout the induction period than PprpB and Ptrc, with PprpB having slightly less background expression than Ptrc. However, the expression levels between Ptrc and PBAD or PprpB cannot be compared directly because they have different sequences at the ribosome-binding site (RBS). Compared to the PBAD-based vectors with the same RBS sequences, PprpB had much higher levels of induced expression. The amount of background expression of all three promoters seems to be correlated with the maximal expression level.
Modulation of PprpB-gfp expression in individual cells.
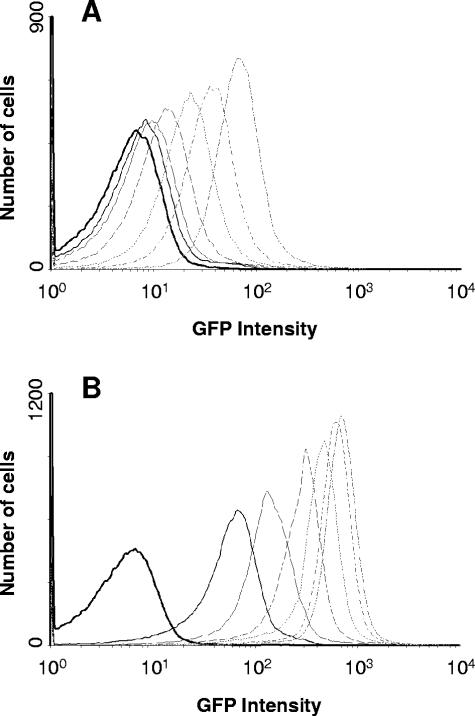
The ability to obtain different levels of expression by partial induction of the promoter is an important feature of a controllable expression system. Flow cytometry was used to examine modulation of PprpB-gfp by measuring the extent of induction from PprpB in single cells at different concentrations of propionate. The results indicate that all cultures were uniformly induced across the population at all propionate concentrations tested, and the level of gene expression in individual cells varied with the propionate concentration (Fig. 3). These results, taken together with the above data, suggest that the variation in population-average expression from PprpB as a function of propionate concentration resulted from partial induction of the PprpB promoter rather than an all-or-none response.
FIG. 3.
Histograms showing the number of cells with a given fluorescence in E. coli BL21(DE3) cultures harboring pPro24-gfp induced with the different concentrations of propionate. Cells grown overnight in LB medium at 37°C were subcultured (1:100) into fresh LB medium (5 ml) with ampicillin (100 μg/ml), grown at 37°C in a shaking incubator until the OD600 reached ca. 0.5, and then exposed to different concentrations of propionate. E. coli harboring pBAD24 empty vector was used as a control. The fluorescence in single cells was determined two (A) and six (B) hours after addition of propionate. Different lines indicate different propionate concentrations added to the medium, from left to right, represent control (heavy solid line), 0 mM (medium solid line), 0.2 mM (light solid line), 0.8 mM (dashed line), 3.2 mM (dotted line), 12.6 mM (− - −), and 50 mM (- - - -).
Basal expression of prpR-PprpB.
In general, a tightly regulated system is desirable because many recombinant products can be toxic to the expression host. This propionate-inducible expression system was found to be slightly “leaky”; background expression of gfp was detected in the absence of exogenous inducer (Fig. 2B and 3B). The basal expression levels depended on the carbon source (acetate, citrate, lactate, galactose, glutamate, and succinate) in minimal medium (data not shown), suggesting that propionate, propionyl-CoA, or 2-MC can be produced from endogenous metabolic pathways. One such pathway, encoded by the E. coli operon sbm-ygfD-ygfG-ygfH, converts succinate/succinyl-CoA to propionate/propionyl-CoA (13). To test the possibility that this pathway produced propionate/propionyl-CoA and increased background expression in the absence of exogenous inducer, a sbm-ygfD-ygfG-ygfH-ygfI deletion was introduced into E. coli BL21(DE3) creating strain JSB. Basal expression of PprpB-gfp in JSB was significantly lower than that obtained with BL21(DE3); however, the induced expression level decreased as well (Fig. 4). Based on this finding, it is not clear whether propionate/propionyl-CoA produced from succinate/succinyl-CoA by the sbm-ygfD- ygfG-ygfH-ygfI gene products contributed to the background. Like PBAD (12), PprpB showed higher background and higher induced expression in minimal medium than in rich medium (data not shown), possibly due to the presence of effector molecules that could induce catabolite repression or that could inhibit expression in some other ways (12).
FIG. 4.
Comparison of PprpB-gfp expression in BL21(DE3) (A) and JSB (B). Cells harboring various plasmids were grown overnight in LB medium at 37°C and subcultured (1:100) in fresh LB medium (5 ml) containing ampicillin (100 μg/ml) and without or with propionate (Prop) and glucose (Glc). The cells were grown at 37°C with shaking. After 15 h of cultivation, the GFP expression level was determined. Strain BL21(DE3) or JSB harboring the pBAD24 empty vector was used as a control. The background fluorescence intensity was not removed by using background subtraction. Error bars show the standard deviation of experiments performed in triplicate.
Basal expression from plasmid pPro33-gfp was generally lower than that from plasmid pPro24-gfp (Fig. 4). This lower basal expression should be a reflection of the decreased plasmid copy number. In E. coli JSB carrying pPro33-gfp, basal expression was similar to background levels observed in JSB harboring the pBAD24 empty vector. PprpB exhibited low basal expression level in the absence of inducer and similar expression level in the presence of inducer, compared to Ptrc (Fig. 2B and C). Since the prpR and prpBCDE promoters are subject to catabolite repression (23), glucose can be used to reduce the background expression to negligible level (Fig. 4).
DISCUSSION
The ability to vary the expression of a gene product at will and to make subtle changes in gene expression levels is important for optimizing metabolic pathways and for examining the role of a particular gene or protein. Regulation also facilitates the study of genes encoding products whose constitutive expression would be detrimental or lethal to cells (21, 22, 32). We have demonstrated that the E. coli propionate-regulated gene expression system can be used to control gene expression in E. coli. The most important features of this system are that it can be used to (i) induce gene expression by using a simple and cost-effective inducer, (ii) obtain high levels of expression in the presence of inducer, and (iii) regulate expression at the single cell level over a wide range of inducer concentrations.
Varying the level of gene expression in individual cells of the culture is necessary for the more subtle approaches of metabolic optimization and determining dose dependence of genes or proteins. However, several available E. coli expression systems suffer from all-or-none gene expression and thus are not regulatable in individual cells (28, 35). These carbohydrate-responsive systems have evolved this all-or-none method of gene expression to allow low-level expression in the absence of the substrate and rapid, high-level response when the substrate is present (22). The all-or-none or autocatalytic phenomenon arises because the transporter for the inducer is under control of the inducer itself (8). Upon exposure to an inducer, the inducer is transported into cell by some minimal number of transporters in the membrane. If a threshold level of inducer accumulates inside the cell, the transporter gene and any other associated genes are induced. The production of more transporter protein and the subsequent import of more inducer rapidly cascades to maximal gene expression. The all-or-none phenomenon can be alleviated by placing the transporter gene under control of an inducer-independent promoter or using a synthetic inducer analogue that does not require active transport across the cell membrane (21, 22). In contrast, bacterial cells are permeable to propionate (6). The propionate is metabolized to 2-MC by PrpEC expressed from the chromosome. Thus, this propionate-inducible expression system can achieve regulatable and consistent induction in all cells of the culture.
Genes whose products severely affect the host cell's growth rate even at low levels are considered toxic and are difficult to maintain in the host. Therefore, cells with an expression defect are selected for in the culture. In these situations, it is highly desirable to use a system that can be efficiently repressed in the absence of inducer. The PBAD promoter system has been used extensively to control and probe cellular processes because the system has a tightly regulated promoter showing a relatively low level of background expression in the absence of arabinose (25, 26). However, as shown in Fig. 2, this system is somewhat weaker in expression than PprpB and Ptrc. The low basal expression from the PBAD system seems to be a result of the weaker PBAD promoter. In contrast, the stronger PprpB and Ptrc showed higher basal expression. Since protein synthesis depends on translational efficiency as well as promoter strength, background expression may be reduced by using a weaker RBS sequence (3) or decreasing the strength of the promoter by introducing nucleotide changes in the consensus promoter sequence or by variations in the spacer sequence (18). However, it is likely that the induced expression level will decrease as well.
In conclusion, we have created a propionate-inducible expression system that shows tight regulation of gene expression in the absence of inducer, regulatable expression at intermediate inducer levels, high maximal expression, and consistent expression in all cells of the culture. This expression system should find wide application in examining the role of genes in physiology and also for production of protein and metabolite products using recombinant cells.
Acknowledgments
This study was supported by a grant from the National Institutes of Health (GM070763-01).
We thank J. D. Newman for helpful discussions.
REFERENCES
- 1.Amann, E., J. Brosius, and M. Ptashne. 1983. Vectors bearing a hybrid trp-lac promoter useful for regulated expression of cloned genes in Escherichia coli. Gene 25:167-178. [DOI] [PubMed] [Google Scholar]
- 2.Baneyx, F. 1999. Recombinant protein expression in Escherichia coli. Curr. Opin. Biotechnol. 10:411-421. [DOI] [PubMed] [Google Scholar]
- 3.Barrick, D., K. Villanueba, J. Childs, R. Kalil, T. D. Schneider, C. E. Lawrence, L. Gold, and G. D. Stormo. 1994. Quantitative analysis of ribosome binding sites in Escherichia coli. Nucleic Acids Res. 22:1287-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beckwith, J. R. 1978. lac: the genetic system, p. 11-30. In W. S. Reznikoff (ed.), The operon. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 5.Blank, L., J. Green, and J. R. Guest. 2002. AcnC of Escherichia coli is a 2-methylcitrate dehydratase (PrpD) that can use citrate and isocitrate as substrates. Microbiology 148:133-146. [DOI] [PubMed] [Google Scholar]
- 6.Blankenhorn, D., J. Phillips, and J. L. Slonczewski. 1999. Acid- and base-induced proteins during aerobic and anaerobic growth of Escherichia coli revealed by two-dimensional gel electrophoresis. J. Bacteriol. 181:2209-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 8.Carrier, T. A., and J. D. Keasling. 1999. Investigating autocatalytic gene expression systems through mechanistic modeling. J. Theor. Biol. 201:25-36. [DOI] [PubMed] [Google Scholar]
- 9.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeLisa, M. P., J. Li, G. Rao, W. A. Weigand, and W. E. Bentley. 1999. Monitoring GFP-operon fusion protein expression during high cell density cultivation of Escherichia coli using an on-line optical sensor. Biotechnol. Bioeng. 65:54-64. [PubMed] [Google Scholar]
- 11.Figge, J., C. Wright, C. J. Collins, T. M. Roberts, and D. M. Livingston. 1988. Stringent regulation of stably integrated chloramphenicol acetyl transferase genes by E. coli lac repressor in monkey cells. Cell 52:713-722. [DOI] [PubMed] [Google Scholar]
- 12.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haller, T., T. Buckel, J. Retey, and J. A. Gerlt. 2000. Discovering new enzymes and metabolic pathways: conversion of succinate to propionate by Escherichia coli. Biochemistry 39:4622-4629. [DOI] [PubMed] [Google Scholar]
- 14.Horswill, A. R., and J. C. Escalante-Semerena. 1997. Propionate catabolism in Salmonella typhimurium LT2: two divergently transcribed units comprise the prp locus at 8.5 centisomes, prpR encodes a member of the sigma-54 family of activators, and the prpBCDE genes constitute an operon. J. Bacteriol. 179:928-940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horswill, A. R., and J. C. Escalante-Semerena. 1999. Salmonella typhimurium LT2 catabolizes propionate via the 2-methylcitric acid cycle. J. Bacteriol. 181:5615-5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horswill, A. R., and J. C. Escalante-Semerena. 1999. The prpE gene of Salmonella typhimurium LT2 encodes propionyl-CoA synthetase. Microbiology 145:1381-1388. [DOI] [PubMed] [Google Scholar]
- 17.Horswill, A. R., and J. C. Escalante-Semerena. 2001. In vitro conversion of propionate to pyruvate by Salmonella enterica enzymes: 2-methylcitrate dehydratase (PrpD) and aconitase enzymes catalyze the conversion of 2-methylcitrate to 2-methylisocitrate. Biochemistry 40:4703-4713. [DOI] [PubMed] [Google Scholar]
- 18.Jensen, P. R., and K. Hammer. 1998. The sequence of spacers between the consensus sequences modulates the strength of prokaryotic promoters. Appl. Environ. Microbiol. 64:82-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jensen, P. R., H. V. Westerhoff, and O. Michelsen. 1993. The use of lac-type promoters in control analysis. Eur. J. Biochem. 211:181-191. [DOI] [PubMed] [Google Scholar]
- 20.Khlebnikov, A., and J. D. Keasling. 2002. Effect of lacY expression on homogeneity of induction from the Ptac and Ptrc promoters by natural and synthetic inducers. Biotechnol. Prog. 18:672-674. [DOI] [PubMed] [Google Scholar]
- 21.Khlebnikov, A., K. A. Datsenko, T. Skaug, B. L. Wanner, and J. D. Keasling. 2001. Homogeneous expression of the PBAD promoter in Escherichia coli by constitutive expression of the low-affinity high-capacity AraE transporter. Microbiology 147:3241-3247. [DOI] [PubMed] [Google Scholar]
- 22.Khlebnikov, A., O. Risa, T. Skaug, T. A. Carrier, and J. D. Keasling. 2000. Regulatable arabinose-inducible gene expression system with consistent control in all cells of a culture. J. Bacteriol. 182:7029-7034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee, S. K., J. D. Newman, and J. D. Keasling. 2005. Catabolite repression of the propionate catabolic genes in Escherichia coli and Salmonella enterica: evidence for involvement of the cyclic AMP receptor protein. J. Bacteriol. 187:2793-2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Magasanik, B. 1972. The lactose operon. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Mertens, N., E. Remaut, and W. Fiers. 1995. Tight transcriptional control mechanism ensures stable high-level expression from T7 promoter-based expression plasmids. Biotechnology 13:175-179. [DOI] [PubMed] [Google Scholar]
- 26.Newman, J. R., and C. Fuqua. 1999. Broad-host-range expression vectors that carry the l-arabinose-inducible Escherichia coli araBAD promoter and the araC regulator. Gene 227:197-203. [DOI] [PubMed] [Google Scholar]
- 27.Nieboer, M., J. Kingma, and B. Witholt. 1993. The alkane oxidation system of Pseudomonas oleovorans: induction of the alk genes in Escherichia coli W3110 (pGEc47) affects membrane biogenesis and results in overexpression of alkane hydroxylase in a distinct cytoplasmic membrane subfraction. Mol. Microbiol. 8:1039-1051. [DOI] [PubMed] [Google Scholar]
- 28.Novick, A., and M. Weiner. 1957. Enzyme induction as an all-or-none phenomenon. Proc. Natl. Acad. Sci. USA 43:553-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palacios, S., and J. C. Escalante-Semerena. 2000. prpR, ntrA, and ihf functions are required for expression of the prpBCDE operon, encoding enzymes that catabolize propionate in Salmonella enterica serovar typhimurium LT2. J. Bacteriol. 182:905-910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palacios, S., and J. C. Escalante-Semerena. 2004. 2-Methylcitrate-dependent activation of the propionate catabolic operon (prpBCDE) of Salmonella enterica by the PrpR protein. Microbiology 150:3877-3887. [DOI] [PubMed] [Google Scholar]
- 31.Panke, S., A. Meyer, C. M. Huber, B. Witholt, and M. G. Wubbolts. 1999. An alkane-responsive expression system for the production of fine chemicals. Appl. Environ. Microbiol. 65:2324-2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pollock, R., R. Issner, K. Zoller, S. Natesan, V. M. Rivera, and T. Clackson. 2000. Delivery of a stringent dimerizer-regulated gene expression system in a single retroviral vector. Proc. Natl. Acad. Sci. USA 97:13221-13226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sambrook, J., and D. W. Russel. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 34.Shine, J., and L. Dalgarno. 1974. The 3′-terminal sequence of Escherichia coli 16S rRNA: complementary to nonsense triplets and ribosome binding sites. Proc. Natl. Acad. Sci. USA 71:1342-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Siegele, D. A., and J. C. Hu. 1997. Gene expression from plasmids containing the araBAD promoter at subsaturating inducer concentrations represents mixed populations. Proc. Natl. Acad. Sci. USA 94:8168-8172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Skerra, A. 1994. Use of the tetracycline promoter for the tightly regulated production of a murine antibody fragment in Escherichia coli. Gene 151:131-135. [DOI] [PubMed] [Google Scholar]
- 37.Smolke, C. D., T. A. Carrier, and J. D. Keasling. 2000. Coordinated, differential expression of two genes through directed mRNA cleavage and stabilization by secondary structures. Appl. Environ. Microbiol. 66:5399-5405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Su, T. Z., H. Schweizer, and D. L. Oxender. 1990. A novel phosphate-regulated expression vector in Escherichia coli. Gene 90:129-133. [DOI] [PubMed] [Google Scholar]
- 39.Textor, S., V. F. Wendisch, A. A. De Graaf, U. Müller, M. I. Linder, D. Linder, and W. Buckel. 1997. Propionate oxidation in Escherichia coli: evidence for operation of a methylcitrate cycle in bacteria. Arch. Microbiol. 168:428-436. [DOI] [PubMed] [Google Scholar]
- 40.Zhang, Q., T. R. Tiersch, and R. K. Cooper. 1998. Inducible expression of green fluorescent protein within channel catfish cells by a cecropin gene promoter. Gene 216:207-213. [DOI] [PubMed] [Google Scholar]