Abstract
In order to get insights into the feedback regulation by tyrosine of the Escherichia coli chorismate mutase/prephenate dehydrogenase (CM/PDH), which is encoded by the tyrA gene, feedback-inhibition-resistant (fbr) mutants were generated by error-prone PCR. The tyrAfbr mutants were selected by virtue of their resistance toward m-fluoro-d,l-tyrosine, and seven representatives were characterized on the biochemical as well as on the molecular level. The PDH activities of the purified His6-tagged TyrA proteins exhibited up to 35% of the enzyme activity of TyrAWT, but tyrosine did not inhibit the mutant PDH activities. On the other hand, CM activities of the TyrAfbr mutants were similar to those of the TyrAWT protein. Analyses of the DNA sequences of the tyrA genes revealed that tyrAfbr contained amino acid substitutions either at Tyr263 or at residues 354 to 357, indicating that these two sites are involved in the feedback inhibition by tyrosine.
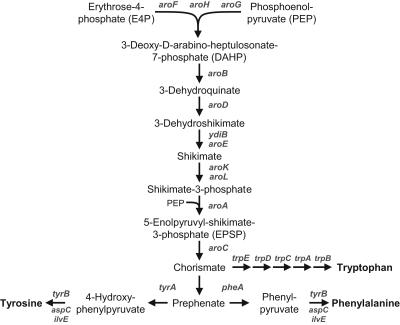
Chorismate is a central intermediate of the shikimate pathway and branch point for five different metabolic pathways in microorganisms (10). The aromatic amino acids l-phenylalanine and l-tyrosine are formed from chorismate via prephenate, which undergoes either decarboxylation/dehydration or decarboxylation/dehydrogenation, followed by a transamination to form the respective amino acids (Fig. 1).
FIG. 1.
Biosynthesis pathway of aromatic amino acids in E. coli.
Chorismate mutase/prephenate dehydrogenase (CM/PDH, TyrA) is a bifunctional enzyme occurring as a homodimer with a molecular weight of approximately 78,000 and is involved in tyrosine biosynthesis in Escherichia coli (13). The rearrangement of chorismate to prephenate is catalyzed by the N-terminal CM domain of TyrA, whereas the C-terminal PDH domain catalyzes the oxidative decarboxylation of prephenate to 4-hydroxyphenylpyruvate. Analogously, the PDT domain of the bifunctional CM/PDT (PheA) yields phenylpyruvate, which is subsequently transaminated to phenylalanine.
Aromatic amino acid biosynthesis is tightly regulated, in particular, at the chorismate branch point and at the first committed step, i.e., at 3-deoxy-d-arabino-heptulosonate-7-phosphate (DAHP) synthase, which catalyzes the condensation of phosphoenolpyruvate and erythrose-4-phosphate. The DAHP synthase occurs as three isoforms, each feedback regulated by either phenylalanine, tyrosine, or tryptophan (20). Furthermore, the TyrR protein, binding any one of the three aromatic amino acids, provides transcriptional control of eight different promoters related to aromatic amino acid biosynthesis (21).
Due to the industrial importance of l-phenylalanine, for example, as a precursor for the sweetener aspartame (l-aspartyl-l-phenylalanine methyl ester), much more effort has been devoted to the investigation of the phenylalanine biosynthesis pathway than to the tyrosine branch (1). For the biotechnological production of phenylalanine, engineered E. coli strains were employed, exhibiting, among other characteristics, alleviated feedback inhibition by the end product (23, 24). As a consequence, different feedback-inhibition-resistant (fbr) mutants of PheA have been characterized and three different domains of PheA were identified: the CM domain (residues 1 to 109), the PDT domain (residues 101 to 285), and the C-terminal regulatory domain (residues 286 to 386) with a spatially distinct allosteric site (19, 22, 27, 28).
In contrast to PheA, no tyrosine-resistant mutants of TyrA have been described so far and the feedback inhibition mechanism of this enzyme is still unknown. It was suggested that tyrosine acts as a competitive inhibitor with respect to prephenate (7) whereas other studies indicated the presence of a distinct allosteric site (25). Binding studies exhibited an increased affinity of TyrA toward tyrosine if NAD+ is present and vice versa, resulting in the formation of an inactive tetramer (12). Recently, a functional CM domain (residues 1 to 88) and PDH domain (residue 94 to 373) were identified by analyzing different TyrA fragments. Interestingly, no regulatory domain could be found and even minor deletions of the C terminus resulted in a complete loss of PDH activity (3).
In this study, we generated and characterized different mutated TyrA proteins in order to understand the feedback inhibition mechanism of the E. coli CM/PDH.
MATERIALS AND METHODS
Bacterial strains and cultivation conditions.
Escherichia coli K-12 (MG1655), E. coli DH5α (Invitrogen), and E. coli BL21(DE3) (Novagen) were used in this study. Cultivations were done at 37°C in Luria-Bertani (LB) or morpholinepropanesulfonic acid (MOPS)-buffered minimal medium (18). For maintenance of plasmids, 20 μg/ml kanamycin was added.
Isolation, manipulation and transfer of DNA.
Plasmid DNA was isolated using the QIAprep Spin Miniprep kit (QIAGEN). Chromosomal DNA from E. coli K-12 was prepared by using the Wizard genomic DNA purification kit (Promega). Agarose gel purification of DNA fragments was done with the Geneclean spin kit (Q-Biogene). Restriction enzymes, ligases, and other DNA-manipulating enzymes were used according to the manufacturer's manual. Plasmid DNA was transferred to chemically competent cells of E. coli DH5α (Invitrogen) and E. coli BL21(DE3) (Novagen), respectively.
Amplification and cloning of tyrAWT.
The wild-type tyrA gene was amplified by PCR from chromosomal DNA of E. coli K-12 using the following primers: tyrA_fw_KpnI (5′-CCG GTA CCA TGG TTG CTG AAT TGA CCG CAT TAC −3′) and tyrA_rev_MluI (5′-CCA CGC GTT TAT TAC TGG CGA TTG TCA TTC GCC-3′). After gel purification and digestion with KpnI and MluI, tyrA was cloned into pZE21-MCS1 (17) via the respective restriction sites, resulting in plasmid pZE21::tyrAWT.
Error-prone PCR and selection of feedback-inhibition-resistant tyrA mutants.
Nucleotide analogue mutagenesis was carried out in the presence of 2 and 20 μM 8-oxo-2′-deoxyguanosine (8-oxo-dGTP) and 6-(2-deoxy-β-d-ribofuranosyl)-3,4-dihydro-8H-pyrimido-(4,5-c)(1,2)oxazin-7-one (dPTP) (26). Using the plasmid pZE21::tyrAWT as template, 10, 20, and 30 amplification cycles with the primers mentioned above were performed using Taq DNA polymerase (New England Biolabs). The 1.1-kbp PCR products were gel purified, and the mutated tyrA genes were amplified in a second PCR under regular conditions. Subsequently, the gel-purified DNA fragments were pooled, digested with KpnI and MluI, ligated into pZE21-MCS1, and transformed to highly competent E. coli DH5α cells (Invitrogen). Putative tyrAfbr mutants were selected on minimal medium agar plates with 20 μg/ml kanamycin and 2 mM m-fluoro-d,l-tyrosine.
Subcloning of tyrA and DNA sequencing.
Putative tyrAfbr genes were amplified by PCR using the following primers: tyrA_LIC_fw (5′-GGT ATT GAG GGT CGC ATG GTT GCT GAA TTG ACC GCA TTA C-3′) and tyrA_LIC_rv (5′-AGA GGA GAG TTA GAG CCT TAT TAC TGG CGA TTG TCA TTC GCC-3′). After gel purification, the PCR products were subcloned into pET-30 Xa/LIC (Novagen) by using ligation-independent cloning (LIC; Novagen) and transformed E. coli DH5α (Invitrogen). The resulting plasmids, pET30::tyrAfbr, were analyzed by DNA sequencing with the following primers: T7_prom (5′-TAA TAC GAC TCA CTA TAG GG-3′), T7_term (5′-GCT AGT TAT TGC TCA GCG G-3′), tyrA_291fw (5′-ACT GCG TCC GGT GGT TAT CG-3′), and tyrA_913rv (5′-GGC GAA GAG AGC GCC AGA AG-3′).
Expression and purification of TyrA.
For high expression levels of the tyrAfbr genes, the respective pET30::tyrAfbr plasmids were transformed to E. coli BL21 (DE3)-competent cells (Novagen). The cells were cultivated in LB medium plus 20 μg/ml kanamycin and 1 mM isopropyl-β-d-thiogalactoside (IPTG). After reaching an optical density at 600 nm of ∼1, the cells were collected by centrifugation, resuspended in binding buffer (20 mM Tris-HCl-0.5 M NaCl-5 mM imidazole, pH 7.9), and disrupted by sonication with a Branson Sonifier 450. The cell extract was centrifuged at 10,000 × g for 15 min, and the supernatant was filtered through a 0.45-μm syringe filter (PALL Gelman Laboratory). The native His6-tagged TyrA protein was purified by precharged His-Bind column chromatography according to the manufacturer's protocol (Novagen). The eluted protein solution was desalted in Econo-Pac10DG columns (Bio-Rad) and concentrated by using CentriprepYM10 centrifugal ultrafiltration devices (Millipore). Expression and purification steps were controlled by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) (16). Protein concentrations were assayed according to the method of Bradford (2).
PDH activity measurement in crude cell extracts.
Cells of E. coli DH5α harboring different pZE21::tyrAfbr derivatives and pZE21::tyrAWT, respectively, were cultivated in 100 ml LB medium plus 20 μg/ml kanamycin for 8 h. The cells were harvested, washed with 50 mM Tris-HCl buffer, pH 8.0, and resuspended in 1.5 ml buffer. After sonication, the disrupted cells were centrifuged at 10,000 ×g for 10 min and the supernatant containing crude cell extract was collected. This NADH-independent PDH assay is based on the formation of the borate complex of 4-hydroxyphenylpyruvate, which has a strong absorption at 330 nm (9). Next, 10 μl of crude extract was added to 490 μl of 2 mM NAD+ and 2 mM prephenate in buffer (50 mM Tris-1 mM EDTA-1 mM dithioerythritol, pH 8.0) and incubated at 37°C for 30 min. The reaction was stopped by the addition of 100 μl of 15% trichloroacetic acid, the samples were chilled on ice for 5 min, and the precipitated proteins were separated by centrifugation for 5 min at 12,000 × g. Each 200 μl of the supernatant was added to (i) 1 ml of 1 M boric acid in 2 M sodium arsenate, pH 6.5 (sample), and (ii) 1 ml of 2 M sodium arsenate, pH 6.5 (blank). After incubation for 10 min at room temperature, the absorbance was measured at 330 nm.
PDH activity measurement of purified TyrA proteins.
PDH activities of purified TyrA proteins were determined spectrophotometrically according to NADH formation (8). A mixture of 1 ml of 0.2 mM prephenate, 2 mM NAD+, 0.1 mg/ml bovine serum albumin, and 10 mM 2-mercaptoethanol in 50 mM Tris-HCl buffer with 1 mM EDTA, pH 8.0, was preheated for 5 min at 37°C. The reaction was started by the addition of approximately 0.1 μg of enzyme which corresponded to 5 to 20 μl of enzyme solution, and the absorbance at 340 nm was followed for 2 min. All PDH activity measurements were done at least in duplicate.
CM activity of purified TyrA proteins.
CM activities were determined spectrophotometrically based on the formation of phenylpyruvate by treatment with HCl (8). The reaction mixture contained 1 mM chorismate, 0.1 mg/ml bovine serum albumin, and 10 mM 2-mercaptoethanol in 50 mM Tris-HCl buffer with 1 mM EDTA, pH 8.0. For the CM activity assay, 0.4 ml of this solution was preheated for 5 min at 37°C in a water bath. After an addition of 5 to 20 μl of enzyme solution, the reaction was incubated at 37°C for 5 min. For the conversion of chorismate to phenylpyruvate, 0.4 ml of 1 M HCl was added and the mixture was further incubated at 37°C for 10 min. The samples were alkalized with 1 ml of 2.5 M NaOH, and the absorbance was measured at 320 nm against a blank sample without enzyme. All CM activity measurements were done in triplicate.
Chemicals.
Chorismate, prephenate, 4-hydroxyphenylpyruvate, and m-fluoro-d,l-tyrosine were obtained from Sigma. Other chemicals, biochemicals, and enzymes were obtained from VWR International, Novagen, Teknova, Invitrogen, Bio-Rad, New England Biolabs, or Sigma.
RESULTS
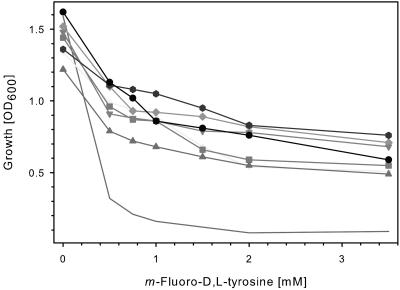
Since it is unknown whether tyrosine acts as a competitive or allosteric inhibitor on the TyrA protein, mutants of the tyrA gene from E. coli K-12 were generated by error-prone PCR and cloned into pZE21-MCS1. After transformation into E. coli DH5α, those strains were selected which were capable of growing in the presence of ≥2 mM m-fluoro-d,l-tyrosine, a tyrosine analogue which inhibits growth of E. coli DH5α harboring plasmid pZE21::tyrAWT. From 621 tyrAfbr mutants obtained, seven mutants were chosen for detailed analyses. Figure 2 shows that the tyrAfbr-containing plasmids enabled growth of the E. coli DH5α host strains in the presence of m-fluoro-d,l-tyrosine, in contrast to the E. coli DH5α strain expressing the tyrAWT gene. Growth of E. coli DH5α without plasmid was inhibited at concentrations of ≥0.1 mM m-fluoro-d,l-tyrosine, and addition of equimolar amounts of l-tyrosine restored growth.
FIG. 2.
Growth of E. coli DH5α strains expressing tyrAfbr genes in the presence of m-fluoro-d,l-tyrosine. Eight strains of E. coli DH5α harboring different plasmids were monitored: —, pZE21::tyrAWT; +, pZE21::tyrAfbr-5; ▪, pZE21::tyrAfbr-7; ▴, pZE21::tyrAfbr-10; ⧫, pZE21::tyrAfbr-13; ▾, pZE21::tyrAfbr-15; , pZE21::tyrAfbr-18; •, pZE21::tyrAfbr-20. The cells were cultivated in MOPS-buffered minimal medium containing m-fluoro-d,l-tyrosine. After incubation at 37°C at 225 rpm on a rotary shaker for 20 h, the optical density at 600 nm (OD600) was measured.
The effect of tyrosine on the mutated TyrA proteins of 20 different E. coli DH5α strains expressing putative tyrAfbr genes was studied by measuring the PDH activities in crude extracts. The PDH activity of E. coli DH5α pZE21::tyrAWT was reduced from 72.4 U/g to 37.6 U/g when 1 mM tyrosine was added to the enzyme assay. In contrast, tyrosine did not affect PDH activities of E. coli DH5α pZE21::tyrAfbr, which were in the range between 40.7 and 84.6 U/g. However, the use of crude extracts in this assay did not reveal a complete inhibition by tyrosine, whereas possible interactions of tyrosine with other proteins or compounds remain unknown.
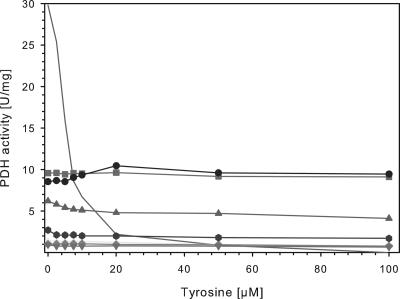
Seven different tyrAfbr and the tyrAWT genes were subcloned into vector pET-30 Xa/LIC, which included an N-terminal His6 tag for one-step purification of the respective fusion proteins. The resulting pET30::tyrAfbr plasmids as well as pET30::tyrAWT were expressed in E. coli BL21(DE3) after induction with 1 mM IPTG. The His6-tagged TyrA proteins were purified to homogeneity and used for NADH-dependent enzyme assays. The specific PDH activity of TyrAWT contributed 29.8 U/mg and decreased significantly in the presence of tyrosine, whereas less than 10% activity was detected at 20 μM tyrosine (Fig. 3). In contrast, all TyrAfbr mutants showed lower activities in the absence of tyrosine, but tyrosine concentrations of up to 100 μM did not significantly decrease the PDH activities. Whereas four mutants exhibited less than 10% activity as compared to TyrAWT, the mutants TyrAmut-7, TyrAmut-10 and TyrAmut-20 showed 21 to 35% of the wild-type PDH activity. Interestingly, the PDH activity of TyrAmut-20 slightly increased with increasing tyrosine concentrations of up to 20 μM (Fig. 3).
FIG. 3.
Effect of increasing tyrosine concentrations on PDH activities of purified TyrAWT and TyrAfbr mutants. The specific PDH activities of eight purified TyrA proteins were measured at different tyrosine concentrations. —, TyrAWT; +, TyrAfbr-5; ▪, TyrAfbr-7; ▴, TyrAfbr-10; ⧫, TyrAfbr-13; ▾, TyrAfbr-15; , TyrAfbr-18; •, TyrAfbr-20.
Regarding the CM activities of the purified TyrA proteins, TyrAmut-7 and TyrAmut-20 showed slightly higher specific acitivities, whereas all other mutants exhibited lower CM activities (Table 1). If 100 μM tyrosine and NAD+ were added, all TyrA proteins revealed reduced CM activity in the range of 43 to 96%, indicating that the effect of tyrosine on the TyrAfbr mutants is similar with TyrAWT.
TABLE 1.
PDH and CM activities of purified TyrAfbr mutants and TyrAWTa
| PDH activity (U/mg) |
CM activity (U/mg) |
|||
|---|---|---|---|---|
| 0 μM tyrosine | 100 μM tyrosine | 0 μM tyrosine | 100 μM tyrosineb | |
| TyrAWT | 29.80 | ND | 1.32 | 1.23 |
| TyrAmut-5 | 1.40 | 0.81 | 0.03 | 0.03 |
| TyrAmut-7 | 9.56 | 9.12 | 1.84 | 0.79 |
| TyrAmut-10 | 6.15 | 4.07 | 0.24 | 0.18 |
| TyrAmut-13 | 1.00 | 0.76 | 0.53 | 0.47 |
| TyrAmut-15 | 0.93 | 0.66 | 0.52 | 0.37 |
| TyrAmut-18 | 2.69 | 1.70 | 1.07 | 0.69 |
| TyrAmut-20 | 8.55 | 9.46 | 1.56 | 1.50 |
The specific enzyme activities of purified TyrA proteins were determined in the presence and absence of 100 μM tyrosine. ND, not detectable.
In the presence of 100 μM NAD+.
All tyrA mutants have been sequenced, and Fig. 4 shows the amino acid sequence alignment of the tyrAfbr mutants with tyrAWT. The mutants revealed one to four amino acid exchanges in the C-terminal PDH domain, whereas none, one or two amino acids were substituted in the N-terminal CM domain. Interestingly, one of two loci in the PDH domain, Tyr263 or residues 354 to 357, was mutated in all analyzed mutants, with an F357L substitution being the most frequently occurring mutation.
FIG. 4.
Comparison of the amino acid sequences of seven tyrAfbr mutants with tyrAWT of E. coli K-12. The deduced amino acid sequences of the generated tyrAfbr mutants (5, 7, 10, 13, 15, 18, 20) are shown in the one-letter code in comparison to the wild-type tyrA gene of E. coli K-12 (WT). Amino acid substitutions of the mutants are shaded.
DISCUSSION
The regulation of aromatic amino acid biosynthesis in E. coli and other bacteria is rather well understood. In particular the feedback inhibition of DAHP synthase (AroG, AroF) and CM/PDT (PheA) has been studied by generation and characterization of fbr mutants (4, 11, 14, 15, 19, 23). Those mutants were of particular interest due to the overproduction of l-phenylalanine by engineered bacteria (1). However, the regulation of TyrA had attracted less interest, possibly due to the lower industrial impact. In this study, we generated mutants of the E. coli tyrA gene by error-prone PCR and selected putative tyrAfbr mutants due to their resistance towards m-fluoro-d,l-tyrosine. Although the growth rate of E. coli expressing tyrAfbr was reduced, it could be well distinguished from that of the strain expressing the wild-type tyrA gene, which was not able to grow in the presence of ≥1 mM m-fluoro-d,l-tyrosine (Fig. 2).
The relation between the ability to grow in the presence of m-fluoro-d,l-tyrosine and the alleviation of the feedback inhibition by tyrosine was confirmed by the biochemical characterization of seven representatives of tyrAfbr mutants. The His6-tagged fusion proteins of TyrA were purified to homogeneity and subjected to PDH and CM activity measurements. Although all TyrAfbr showed reduced PDH activities in the absence of tyrosine, addition of tyrosine did not inhibit the enzyme activity, in contrast to TyrAWT (Fig. 3). Hence, all seven TyrAfbr proteins are feedback-resistant for tyrosine, which was demonstrated for the first time.
In contrast to the PDH activities, the CM activities of the TyrAfbr mutants were reduced in the presence of tyrosine and NAD+, exhibiting similar sensitivity to the CM of TyrAWT (Table 1). This is in agreement with previous studies, where the CM activity was found to be reduced to approximately 60%, but not further (12). Thus, the feedback regulation of the tyrAfbr mutants affects only the PDH and not the CM domain of TyrA.
Since data on the three-dimensional structure of the TyrA protein are not available, little is known about the enzymatic mechanism and in particular the regulation of this enzyme. It has been shown that His197 plays an important role in the catalytic activity of PDH, and Arg294 is essential for prephenate binding (5, 6). However, attempts to identify a discrete regulatory domain of TyrA have failed so far, nor have any amino acid residues involved in the binding of tyrosine been identified. Recent studies on the identification of domain substructures showed that deletion of C-terminal residues caused a complete loss of PDH activity, and therefore the authors suggested the absense of a discrete regulatory domain (3). The DNA sequences of the tyrAfbr genes generated in this study have been analyzed and compared to the tyrAWT gene (Fig. 4). The number of amino acid substitutions varied from two of tyrAmut-20 to six of tyrAmut-5, whereas TyrAmut-20 revealed the highest PDH activity, comprising 35% of the TyrAWT activity, and even a slightly increased CM activity (Table 1). Obviously, the residues 354 to 357 are involved in the inhibitory binding of tyrosine, because A354V and F357L substitutions alleviated feedback inhibition of the PDH domain. Although F357L was the most frequent mutation, Q355R substitutions were also identified in tyrAfbr mutants (data not shown). Those mutants which did not possess amino acid alterations at these residues revealed either a Y263H or a Y263C substitution (data not shown), strongly indicating that Tyr263 is also involved in the feedback inhibition mechanism. However, there were no obvious relations between the mutations at these two sites and the corresponding PDH activities, i.e., mutants with either amino acid exchange did not show generally higher or lower PDH activity. Furthermore, the effect of other mutations on enzyme activities remains unknown.
In conclusion, the characterization of tyrAfbr mutants confirmed that the regulatory site of TyrA is located in the PDH domain with residues 263 and 354 to 357 involved in the tyrosine binding. Due to the lower PDH activities of the TyrAfbr proteins as compared to TyrAWT, the feedback inhibition site seemed to be closely associated with the catalytic site of the C-terminal PDH domain, rather than a spatially distinct regulatory site.
Acknowledgments
We thank the DuPont-MIT Alliance for financial support of this work and the Deutsche Forschungsgemeinschaft (DFG) for providing the research fellowship LU 893/2-1.
REFERENCES
- 1.Bongaerts, J., M. Krämer, U. Müller, L. Raeven, and M. Wubbolts. 2001. Metabolic engineering for microbial production of aromatic amino acids and derived compounds. Metab. Eng. 3:289-300. [DOI] [PubMed] [Google Scholar]
- 2.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 3.Chen, S., S. Vincent, D. B. Wilson, and B. Ganem. 2003. Mapping of chorismate mutase and prephenate dehydrogenase domains in the Escherichia coli T-protein. Eur. J. Biochem. 270:757-763. [DOI] [PubMed] [Google Scholar]
- 4.Choi, Y. J., and D. E. Tribe. 1982. Continous production of phenylalanine using an Escherichia coli regulatory mutant. Biotechnol. Lett. 4:223-228. [Google Scholar]
- 5.Christendat, D., V. C. Saridakis, and J. L. Turnbull. 1998. Use of site-directed mutagenesis to identify residues specific for each reaction catalyzed by chorismate mutase-prephenate dehydrogenase from Escherichia coli. Biochemistry 37:15703-15712. [DOI] [PubMed] [Google Scholar]
- 6.Christendat, D., and J. L. Turnbull. 1999. Identifying groups involved in the binding of prephenate to prephenate dehydrogenase from Escherichia coli. Biochemistry 38:4782-4793. [DOI] [PubMed] [Google Scholar]
- 7.Christopherson, R. I. 1985. Chorismate mutase-prephenate dehydrogenase from Escherichia coli: cooperative effects and inhibition by L-tyrosine. Arch. Biochem. Biophys. 240:646-654. [DOI] [PubMed] [Google Scholar]
- 8.Davidson, B. E., and G. S. Hudson. 1987. Chorismate mutase-prephenate dehydrogenase from Escherichia coli. Methods Enzymol. 142:440-450. [DOI] [PubMed] [Google Scholar]
- 9.Dayan, J., and D. B. Sprinson. 1970. Determination of prephenate dehydrogenase activity. Methods Enzymol. 17:562-563. [Google Scholar]
- 10.Dosselaere, F., and J. Vanderleyden. 2001. A metabolic node in action: chorismate-utilizing enzymes in microorganisms. Crit. Rev. Microbiol. 27:75-131. [DOI] [PubMed] [Google Scholar]
- 11.Hu, C., P. Jiang, J. Xu, Y. Wu, and W. Huang. 2003. Mutation analysis of the feedback inhibition site of phenylalanine-sensitive 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase of Escherichia coli. J. Basic Microbiol. 43:399-406. [DOI] [PubMed] [Google Scholar]
- 12.Hudson, G. S., G. J. Howlett, and B. E. Davidson. 1983. The binding of tyrosine and NAD+ to chorismate mutase/prephenate dehydrogenase from Escherichia coli K12 and the effects of these ligands on the activity and self-association of the enzyme. Analysis in terms of a model. J. Biol. Chem. 258:3114-3120. [PubMed] [Google Scholar]
- 13.Hudson, G. S., V. Wong, and B. E. Davidson. 1984. Chorismate mutase/prephenate dehydrogenase from Escherichia coli K12: purification, characterization, and identification of a reactive cysteine. Biochemistry 23:6240-6249. [DOI] [PubMed] [Google Scholar]
- 14.Jossek, R., J. Bongaerts, and G. A. Sprenger. 2001. Characterization of a new feedback-resistant 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase AroF of Escherichia coli. FEMS Microbiol. Lett. 202:145-148. [DOI] [PubMed] [Google Scholar]
- 15.Kikuchi, Y., K. Tsujimoto, and O. Kurahashi. 1997. Mutational analysis of the feedback sites of phenylalanine-sensitive 3-deoxy-d-arabino-heptulosonate-7-phosphate synthase of Escherichia coli. Appl. Environ. Microbiol. 63:761-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 17.Lutz, R., and H. Bujard. 1997. Independent and tight regulation of transcriptional units in Escherichia coli via the lacR/O, the tetR/O, and araC/I1-I2 regulatory elements. Nucleic Acid Res. 25:1203-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neidhardt, F. C., P. L. Bloch, and D. F. Smith. 1974. Culture medium for enterobacteria. J. Bacteriol. 119:736-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nelms, J., R. M. Edwards, J. Warwick, and I. Fotheringham. 1992. Novel mutations in the pheA gene of Escherichia coli K-12 which result in highly feedback inhibition-resistant variants of chorismate mutase/prephenate dehydratase. Appl. Environ. Microbiol. 58:2592-2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pittard, A. J. 1996. Biosynthesis of aromatic amino acids, p. 458-484. In F. C. Neidhardt (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology, vol. 1. American Society for Microbiology, Washington, D.C. [Google Scholar]
- 21.Pittard, J., H. Camakaris, and J. Yang. 2005. The TyrR regulon. Mol. Microbiol. 55:16-26. [DOI] [PubMed] [Google Scholar]
- 22.Pohnert, G., S. Zhang, A. Husain, D. B. Wilson, and B. Ganem. 1999. Regulation of phenylalanine biosynthesis. Studies on the mechanism of phenylalanine binding and feedback inhibition in the Escherichia coli P-protein. Biochemistry 38:12212-12217. [DOI] [PubMed] [Google Scholar]
- 23.Sugimoto, S., M. Yabuta, N. Kato, T. Seki, T. Yoshida, and H. Taguchi. 1987. Hyperproduction of phenylalanine by Escherichia coli: application of a temperature-controllable expression vector carrying the repressor-promotor system of bacteriophage lambda. J. Biotechnol. 5:237-253. [Google Scholar]
- 24.Tribe, D. E. July1987. Novel microorganism and method. U.S. patent 4,681,852.
- 25.Turnbull, J., J. F. Morrison, and W. W. Cleland. 1991. Kinetic studies on chorismate mutase-prephenate dehydrogenase from Escherichia coli: models for feedback inhibition of prephenate dehydrogenase by L-tyrosine. Biochemistry 30:7783-7788. [DOI] [PubMed] [Google Scholar]
- 26.Zaccolo, M., D. M. Williams, D. M. Brown, and E. Gherardi. 1996. An approach to random mutagenesis of DNA using mixtures of triphosphate derivatives of nucleoside analogues. J. Mol. Biol. 255:589-603. [DOI] [PubMed] [Google Scholar]
- 27.Zhang, S., G. Pohnert, P. Kongasaeree, D. B. Wilson, J. Clardy, and B. Ganem. 1998. Chorismate mutase-prephenate dehydratase from Escherichia coli. Study of catalytic and regulatory domains using genetically engineered proteins. J. Biol. Chem. 273:6248-6253. [DOI] [PubMed] [Google Scholar]
- 28.Zhang, S., D. B. Wilson, and B. Ganem. 2000. Probing the catalytic mechanism of prephenate dehydratase by site-directed mutagenesis of the Escherichia coli P-protein dehydratase domain. Biochemistry 39:4722-4728. [DOI] [PubMed] [Google Scholar]