Abstract
Temperature-sensitive (TS) plasmids were generated through chemical mutagenesis of a derivative of the streptomycin resistance parent plasmid pD70, isolated from Mannheimia hemolytica serotype 1. Three TS plasmids which failed to replicate at or above 42°C in M. hemolytica but which were fully functional below 31°C were selected for further analysis. Two of the TS plasmids were shown by sequencing to possess unique single-base-pair mutations. The third TS plasmid contained a unique base pair substitution and a second mutation that had been previously identified. These mutations were clustered within a 200-bp region of the presumed plasmid origin of replication. Site-directed single-nucleotide substitutions were introduced into the wild-type pD70 origin of replication to confirm that mutations identified by sequencing had conferred thermoregulated replication. Deletion analysis on the wild-type pD70 plasmid replicon revealed that approximately 720 bp are necessary for plasmid maintenance. Replication of the TS plasmids was thermoregulated in Pasteurella multocida and Haemophilus somnus as well. To consistently transform H. somnus with TS plasmid, in vitro DNA methylation with commercially available HhaI methyltransferase was necessary to protect against the organism's restriction enzyme HsoI (recognition sequence 5′-GCGC-3′) characterized herein.
The family Pasteurellaceae (23) includes a number of pathogens that are the principal etiologic agents of many animal diseases. Some important species of this family that cause disease in cattle include Mannheimia hemolytica (10), Pasteurella multocida (5), and Haemophilus somnus (14). The genome sequence of P. multocida has been completed (22), and those of H. somnus and M. hemolytica are in progress; thus, our comprehensive knowledge of these organisms is rapidly expanding. The availability of genome sequences for these organisms will provide new insights for future areas of research; but to fully capitalize on this information, improved tools and systems for performing defined mutations in these organisms are needed.
With the development of transformation methods such as electroporation and conjugation, targeted mutants for M. hemolytica, P. multocida, and H. somnus have been constructed using suicide plasmids to generate allelic mutants (16, 27, 31). Also, the transposon elements Tn10 (20) and Tn916 (8, 11) were used to produce mutant libraries which to date have been limited to strains of P. multocida. Mutant generation by current methods is cumbersome and inefficient because the nonreplicative vectors used in their construction are rapidly destroyed by active restriction systems possessed by these organisms (4, 17, 26). Furthermore, despite the merits of these classes of mutants for investigating mechanisms of pathogenesis, insertion and transposon mutants carrying antibiotic markers can exert unintended effects on neighboring genes. These include polar termination of distal operon expression (19), altered regulation of adjacent genes through promotion associated with the insertion marker (2, 7), and production of fusion products (18). The inherent problems with insertion mutants can make assignment of the effect resulting from inactivation of the target gene equivocal. An additional drawback is that mutant strains carrying exogenous antibiotic resistance genes may be generally unsuitable for commercialization as vaccines.
Most of the problems described above, however, can be reduced or avoided through the application of temperature-sensitive (TS) replicons to produce unmarked in-frame deletion mutants which carry no exogenous DNA. A method reported previously by Hamilton et al. (13) details a straightforward, stepwise process for generating in-frame deletion mutants in Escherichia coli. This procedure involves introducing a TS plasmid carrying chromosomal sequences with an in-frame deletion along with a selectable antibiotic resistance marker into host cells. The transformed cells are propagated with antibiotic selection at the permissive temperature for plasmid replication. Cells are then transferred onto selective solid medium and incubated at a nonpermissive temperature for plasmid replication to obtain single-crossover mutants. To induce plasmid resolution and double-crossover mutant formation, single-crossover mutants are passed in nonselective broth at the permissive temperature for plasmid replication. Depending upon where the second crossover occurs, either wild-type or mutant products are produced. Others have modified this basic scheme for performing gene replacements in E. coli using a TS plasmid containing the sacB gene of Bacillus subtilis which provides positive selection for the loss of excised plasmid (3,21). A similar method for generating unmarked mutants of Saccharomyces cerevisiae, referred to as “pop in/pop out,” has been described previously (25).
The purpose of this study was to develop a TS shuttle plasmid(s) that is broadly applicable for generating unmarked mutants of Pasteurellaceae species. In this communication, we describe the mutagenesis, selection, and genetic characterization of three TS replicons that were derived from plasmid pD70 (4) of M. hemolytica. Deletion mapping of the origin of replication of the wild-type plasmid was performed to delineate those sequences necessary for plasmid maintenance. Derivative TS plasmids, convenient for engineering purposes, were constructed, and their performances were evaluated in strains of M. hemolytica, P. multocida, and H. somnus, which are known to cause respiratory disease in cattle. A restriction barrier preventing the consistent transformation of H. somnus was investigated, and a new restriction enzyme, HsoI, was characterized. In vitro methylation of plasmid by HhaI greatly improved transformation of H. somnus.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used are listed in Table 1.
TABLE 1.
Strains and plasmids used
| Strain or plasmid | Description | Source |
|---|---|---|
| Strains | ||
| E. coli DH10B | F−mcrA Δ(mrr-hsd RMS-mcrBC) φ80lacZΔM15 ΔlacX74 endA1 recA1 deoR Δ(ara-leu)7697 araD139 galU galK nupG rpsLλ | Gibco BRL |
| Mannheimia haemolytica serotype 1 NADC D153 | NADC (R. Briggs) | |
| Pasteurella multocida NADC TT94 | Bovine lung isolate A:3 | NADC (R. Briggs) |
| Haemophilus somnus | ||
| NADC HS 91 | Bovine lung isolate | NADC (R. Briggs) |
| Strain 2336 | Bovine lung isolate | UCSD (L. Corbeil) |
| Plasmids | ||
| pBluescript SK | Cloning vector (Ampr) | Stratagene |
| pD70 | 4.3-kb plasmid (Smr) | NADC (R. Briggs) |
| pD70KanR | 5.6-kb plasmid, Kanr gene in BamHI site of pD70 | This work |
| pCT109 T | TS derivative of pD70KanR | This work |
| pGA301 T | TS derivative of pD70KanR | This work |
| pCT109GA301 T | TS derivative of pD70KanR | This work |
| pD70oriKanR | 2.5-kb, 1.2-kb ori of pD70 + 1.6-kb Kanr | This work |
| pD70-5′Δ0oriKanR | bp 0 deleted in ori | This work |
| pD70-5′Δ76oriKanR | bp 34-110 deleted in ori | This work |
| pD70-3′Δ0oriKanR | bp 0 deleted in ori | This work |
| pD70-3′Δ100oriKanR | bp 1021-1119 deleted in ori | This work |
| pD70-3′Δ200oriKanR | bp 921-1119 deleted in ori | This work |
| pD70-3′Δ300oriKanR | bp 831-1119 deleted in ori | This work |
| pD70-3′Δ400oriKanR | bp 721-1119 deleted in ori | This work |
| pD70-3′Δ500oriKanR | bp 611-1119 deleted in ori | This work |
| pD70-3′Δ600oriKanR | bp 523-1119 deleted in ori | This work |
| pD70-3′Δ700oriKanR | bp 421-1119 deleted in ori | This work |
Plasmid construction and M. hemolytica transformation.
Plasmid pD70, an endogenous 4.25-kb plasmid of M. hemolytica (Fig. 1) which encodes streptomycin resistance, was linearized by HindIII digestion and made blunt by treatment with deoxynucleoside triphosphates and Klenow fragment. A Tn903 kanamycin resistance cassette (Geneblock; Pharmacia Biotech Inc., Piscataway, NJ) previously digested with BamHI (Gibco BRL, Gaithersburg, MD) and made blunt as described above was ligated into the treated site on pD70 to produce pD70KanR (Fig. 1). Plasmid pD70KanR was amplified in E. coli DH10B containing PhaI methyltransferase on a cosmid (4). Specifically methylated pD70KanR plasmid was introduced into M. hemolytica serotype 1 strain NADC-D153 by electroporation. Cells were made electrocompetent by growing them to logarithmic phase in 100 ml of Columbia broth (Difco Laboratories, Detroit, MI) at 37°C with gentle shaking. The cells were pelleted by centrifugation at 5,000 × g and washed in 100 ml of 272 mM sucrose at 0°C. The pellet was suspended in an equal volume of 272 mM sucrose at 0°C. Competent cells (100 μl) were placed into 0.1-cm electroporation cuvettes and mixed with 100 ng of the treated DNA. The cells were electroporated (Gene pulser; Bio-Rad, Richmond, CA) at 18,000 V/cm and 800 Ω, yielding time constants ranging from 11 to 12 ms. Immediately after electroporation, the cells were resuspended in 1.0 ml Columbia broth at 0°C. Recovery was performed for 2 h at 30°C. The suspension was spread (100 μl/plate) onto Columbia agar (Difco Laboratories) plates containing 50 μg/ml kanamycin.
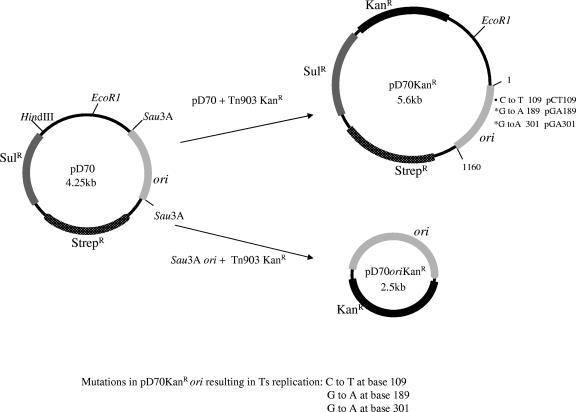
FIG. 1.
Plasmids used in this study. Plasmid pD70 is an endogenous plasmid of M. hemolytica. Plasmid pD70KanR was constructed by inserting the kanamycin resistance cassette into the modified HindIII site of pD70. Plasmid pD70oriKanr was constructed by digesting pD70 with Sau3A. The resulting 1,160-bp Sau3A fragment containing the origin of replication was joined to the Tn903 kanamycin resistance cassette. Ts, temperature sensitive.
Plasmid mutagenesis and selection of TS plasmids.
Plasmid DNA was recovered from kanamycin-resistant M. hemolytica by the sodium dodecyl sulfate alkaline lysis method and was CsCl purified as described elsewhere previously (1). One microgram of purified plasmid was mutagenized with hydroxylamine for 90 min at 70°C as described previously by Thomas (28). The treated plasmid DNA was dialyzed against TE (10 mM Tris, 1mM EDTA, pH 8.0), ethanol precipitated, and resuspended in TE. Plasmid DNA was introduced into M. hemolytica serotype 1 by electroporation as described above and, after recovery in broth cells, was spread onto Columbia agar plates containing 50 μg/ml kanamycin. Plates were incubated for 28 h at 30°C and then transferred to 42°C for 6 h. Colonies smaller than typical colonies were picked and dotted onto kanamycin plates. After overnight incubation at 30°C, growth from each selected colony was duplicated onto Columbia agar plates with and without kanamycin and then incubated overnight at 42°C. Growth from nonselective plates was transferred to kanamycin plates which were incubated overnight at 30°C. Clones which failed to grow with selection at 42°C but which grew well on selective medium at 30°C after passage without selection at 42°C were presumed to be temperature sensitive for kanamycin expression. Similar clones which failed to grow on selective plates at 30°C after passage without selection at 42°C were presumed to be temperature sensitive for plasmid maintenance. Three of the latter clones were selected for further study.
Sequencing of the TS mutations.
Origins of replication of parent and TS plasmids were amplified by PCR using the custom-synthesized primer pair pD70ori (Table 2). All primers described in this paper were custom synthesized with an oligonucleotide synthesizer (Applied Biosystems Inc., Integrated DNA Technologies, Inc., Coralville, IA). M. hemolytica cells carrying the TS plasmids were used as templates, and PCRs were done using the EasyStart PCR mix using a tube protocol of Molecular BioProducts (San Diego, CA). Reactions were performed for 30 cycles with 30 s at 95°C, 30 s annealing at 50°C using the pD70ori primer pair, and 60 s at 72°C per cycle. The PCR products were purified with QIAquick spin columns (QIAGEN Inc., Valencia, CA) and sequenced using the forward and reverse pD70ori primers (Fig. 2). Automated sequencing was performed with fluorescent terminators by cycle sequencing with an Applied Biosystems model 373 DNA sequencer (DNA facility at Iowa State University, Ames, IA). To minimize the possibility of PCR-introduced errors, each origin was amplified and sequenced independently two times using specific primers shown in Table 1.
TABLE 2.
Oligonucleotides used
| Oligonucleotide | Sequencea | Product |
|---|---|---|
| D70ori F | 5′-GATCCCTTTTTCTGTAATCTG-3′ | pD70ori (1,160 bp) |
| D70ori R | 5′-GATCATAGGCTCAATTCTCGC-3′ | |
| CT109 F | 5′-CCTACTACCAAAAAACCATTCAA-3′ | TS plasmid, pCT109PCR |
| CT109 R | 5′-AACGCTTCGCAATCTCTTG-3′ | |
| GA301 F | 5′-ATAAGCGAGATTTAAGGATAACAG-3′ | TS plasmid, pGA301PCR |
| GA301 R | 5′-TAGGTGTGCTTGTATTTCTTG-3′ | |
| 5′Δ0ori F | 5′-GAACGCACGAAACAGATTACAGA-3′ | pD70-5′Δ0oriKanR |
| 5′Δ0ori R | 5′-TTTGCTAAGATACAGACCCTAG-3′ | 0 bases deleted |
| 5′Δ0ori F | 5′-GAACGCACGAAACAGATTACAGA-3′ | pD70-5′Δ76oriKanR |
| 5′Δ76ori R | 5′-CTACTACCAAAAAACCATTCAAC-3′ | 34-110 bases deleted |
| 3′Δ0ori R | 5′-GAAAAAAATCTCAAAAAATTGCGAG-3′ | pD70-3′Δ0oriKanR |
| 3′Δ0ori F | 5′-AATTTCTCGCAAGGTCGCC-3′ | 0 bases deleted |
| 3′Δ0ori R | 5′-GAAAAAAATCTCAAAAAATTGCGAG-3′ | pD70-3′Δ100oriKanR |
| 3′Δ100ori F | 5′-CAGCTCGAACAGTAACG-3′ | 1,021-1,119 bases deleted |
| 3′Δ0ori R | 5′-GAAAAAAATCTCAAAAAATTGCGAG-3′ | pD70-3′Δ200oriKanR |
| 3′Δ200ori F | 5′-TTTCTCTCTAACAAGGCTTG-3′ | 921-1,119 bases deleted |
| 3′Δ0ori R | 5′-GAAAAAAATCTCAAAAAATTGCGAG-3′ | pD70-3′Δ300oriKanR |
| 3′Δ300ori F | 5′-TGACCGCTGAAAGCG-3′ | 831-1,119 bases deleted |
| 3′Δ0ori R | 5′-GAAAAAAATCTCAAAAAATTGCGAG-3′ | pD70-3′Δ400oriKanR |
| 3′Δ400ori F | 5′-ACACCAAAAGCCCCG-3′ | 721-1,119 bases deleted |
| 3′Δ0ori R | 5′-GAAAAAAATCTCAAAAAATTGCGAG-3′ | pD70-3′Δ500oriKanR |
| 3′Δ500ori F | 5′-TTATTATTAGGGAAGTGGTTATTG-3′ | 611-1,119 bases deleted |
| 3′Δ0ori R | 5′-GAAAAAAATCTCAAAAAATTGCGAG-3′ | pD70-3′Δ600oriKanR |
| 3′Δ600ori F | 5′-CATCAGCTTTTCAATGGCT-3′ | 523-1,119 bases deleted |
| 3′Δ0ori R | 5′-GAAAAAAATCTCAAAAAATTGCGAG-3′ | pD70-3′Δ700oriKanR |
| 3′Δ700ori F | 5′-TTTCTATTCGGTTCTCAAATGTT-3′ | 421-1,119 bases deleted |
Boldface indicates single-nucleotide substitutions for reverse engineering of temperature-sensitive origins of replication.
FIG. 2.
Sequence of the pD70 Sau3A fragment containing the origin of replication. Sites where nucleotide substitutions were found are shown in boldface type, repeats are underlined, a direct repeat within an inverted repeat is double underlined, and a small open reading frame is underlined in boldface.
Kinetics of TS plasmid loss in M. hemolytica.
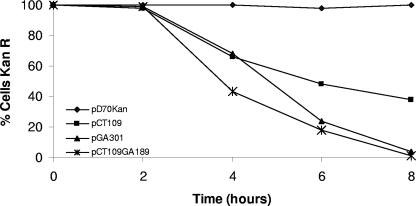
M. hemolytica cultures carrying pD70KanR or the putative TS plasmids pCT109, pGA301, or pCT109GA189 were grown overnight in Columbia broth containing 25 μg/ml kanamycin at 30°C. Five microliters from each culture grown overnight was transferred in duplicate to fresh 10-ml broth cultures without antibiotic (dilute antibiotic present from the inoculum). The duplicate cultures were incubated at 30°C and 41°C. At 0, 2, 4, 6, and 8 h, a loop of broth from each culture was streaked onto a Columbia agar plate without selection and then incubated at 30°C. One hundred colonies off each plate were picked in duplicate and transferred onto fresh Columbia agar plates either with or without kanamycin and then incubated at 30°C for 24 h. Plasmid loss was determined by the percentage of colonies unable to grow on the replica plates containing kanamycin (Fig. 3).
FIG. 3.
Kinetics of plasmid loss from M. hemolytica. Cells carrying plasmids pD70oriKanr, pCT109, pGA301, and pCT109GA189 were grown in broth without antibiotic at 30°C or 41°C. Samples were removed at the indicated times and spread onto nonselective Columbia agar plates and then grown at 30°C. One hundred colonies were transferred to Columbia plates (kanamycin, 25 μg/ml) and grown at 30°C to estimate the proportion of cells retaining plasmid.
Host range of the TS plasmids.
Previously, we had determined that the wild-type pD70KanR plasmid could be introduced into P. multocida and H. somnus by means of electroporation using conditions essentially the same as those used for M. hemolytica (4). The TS plasmids were electroporated into P. multocida TT94 and H. somnus HS91 using conditions identical to those used for M. hemolytica. Transformants were propagated with or without antibiotic selection at 30°C and 40°C to assess bacterial growth and plasmid yield under the varying conditions.
Generation of TS plasmids by site-specific PCR mutagenesis of wild-type plasmid.
Specific base pair mutations were introduced into the derivative plasmid pD70oriKanr (Fig. 1) by PCR. The derivative plasmid pD70oriKanr was constructed by ligating a 1,160-bp Sau3A fragment of the pD70 origin of replication with a Tn903 Kanr cassette possessing BamHI ends. The resistance cassette was obtained by BamHI digestion of pBluescript SK II Kanr. The ligation mixture was introduced into P. multocida by electroporation, and pD70oriKanr was isolated from kanamycin-resistant colonies by the sodium dodecyl sulfate alkaline method and CsCl purification. The point mutations of C to T at base 109 and G to A at base 301 were introduced into the template plasmid pD70oriKanr by PCR using the primer sets CT109 and GA301, respectively (Table 2). Degradation of template pD70oriKanr in the PCRs was accomplished by digestion with the restriction enzyme DpnI, whose substrate is dam-methylated DNA. Fragment ends of the PCR products were phosphorylated with polynucleotide kinase (GibcoBRL) and dATP according to recommendations of the manufacturer and then circularized with T4 ligase. Each of the PCR-mutagenized products was individually introduced into P. multocida by electroporation. After recovery, cells were spread onto Columbia agar plates containing 25 μg/ml kanamycin and then incubated at 30°C. Kanr colonies that arose were replica plated onto Columbia agar plates containing kanamycin and incubated at 30°C and 40°C. Plasmid origins of clones unable to grow at 40°C were amplified by PCR using the pD70ori primer sets (Table 2). The resultant PCR products were purified with QIAquick spin columns and sequenced using the D70ori forward primer.
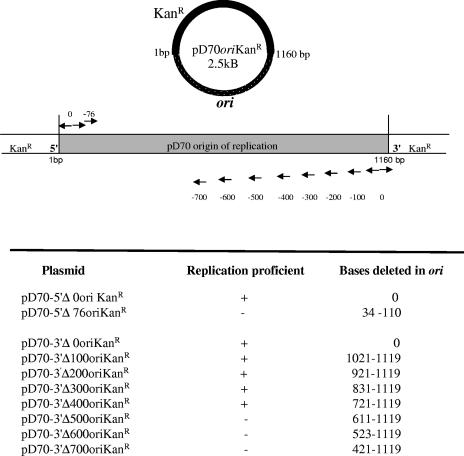
Deletion analysis of the pD70 origin of replication.
A series of sequential deletions was introduced into the origin of plasmid pD70oriKanr by PCR. The primer sets used to generate deletions in the 5′ end of the plasmid origin are seen in Table 2. The latter primer set produced a 76-bp deletion at nucleotides 34 to 110 in pD70oriKanr (Fig. 4). The primer sets creating the defined deletions in the 3′ end of the plasmid origin are shown in Table 2, and the extent of deletions are depicted in Fig. 4. As described above, template plasmid DNA was inactivated by digestion with DpnI, and all PCR-derived products were phosphorylated and then circularized by ligation. Each deletion plasmid was introduced into P. multocida by electroporation, and cells were spread onto Columbia plates containing 25 μg/ml kanamycin and then incubated at 37°C. Plasmid origins were amplified by PCR from Kanr cells with the D70ori primer set, and products were purified for sequencing as described above.
FIG. 4.
Diagram of the primer sets used to produce PCR products containing deletions within origin of replication of plasmid pD70oriKanr. Each PCR fragment was circularized and introduced into P. multocida. Plasmids were recovered from kanamycin-resistant colonies possessing deletions and sequenced.
Isolation of restriction endonuclease HsoI from H. somnus.
Haemophilus somnus strain 2336 (kindly supplied by Lynette Corbeil, San Diego, CA) was grown for 16 h on eight chocolate agar plates (Columbia blood agar base [Difco] supplemented with 5% defibrinated bovine blood at 90°C [200-ml total volume]). The cells were harvested in TE (10 mM Tris, 1 mM EDTA, pH 8.0), pelleted by centrifugation at 16,000 × g for 5 min at 4°C, and washed once in TE. The washed pellet was resuspended in 12 ml chromatography running buffer (20 mM sodium phosphate, 10 mM 2-mercaptoethanol, pH 8.0, 0°C) and placed on ice. The bacterial cells were disrupted by sonication for 2 min in 15-s bursts. Debris and unbroken cells were removed by centrifugation at 16,000 × g for 10 min, and the supernatant was filtered through a 0.45-μm-pore-size membrane (Millex-HA; Millipore Corp., Bedford, MA). No further treatment of the crude extract was performed prior to chromatography.
All chromatographic procedures were performed at room temperature. Prepacked heparin-Sepharose columns (Econo-pac heparin columns; Bio-Rad) were equilibrated as recommended by the manufacturer. A flow rate of 1.0 ml/min was used for separation by using a gradient low-pressure automated chromatography system (Automated Econo-System; Bio-Rad). Five milliliters of crude extract was injected, and a linear gradient from 0 to 1.0 M NaCl in 60 ml of running buffer was used to elute proteins. Fractions (1 ml) were stored on ice prior to activity assay. A second identical chromatographic separation was performed with a new column from which active fractions were collected and pooled for storage. Aliquots (5 μl) of the chromatographic fractions were incubated with 1 μl of React 1 (Gibco BRL), 0.5 μl of unmethylated bacteriophage lambda DNA (0.5 μg/μl; New England Biolabs, Beverly, MA), and 35 μl water at 37°C for 2 h. After the addition of tracking dye and electrophoresis on a 1% agarose gel in Tris-borate-EDTA buffer, the banding patterns were visualized by ethidium bromide staining and UV illumination. The fractions corresponding with DNA cleavage activity were pooled from the second chromatographic separation, concentrated 20-fold on 30,000-molecular-weight-cutoff ultrafilters, and brought to final concentrations of 150 mM NaCl, 10 mM sodium phosphate, 0.1 mM EDTA, 5 mM 2-mercaptoethanol, 0.25 μg of bovine serum albumin per ml, and 50:50 (vol/vol) glycerol, pH 8.0, for storage at −20°C. The concentrated preparation was designated HsoI.
Determination of the sequence recognition site for HsoI.
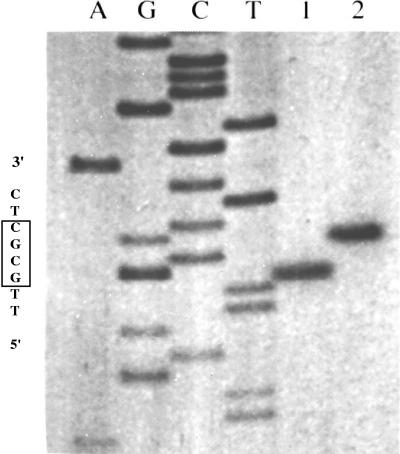
The recognition sequence of HsoI was identified by digestion of pBluescript SK (Stratagene Inc., La Jolla, CA) and of unmethylated bacteriophage lambda DNA (New England Biolabs) using conditions described above. The digestion products were analyzed by electrophoresis on an agarose gel as described above. The cleavage site was identified by digestion of a primed-synthesis reaction on pBluescript SK (Fig. 5). An oligonucleotide primer which is complementary with sequences 3′ from an HsoI site of pBluescript SK was synthesized. Single-stranded DNA was used for the template. Standard dideoxy DNA sequencing reactions were performed, and an additional reaction containing no dideoxy terminator was extended through the HsoI site with the Klenow fragment of DNA polymerase I by using 32P-end-labeled primer. The extension reaction was stopped by phenol-chloroform extraction followed by ethanol precipitation. HsoI or HhaI (New England Biolabs) was added to the end-labeled reaction and was allowed to digest the DNA for 2min. The digestions were stopped by the addition of gel loading buffer and heating to 80°C for 3 min.
FIG. 5.
Determination of the HsoI cleavage site alongside that of isoschisomer HhaI. The cleavage sites were determined by digestion of a primed-synthesis reaction containing the recognition site on pBluescript SK. A standard dideoxy DNA sequencing reaction done with SK primer is shown in the left four lanes. An additional reaction containing no dideoxy terminator was extended through the recognition site of the two restriction enzymes with the Klenow fragment by using 32P-end-labeled SK primer. Lane 1, end-labeled double-stranded plasmid DNA cut with HsoI; lane 2, the same plasmid cut with HhaI.
In vitro methylation of plasmid DNA by HhaI methyltransferase and protection from HsoI digestion.
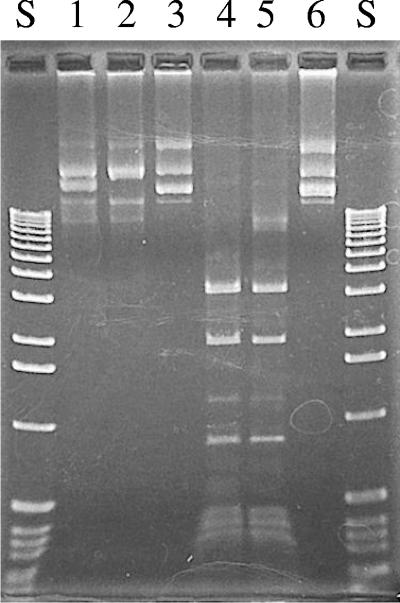
By using conditions recommended by the manufacturer, plasmid DNA (a hybrid replacement plasmid recovered from E. coli DH10B) was methylated in vitro by HhaI methyltransferase (New England Biolabs) using S-adenosylmethionine as the methyl donor. Following HhaI methyltransferase treatment, plasmid DNA was phenol-chloroform extracted, ethanol precipitated, and resuspended in TE. Both HhaI-methylated and untreated control plasmids were assessed for resistance to restriction enzyme digestion by HsoI or by HhaI (Fig. 6). Specifically methylated TS and parent plasmids were then used to transform H. somnus.
FIG. 6.
Protection of DNA against HsoI and HhaI cleavage by HhaI methyltransferase. Plasmid DNAs in lanes 1 to 3 were protected by in vitro methylation with HhaI methyltransferase. Plasmid DNAs in lanes 4 to 6 were not methylated. The plasmids in lanes 1 and 4 were digested with HhaI, the plasmids in lanes 2 and 5 were digested with HsoI, and DNAs in lanes 3 and 6 were not treated with restriction enzymes.
Nucleotide sequence accession number.
The sequence of parent plasmid pD70has been deposited in the GenBank database under accession number DQ125466.
RESULTS
Generation and properties of replication-conditional plasmids.
The plasmid used in the mutagenesis experiment is a derivative of pD70 (Fig. 1), an endogenous plasmid of M. hemolytica which encodes streptomycin resistance. Because colonies arising with streptomycin selection are not reliably transformed, the Tn903 kanamycin resistance marker was inserted into pD70 to produce pD70KanR (Fig. 1). Plasmid pD70KanR was specifically methylated by passing it through E. coli containing PhaI methyltransferase on a cosmid vector (4). Afterwards, 1 μg of pD70KanR was mutagenized in vitro by hydroxylamine (28). Plasmid DNA was dialyzed to eliminate the mutagenic agent and introduced into M. hemolytica by electroporation. M. hemolytica was transformed to kanamycin resistance by the mutagenized plasmid at an efficiency of approximately 6 × 104 CFU/μg DNA. Of the transformants, about 1% (360) formed atypically small colonies after incubation at 42°C. Passage of atypical colonies at either 30°C or 42°C on plates with or without kanamycin revealed that about 90% of the transformants were temperature sensitive for expression of kanamycin resistance (about 10% failed to grow on the first passage and were not tested further). These clones formed colonies on selective or nonselective plates at 30°C but failed to grow on selective plates at 42°C. Passage of clones from nonselective plates at 42°C to selective plates at 30°C resulted in heavy growth, indicating that plasmid was still present. A 5.5-kb plasmid was detected in plasmid preparations out of representatives of these cultures.
Ten of the 360 colonies behaved similar to the colonies containing temperature-sensitive kanamycin genes on passage, except that their growth was reduced on selective medium at 30°C after passage without selection at 42°C. These colonies appeared to vary in percent resistant to kanamycin after passage without selection at 42°C, indicating possible differences in their degrees of instability at a nonpermissive temperature (42°C). Three plasmids were selected for further characterization based on their low yield of kanamycin-resistant colonies after passage without selection at the nonpermissive temperature.
Sequencing of the TS mutations.
To identify a mutation(s) producing temperature-sensitive replication in the three plasmids chosen for further study, a 1,160-bp region that supports replication was amplified by PCR from each plasmid and sequenced (Fig. 2). This analysis revealed that the genetic lesions were clustered within a 200-bp area. The first plasmid contained a C-to-T substitution at base 109 and was designated pCT109, the second plasmid had a G-to-A substitution at base 301 and was designated pGA301, and the third plasmid had C-to-T and G-to-A substitutions at bases 109 and 189, respectively, and was designated pCT109GA189. The three substitutions identified correspond to the known mutagenic effect of hydroxylamine, which is a transition of G or C to A or T (28).
Kinetics of TS plasmid curing in M. hemolytica.
The three TS plasmids as well as the parent, pD70KanR, were maintained in M. hemolytica without significant loss (>99% retention for each) for 24 h at 30°C without selection. In contrast, at 41°C, the three TS plasmids were lost from M. hemolytica at varying rates while the parent plasmid was stably maintained (Fig. 3). After 8 h at 41°C, cells transformed with plasmid pCT109, pGA301, and pCT109GA189 were 35%, 4%, and >1% Kanr, respectively (Fig. 3). The single transition in pGA301 resulted in quicker curing at temperature than did pCT109. However, the additional mutation of G to A present in plasmid pCT109GA189 resulted in its more rapid curing than that of pGA301. The copy numbers of the TS plasmids and their parent plasmid were similar after 8 h at 30°C with or without antibiotic selection, as determined by double-stranded Miniprep analysis obtained from comparable numbers of cells. In contrast, after 8 h at 41°C without selection, TS plasmid levels were reduced or undetectable and corresponded with their rates of Kanr loss.
Host range of the TS plasmids.
The pD70KanR parent and the three TS plasmids were readily introduced into P. multocida strain TT94, albeit at lower levels than with M. hemolytica. Transformation efficiency of the wild-type plasmid into P. multocida was approximately 1 × 104 CFU/μg DNA, and that with the TS plasmid pGA301 was approximately 1 × 103 CFU/μg DNA. P. multocida transformed with wild-type pD70KanR grew well under selection at 40°C and yielded plasmid DNA from nonselective broth cultures at 40°C. The TS plasmids performed in P. multocida as they did in M. hemolytica, and no growth was observed on selective plates incubated at 40°C and no plasmid was detected in broth cultures grown without selection at 40°C. P. multocida yielded TS plasmid when grown with or without selection at 30°C. In contrast, transformation efficiency of wild-type pD70KanR into H. somnus was approximately 1 × 102 CFU/μg DNA, and transformation with the TS plasmid pGA301 was often unsuccessful. Transformants from specifically methylated plasmids are described in the following paragraphs.
Reverse engineering of the TS plasmids by PCR.
The substitutions of C to T at base 109 and G to A at base 301 were introduced separately in pD70oriKanr by PCR using the mismatched primer sets shown in Table 2. When introduced into P.multocida, both were thermoregulated and supported growth at 30°C on selective plates but did not support growth at 40°C. Sample origins of plasmids pCT109PCR and pGA301PCR were obtained by PCR and sequenced to confirm that the mutations had occurred as intended.
Deletion mapping of the origin of replication.
To better characterize the minimal replicating unit of the parent and TS plasmids, a series of increasingly larger deletions at the 5′ and 3′ ends of the origin of plasmid pD70oriKanr was produced by PCR using the various primer sets shown in Table 2. No transformants containing the approximately 80-bp deletion at the 5′ end of the 1,160-bp fragment were detected. Sequential deletions in the 3′ end of the fragment demonstrated that bp 721 to 1119 could be removed without elimination of plasmid replication (Fig. 4). No transformants containing larger deletions at the 3′ end were detected. The deleted origins of the functioning plasmids were sequenced to verify that the intended deletion had occurred.
Isolation and characterization of the restriction endonuclease HsoI from Haemophilus somnus.
Failure to consistently transform H. somnus with exogenous plasmid DNAs by electroporation suggested the existence of a restriction barrier. We investigated that possibility and discovered a new restriction enzyme, HsoI. The restriction enzyme was purified from H. somnus by standard chromatographic procedures. Chromatographic fractions exhibiting endonuclease activity were eluted from heparin-Sepharose columns by 680 to 760 mM NaCl (960 to 1,060 μS). A single pass through these columns was sufficient to identify both the recognition specificity and the cleavage site. Digestion of lambda DNA and pBluescript SK DNA with the concentrated HsoI preparation resulted in distinctive restriction fragment patterns consistent with the recognition sequence of 5′-…GCGC…-3′, which is identical to HhaI (Fig. 5), a commercially available restriction endonuclease isolated from Haemophilus haemolyticus (New England Biolabs). The cleavage site of HsoI (5′-…G↓CGC…-3′) produces 5′ overhangs and differs from HhaI, which cleaves 5′-…GCG↓C…-3′ to produce 3′ overhangs (Fig. 5).
In vitro methylation of plasmid DNA by HhaI methyltransferase protects against HsoI digestion and increases transformation efficiency.
In vitro HhaI-methylated plasmid DNA was protected against digestion by both HsoI and HhaI (Fig. 6). Methylated wild-type pD70KanR was electroporated into H.somnus at an efficiency of 1 × 106 cells per μg of plasmid. This treatment increased transformation efficiency by approximately 4 orders of magnitude. The methylated TS plasmids also were successfully introduced into H. somnus but at an approximately 100-fold lower efficiency than that of the parent plasmid. No plasmid was detected in broth cultures grown without selection at 40°C. When H. somnus was transformed with wild-type plasmid, however, colonies grew well under selection at 40°C and yielded plasmid DNA from nonselective broth cultures. All H. somnus transformants yielded plasmid when grown with or without selection at 30°C.
DISCUSSION
The streptomycin resistance plasmid pD70 of M. hemolytica can be used to construct plasmid derivatives that will replicate in a wide range of gram-negative organisms including E. coli (29). In this study, we report that in vitro mutagenesis of pD70 was successfully used to produce TS plasmids from M. hemolytica. Three plasmids were selected for further analysis, and their curing from M. hemolytica at or above 40°C was attributed to specific base pair mutations clustered within a 200-bp region in their origins of replication. The kinetics of plasmid loss at high temperature differed between the TS plasmids, ranging from moderate (pCT109) to intermediate (pGA301) to rapid (pCT109GA189). Potentially, this variation in replication by the TS plasmids may be tailored to suit specific applications. For example, pGA301, with an intermediate rate of plasmid curing, may be better for generating deletion mutants since its slightly more active origin may cause Campbell-like integrates to resolve via double crossover more rapidly. Plasmid pCT109GA189, with almost complete shutdown of replication at a high temperature, may be more suited for transposon introduction.
A potential problem with the broad application of this type of system is the inability of a TS plasmid to stably replicate in other species of interest. The TS plasmid pGA301 was stably maintained at a permissive temperature and cured at a high temperature in the three bacterial species we examined here.
Plasmid curing at a nonpermissive temperature is likely due to a defect in replication rather than failure of partitioning. The copy number of pD70 is estimated to be approximately 10 per cell (Sarah Highlander, personal communication). With the wild-type plasmid, an approximately 10% plasmid loss was observed after 100 generations without selection, consistent with random inheritance of plasmid with such a copy number (our unpublished observations). It is unlikely, therefore, that an effective partitioning system is acting on pD70. Loss kinetics of the temperature-sensitive derivatives, too, are far too rapid to be accounted for by the failure of partitioning alone. Furthermore, we found that plasmid integrated into chromosome was stable at a nonpermissive temperature for plasmid replication but was markedly unstable at a permissive temperature. This observation is consistent with temperature-regulated plasmid replication.
Deletion mapping of the wild-type origin indicated that approximately 720 bp are required for sustained replication of plasmid pD70 in P. multocida. This region shares >99% identity with the P. multocida pIGI replicon (30), the Haemophilus ducreyi pLS88 replicon (9), and the Actinobacillus pleuropneumoniae replicon pKMA2425 (GenBank accession number AJ830714); 97% identity with the M. hemolytica replicon pAB2 (6); and 91% identity with the Actinobacillus actinomycetemcomitans pVT745 replicon (12). The high degree of similarity indicates that there is a common ancestry between the family of plasmids found in the Haemophilus-Actinobacillus-Pasteurella group. Moreover, this close relationship is further underscored by the ability of the TS plasmid pGA301 to transform and replicate in a temperature-dependent fashion in the three species of bacteria studied herein.
It has been reported that this family of plasmids relies entirely on the proteins of the host bacterium for replication (9,30). There is a small open reading frame extending from nucleotides 88 to 204 of the 1,160-bp pD70 Sau3A fragment, within which are two of the three nucleotide substitutions associated with a temperature-conditional phenotype (Fig. 2). Evidence against this hypothetical protein's role in plasmid replication, however, is the much greater temperature sensitivity exhibited by pCT109GA189 than that exhibited by pCT109, despite the fact that the lesion at nucleotide 189 is predicted to be silent with respect to the protein.
Nucleotide sequence analysis of pIG1 (30) revealed that the origin of replication contains three inverted repeats, IR20, IR16, and IR38. Two inverted repeats and two direct repeats are present in the 720-bp functional origin of pD70 (Fig. 2). Interestingly, one direct repeat is nested within the larger of the two inverted repeats. How these features may be involved in plasmid replication is unknown.
In the course of this work, we discovered that H. somnus contains a restriction modification system, HsoI. Introduction of plasmid DNA into H. somnus was increased by approximately 4 orders of magnitude by in vitro methylation with HhaI methyltransferase, although the efficiency of transformation dropped as the size of the plasmid increased. It is possible that a second restriction system may be responsible for this decrease as plasmid size increased. The possibility of systems analogous to mcr (24) and mrr (15) in E. coli was not investigated. Also, partial rather than complete protection conferred by in vitro methylation could account for the reduction in efficiency seen with increasing plasmid size.
With the availability of genome sequences for many species of Pasteurellaceae, efficient methodologies to generate targeted gene disruptions in these organisms are needed to fully exploit this new wealth of information. Described in this work are three distinct TS plasmids and a TS derivative plasmid that is intended for the genetic engineering of unmarked deletion mutants in members of Pasteurellaceae.
Acknowledgments
This study was supported in part by grant 18-7-512 from the Biotechnology and Research Development Corporation.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. Greene Publishing Associates and Wiley Interscience, New York, N.Y.
- 2.Berg, D. E., A. Weiss, and L. Crossland. 1980. Polarity of Tn5 insertion mutations in Escherichia coli. J. Bacteriol. 142:439-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blomfield, I. C., V. Vaughn, R. F. Rest, and B. I. Eisenstein. 1991. Allelic exchange in Escherichia coli using the Bacillus subtilis sacB gene and a temperature-sensitive pSC101 replicon. Mol. Microbiol. 5:1447-1457. [DOI] [PubMed] [Google Scholar]
- 4.Briggs, R. E., F. M. Tatum, T. A. Casey, and G. H. Frank. 1994. Characterization of a restriction endonuclease, PhaI, from Pasteurella haemolytica serotype A1 and protection of heterologous DNA by a cloned PhaI methyltransferase gene. Appl. Environ. Microbiol. 60:2006-2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carter, G. R. 1967. Pasteurellosis: Pasteurella multocida and Pasteurella haemolytica. Adv. Vet. Sci. 11:321-338. [PubMed] [Google Scholar]
- 6.Chang, Y. F., D. P. Ma, H. Q. Bai, R. Young, D. K. Struck, S. J. Shin, and D. H. Lein. 1992. Characterization of plasmid with antimicrobial resistant genes in Pasteurella haemolytica A1. J. DNA Seq. Mapping 3:89-97. [DOI] [PubMed] [Google Scholar]
- 7.Ciampi M. S., M. B. Schmid, and J. R. Roth. 1982. Transposon Tn10 provides a promoter for transcription of adjacent sequences. Proc. Natl. Acad. Sci. USA 79:5016-5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeAngelis, P. L. 1998. Transposon Tn916 insertional mutagenesis of Pasteurella multocida and direct sequencing of disruption site. Microb. Pathog. 24:203-209. [DOI] [PubMed] [Google Scholar]
- 9.Dixon, L. G., W. L. Albritton, and P. J. Willson. 1994. An analysis of the complete nucleotide sequence of the Haemophilus ducreyi broad-host range plasmid, pSL88. Plasmid 32:228-232. [DOI] [PubMed] [Google Scholar]
- 10.Frank, G. H. 1989. Pasteurellosis of cattle, p. 197-222. In C. Adlam and J. M. Rutter (ed.), Pasteurella and pasteurellosis. Academic Press Ltd., London, United Kingdom.
- 11.Fuller, T. E., M. J. Kennedy, and D. E. Lowery. 2000. Identification of Pasteurella multocida virulence genes in a septicemic mouse model using signature-tagged mutagenesis. Microb. Pathog. 29:25-38. [DOI] [PubMed] [Google Scholar]
- 12.Galli, D. M., J. Chen, K. F. Novak, and D. J. LeBlanc. 2001. Nucleotide sequence and analysis of conjugative plasmid pV745. J. Bacteriol. 183: 1585-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hamilton, C. M., M. Aldea, B. K. Washburn, P. Babitzke, and S. R. Kushner. 1988. New method for generating deletions and gene replacements in Escherichia coli. J. Bacteriol. 171:4617-4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harris, W. F., and E. D. Janzen. 1989. The Haemophilus somnus disease complex (hemophilosis): a review. Can. Vet. J. 30:816-822. [PMC free article] [PubMed] [Google Scholar]
- 15.Heitman, J., and P. Model. 1987. Site-specific methylases induce the SOS DNA repair response in Escherichia coli. J. Bacteriol. 169:3243-3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Homchampa, P., R. A. Strugnell, and B. Adler. 1992. Molecular analysis of the aroA gene of Pasteurella multocida and vaccine potential of an aroA mutant. Mol. Microbiol. 23:3585-3594. [DOI] [PubMed] [Google Scholar]
- 17.Hoskins, I. C., and A. J. Lax. 1997. Identification of restriction barriers in Pasteurella multocida. FEMS Microbiol. Lett. 156:223-226. [DOI] [PubMed] [Google Scholar]
- 18.Ito, K., P. J. Bassford, Jr., and J. Beckwith. 1981. Protein localization in E. coli: is there a common step in the secretion of periplasmic and outer-membrane proteins? Cell 24:707-717. [DOI] [PubMed] [Google Scholar]
- 19.Kleckner, N. 1981. Transposable elements in prokaryotes. Annu. Rev. Genet. 15:341-404. [DOI] [PubMed] [Google Scholar]
- 20.Lee, M. D., and A. D. Henk. 1996. Tn10 insertional mutagenesis in Pasteurella multocida. Vet. Microbiol. 50:143-148. [DOI] [PubMed] [Google Scholar]
- 21.Link, A. J., D. Phillips, and G. M. Church. 1997. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J. Bacteriol. 17:6226-6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.May, B. J., Q. Zhang, L. L. Li, M. L. Paustian, T. S. Whittam, and V. Kapur. 2001. Complete genomic sequence of Pasteurella multocida, Pm70. Proc. Natl. Acad. Sci. USA 98:3460-3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pohl, S. 1981. DNA relatedness among members of Actinobacillus, Haemophilus and Pasteurella, p. 245-254. In M. Kilian, W. Frederiksen, and E. L. Biberstein (ed.), Haemophilus, Pasteurella and Actinobacillus. Academic Press, London, United Kingdom.
- 24.Raleigh, E. A., and G. Wilson. 1986. Escherichia coli K-12 restricts DNA containing 5-methylcytosine. Proc. Natl. Acad. Sci. USA 83:9070-9074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rothstein, R. 1991. Targeting, disruption, replacement, and allelic rescue: integrative DNA transformation in yeast. Methods Enzymol. 175:3303-3317. [DOI] [PubMed] [Google Scholar]
- 26.Sanders, J. D., Y. Tagawa, R. E. Briggs, and L. B. Corbeil. 1997. Transformation of a virulence associated gene of Haemophilus somnus into a strain lacking the gene. FEMS Microbiol. Lett. 154:251-258. [DOI] [PubMed] [Google Scholar]
- 27.Tatum, F. M., R. E. Briggs, and S. M. Halling. 1994. Molecular cloning and nucleotide sequencing and construction of an aroA mutant of Pasteurella haemolytica serotype A1. Appl. Environ. Microbiol. 60:2011-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thomas, C. M. 1987. Plasmid replication, p. 7-36. In K. G. Hardy (ed.), Plasmids: a practical approach. IRL Press, Oxford, United Kingdom.
- 29.Wood, A. R., F. Wright, G. D. Baird, and W. Donachie. 1995. A native plasmid of Pasteurella haemolytica serotype A1: DNA sequence analysis and investigation of its potential as a vector. Res. Vet. Sci. 58:163-168. [DOI] [PubMed] [Google Scholar]
- 30.Wright, C. L., R. A. Strugnell, and A. L. Hodgson. 1997. Characterization of a Pasteurella multocida plasmid and its use to express recombinant proteins in P. multocida. Plasmid 37:65-79. [DOI] [PubMed] [Google Scholar]
- 31.Wu, Y., J. H. McQuiston, A. Cox, T. D. Pack, and T. J. Inzana. 2000. Molecular cloning and mutagenesis of a DNA locus involved in lipooligosaccharide biosynthesis in Haemophilus somnus. Infect. Immun. 68:310-319. [DOI] [PMC free article] [PubMed] [Google Scholar]