Abstract
A temperature-sensitive (TS) plasmid was generated from the endogenous streptomycin resistance plasmid of Mannheimia hemolytica and used to engineer in-frame aroA deletion mutants of Mannheimia hemolytica, Pasteurella multocida, and Haemophilus somnus. TS replacement plasmids carrying in-frame aroA deletions were constructed for each target species and introduced into host cells by electroporation. After recovery in broth, cells were spread onto plates containing antibiotic and incubated at 30°C, the permissive temperature for autonomous plasmid replication. Transfer of transformants to selective plates cultured at a nonpermissive temperature for plasmid replication selected for single-crossover mutants consisting of replacement plasmids that had integrated into host chromosomes by homologous recombination. Transfer of the single-crossover mutants back to a permissive temperature without antibiotic selection drove plasmid resolution, and, depending on where plasmid excision occurred, either deletion mutants or wild-type cells were generated. The system used here represents a broadly applicable means for generating unmarked mutants of Pasteurellaceae species.
The leading cause of morbidity and mortality sustained by the cattle feedlot industry in North America is bovine respiratory disease (BRD). In the United States, Mannheimia hemolytica is the organism most commonly isolated from feedlot calves with pneumonia (12). Other etiologic agents causing substantial losses to both the dairy and beef cattle industries are Pasteurella multocida and Haemophilus somnus (8, 24). The pathogenesis of BRD is multifactorial, with nutrition, stress, and viral and bacterial infections acting in combination to cause clinical disease. It is typically the bacterial components, however, that cause the serious economic losses. Whereas these bacteria are relatively common inhabitants of the nasopharyngeal mucosal surfaces, under stressful conditions, they often spread to the lung to establish pneumonia and pleuritis.
While there has been considerable effort in the development and use of vaccines and therapeutic treatments to mitigate BRD, economic losses are estimated to exceed one billion dollars annually in the United States (15). Currently, most bacterial vaccines to control BRD in cattle are of the killed variety. However, live bacterial vaccines have been shown in many cases to be more effective at eliciting broad immune responses involving both cell- and antibody-mediated effectors. Also, live vaccines are often superior at stimulating mucosal immunity which may be crucial for controlling BRD. Other advantages of live vaccines include the expression of all relevant antigens including those proteins produced only in vivo upon proliferation, potential for delivery by parenteral or nonparenteral routes such as the oral or nasal route, and low production costs.
The development of molecular genetic techniques has enabled the construction of stable genetically defined attenuated strains of bacteria for use as vaccines. If detailed knowledge of the pathogen is available, attenuation can be achieved through inactivation of one or more virulence factors. An alternative strategy, referred to as “rational” attenuation, may be employed when no obvious virulence factors are known. This approach involves inactivating genes in metabolic pathways which are necessary for bacterial growth and survival in vivo. Examples of biochemical pathways targeted to produce attenuated strains include the following: aromatic amino acid biosynthesis (18, 19, 22), purine biosynthesis (26, 27), capsule biosynthesis (9, 25), galactose epimerase (11, 13), and adenylate cyclase (39).
Frequently, gene replacement mutagenesis is easy to perform, but with some bacteria, it can be difficult or impractical due in part to an insufficient understanding of their genetic systems. For allelic exchange mutagenesis to be efficient with suicide replacement plasmid, high transformation efficiency is required because homologous recombination is a rare event. Mannheimia hemolytica, P. multocida, and H. somnus have stringent restriction-modification systems (7, 23, 30), and for this reason, mutant production with replacement plasmids is made more difficult. Native plasmids of P. multocida and M. hemolytica such as pIG1 (37), pYFC1 (10), pPH843 (2), and pBAC64 (5) have been engineered primarily as shuttle vectors intended for gene expression. For targeted mutagenesis, nonreplicative replacement plasmids have been overwhelmingly used to engineer allelic mutants of Pasteurellaceae organisms (19, 20, 38).
It is desirable, if not mandatory, that live vaccines are devoid of exogenous antibiotic resistance markers. Unmarked mutations carrying no foreign DNA can be generated in gram-negative bacteria using counterselectable marker systems in which the rare desired products are identified by means of positive selection. For example, the Bacillus subtilis sacB gene, coding for levansucrase, is a widely used counterselectable marker system (14). In gram-negative bacteria, the sacB product is toxic when cells are grown in the presence of sucrose, and single-crossover mutants harboring integrated replacement plasmid are eliminated from the population upon selection. Thus, only cells having undergone a second crossover to form either the wild type (wt) or mutants remain viable. Unfortunately, both the sacB system and another commonly used counterselection system involving tetracycline-fusaric acid (6) have been reported to be ineffective in M. hemolytica (17).
To date, defined unmarked aroA mutants of avian strains of P. multocida A:1 and A:3 were produced by auxotrophic enrichment in the presence of antibiotic (21). An lktC deletion mutant of M. hemolytica lacking an antibiotic resistance element was constructed using a Cre-lox system which inserted a 34-bp loxP site in the target gene (17). No unmarked H. somnus mutants have been reported in the literature. Neither of the published methods for generating mutants of M. hemolytica or P. multocida that are free of antibiotic resistance markers furnishes the flexibility or ease of application as does the technique, described previously by Hamilton et al. (16), involving a temperature-sensitive (TS) plasmid. This method proceeds by homologous recombination between the bacterial chromosome and a temperature-regulated replacement plasmid carrying mutated genes. At the nonpermissive temperature, cells maintain antibiotic resistance only if the replacement plasmid integrates into the chromosome. Plasmid resolution is rapidly achieved by transferring single-crossover mutants to the permissive temperature for plasmid replication, and, depending on the position of the second crossover, either the wild type or mutants are generated.
Despite the merits of the above-mentioned systems for engineering species of Pasteurellaceae, a widely applicable method for generating unmarked mutations in this family was sought. Herein, a new TS plasmid, derived from an endogenous plasmid of M. hemolytica pD70, was used to produce gene replacements in the three species primarily associated with BRD. The broad utility of this system was demonstrated by generating unmarked, in-frame aroA mutants of M. hemolytica, P. multocida, and H. somnus.
MATERIALS AND METHODS
Bacterial strains and cosmid libraries.
Mannheimia hemolytica strain NADC-D60 is a bovine lung isolate of serotype 1. Pasteurella multocida strain NADC-TT94 is a bovine lung isolate of Carter Heddleston type A:3. Haemophilus somnus strain 2336 was a bovine lung isolate kindly provided by Lynette Corbeil, University of California San Diego Medical Center, San Diego, CA. H. somnus was cultured on chocolate agar plates (Columbia blood agar base; Difco Laboratories, Detroit, MI) supplemented with 5% defibrinated bovine blood added to medium at 90°C. M. hemolytica and P. multocida were grown on Columbia blood agar base plates (Difco Laboratories). Escherichia coli strain DH11S (Life Technologies, Inc., Rockville, MD) was used for plasmid propagation and cloning. Genomic DNAs were purified and cosmid libraries were constructed for H. somnus and P. multocida by methods identical to those previously described elsewhere for M. hemolytica (7). Cosmid libraries were generated using the cosmid vector pLAFRX (7), which was derived from cosmid c2XB (3).
Construction of the TS plasmids pGA301ori and pGA301oriC.
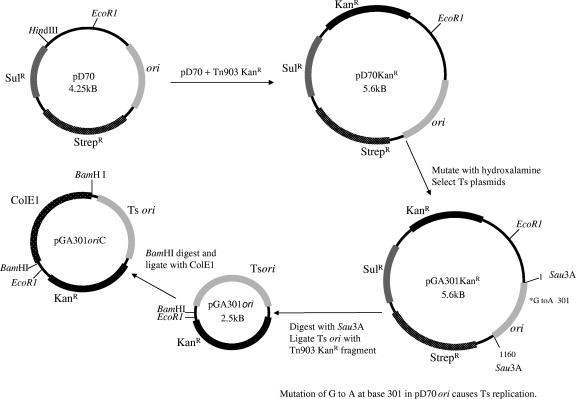
The construction of the TS plasmid pGA301 has been described elsewhere (7a). Briefly, the TS plasmids pGA301ori and pGA301oriC were derived from endogenous plasmid pD70 of M. hemolytica (Fig. 1). The TS origin was ligated to a kanamycin resistance cassette, Tn903 Kanr, which was excised from pBluescript SK (Stratagene Inc., La Jolla, CA) by BamHI digestion. The resultant plasmid, pGA301ori, was introduced into P. multocida by electroporation and amplified. Plasmid was isolated from cells by the alkaline sodium dodecyl sulfate method and purified by CsCl gradient centrifugation (1). The ColE1 origin of pUC was amplified by PCR using the ColE1 primer set shown in Table 1. The PCR-derived ColE1 fragment was treated with BamHI and inserted into the BamHI site of pGA301ori to generate pGA301oriC.
FIG. 1.
Plasmid used in this study. Plasmid pD70 is an endogenous plasmid of M. hemolytica. Plasmid pD70KanR was constructed by inserting the kanamycin resistance cassette into the modified HindIII site of pD70. Plasmid pD70KanR was mutagenized with hydroxylamine, and the TS (Ts) plasmid pGA301 was selected after screening in M. hemolytica. Plasmid pGA301oriKanr was constructed by digesting pGA301 with Sau3A. The resulting 1,160-bp Sau3A fragment containing the TS origin of replication was joined to the kanamycin resistance cassette of Geneblock. Plasmid pGA301oriKanC was constructed by inserting the ColE1 origin of replication into pGA301oriKanr.
TABLE 1.
Oligonucleotide primers
| Oligonucleotide | Sequence | Product (size [kb]) |
|---|---|---|
| UnivaroA For | 5′-AA(A,G)CCNTATATT(T,C)AC-3′ | aroA (approx. 0.9) |
| UnivaroA Rev | 5′-AA(A,G)CACATNC(G,T)(A,G)TG(A,G)TC-3′ | |
| MharoA For | 5′-TTACTGCGTGAAGGCGTGATTG-3′ | M. haemolytica aroA (1.2) |
| MharoA Rev | 5′-GGTTTCAATTTCAGCGTG-3′ | |
| PmaroA For | 5′-TTACTCTCAATCCCATCAGCTATA-3′ | P. multocida aroA (1.3) |
| PmaroA Rev | 5′-CTATCTGTAGGCTACTTCGCGTG-3′ | |
| HsaroA For | 5′-AAAAGTGTTAGGCGAGGTCTTTG-3′ | H. somnus aroA (1.8) |
| HsaroA Rev | 5′-TTAACAGGTTGTCCTGATTGG-3′ | |
| ColE1 For | 5′-AAAGGATCCTTCGGTCGAAAAGGA-3′ | ColE1 origin of pUC (0.8) |
| ColE1 Rev | 5′-AAAGGATCCAAACGGTATTGGG-3′ | |
| pD70ori For | 5′-GATCCCTTTTTCTGTAATCTG-3′ | TS origin (1.16) |
| pD70ori Rev | 5′-GATCATAGGCTCAATTCTCGC-3′ | |
| KanR For | 5′-ATGAGCCATATTCAACGG-3′ | Tn903 Kan resistance casette (0.9) |
| KanR Rev | 5′-TCAGAAAAACTCATCGAGCATC-3′ |
Construction of cosmid libraries: cloning of M. hemolytica, P. multocida, and H. somnus aroA genes and sequence determination.
Cloning of the M. hemolytica aroA gene was described previously (32). P. multocida and H. somnus aroA gene fragments were amplified using the degenerate custom primer set UnivaroA (Table 1), which anneals to conserved regions of bacterial aroA genes. The primers were synthesized with an oligonucleotide synthesizer (Applied Biosystems Inc., Foster City, CA) by Integrated DNA Technologies, Inc., Coralville, IA. P. multocida and H. somnus cells were used as templates, and PCRs were done using the using the EasyStart PCR mix using a tube protocol of Molecular BioProducts (San Diego, CA). Reaction conditions were 30 cycles with 30 s at 95°C, 45 s at 45°C, and 60 s at 72°C per cycle. Each reaction produced fragments of approximately 900 bp that were inserted into pCR2.1 (Invitrogen Inc., Carlsbad, CA), and fragments were sequenced to verify their authenticity. To obtain complete aroA genes for sequence determination and also to provide replacement plasmids with sufficient length to efficiently produce mutants via homologous recombination, aroA-containing cosmid clones were isolated from P. multocida and H. somnus cosmid libraries by colony hybridization using labeled PCR-derived aroA sequences as probes (29). EcoRI-derived fragments of 8.0 kb and 4.0 kb from the H. somnus and P. multocida cosmid clones were subcloned into pBluescript SK, and the complete nucleotide sequences for each aroA gene were deduced using the Dye Terminator Chemistry kit from PE Applied Biosystems and an ABI Prism 377 DNA sequencer. This work was performed by the Nucleotide Acids Facility, Iowa State University, Ames, IA. E. coli strain AB2829 (28) (an aroA mutant) was used to determine if the aroA genes in pBluescript SK would complement growth on minimal medium plates.
Construction of the TS replacement plasmids. (i) M. hemolytica.
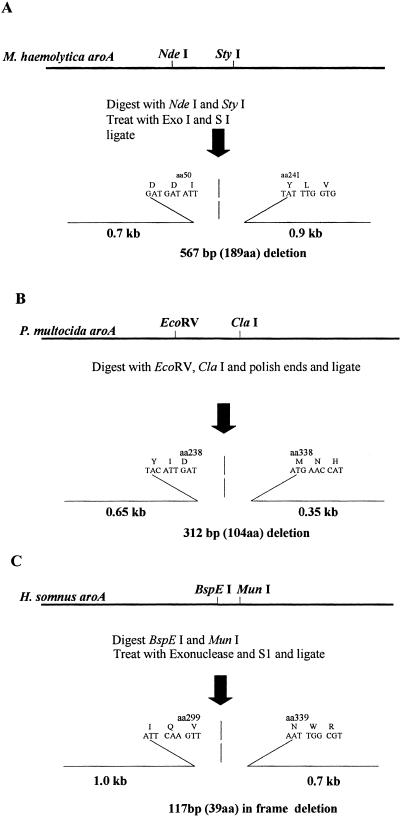
A 2.2-kb fragment containing M. hemolytica aroA was cloned into pBluescript SK as described previously (32). The aroA clone was cleaved at unique NdeI and StyI restriction sites within the coding region to excise a fragment of approximately 470 bp. Following digestion, the fragment was briefly treated with ExoIII and S1 nuclease using an Erase-a-base kit (Promega Corp., Madison, WI). The ends were polished by treatment with the Klenow fragment and nucleoside triphosphates and then circularized with ligase. Following transformation into E. coli DH11, plasmid DNA was isolated from colonies and sequenced. A clone possessing an in-frame aroA deletion was selected, and the deleted aroA fragment was excised by double digestion with XhoI and EcoRV (Fig. 2A). The fragment ends were made blunt as described above. The end-polished fragment was inserted by blunt-end ligation into the EcoRI site of pGA301oriC that also was filled in using the Klenow fragment as described above to generate the replacement plasmid pMhΔaroA.
FIG. 2.
(A) M. hemolytica aroA replacement plasmid consisting of a deleted in-frame aroA fragment incorporated into pGA301oriKanr. The replacement plasmid deletion was determined by sequencing with primers adjacent to the deletion site. (B) P. multocida aroA replacement plasmid consisting of deleted in-frame aroA fragment incorporated into the TS plasmid. (C) H. somnus aroA replacement plasmid containing the indicated deletion in aroA. aa, amino acid.
(ii) P. multocida.
A 1.3-kb P. multocida aroA product was generated by PCR using the aroA cosmid clone as a template and the PmaroA primer set (Table 1). The primer set PmaroA hybridized to nucleotides approximately 20 bp downstream of the aroA start and those which spanned the stop codon, respectively. Initially, this fragment was inserted into pCR2.1 (Invitrogen Inc.) and then subcloned into the EcoRI site of pUC8. The aroA fragment was digested at unique EcoRV and ClaI sites to remove an approximately 300-bp fragment within the coding region of the gene. The end generated after ClaI digestion was made blunt using the 3′→5′exonuclease activity of the Klenow fragment of DNA polymerase I. The plasmid was circularized with T4 ligase, and the mixture was transformed into E. coli DH10B cells (Life Science Inc.). Plasmids were isolated from randomly selected clones for sequencing, and a plasmid that contained an in-frame deleted P. multocida aroA was identified (Fig. 2B). The deleted aroA fragment was excised from purified plasmid with EcoRI and was then subcloned into the EcoRI site of pGA301ori to generate the replacement plasmid pTsPmΔaroA.
(iii) H. somnus.
A 1.8-kb H. somnus aroA fragment was generated by PCR using the aroA-containing cosmid clone and the primer set HsaroA (Table 1) that hybridizes to nucleotides approximately 80 bp upstream of the start codon and 440 bp downstream from the stop codon of aroA, respectively. The resultant PCR fragment was ligated into pCR2.1 and then excised by EcoRI digestion and transferred to the EcoRI site of pGA301oriC to generate replacement plasmid pTsHsaroC. A deletion was introduced within the gene's coding region by digestion with BspEI followed by exonuclease III and S1 nuclease treatment using an Erase-a-base kit (Promega Corp., Madison, WI). The plasmid was then digested at a unique MunI site within aroA and treated with the Klenow fragment and nucleoside triphosphates to make the fragment ends blunt. DNA was recirularized with T4 ligase and transformed into E. coli DH10B. Plasmid DNAs from clones were isolated by the alkaline lysis method, CsCl purified (1), and sequenced to identify a replacement plasmid, pTsHsΔaroAC, possessing an in-frame aroA deletion (Fig. 2C).
Generation and characterization of the aroA mutants. (i) M. hemolytica.
Plasmid pMhΔaroA was first transformed into E. coli strain DH10B, PhaI Mtase, which contains the PhaI methyltransferase gene on cosmid vector pLAFRX. The methyl-modified replacement plasmid was isolated by the alkaline lysis method and CsCl purified (1). Mannheimia hemolytica was electroporated with 200 ng plasmid DNA as previously described (7). The cells were then recovered at 30°C for 2 h and plated onto Columbia blood agar plates containing 50 μg/ml kanamycin. Colonies were visible after 24 h of incubation at 30°C. Individual colonies were transferred to 5 ml Columbia broth containing kanamycin and incubated at 30°C for 24 h. To confirm that cells were transformed with replacement plasmid, 4 ml of culture was pelleted and plasmid DNA was isolated by the alkaline lysis method (28). Plasmid DNA was digested with restriction enzymes and electrophoresed on a 1% agarose gel in Tris-borate-EDTA buffer, and banding patterns were visualized by ethidium bromide staining and UV illumination. Approximately 10 μl of broth from the transformed cell cultures was spread onto Columbia blood agar plates with 50 μg/ml kanamycin and grown at 41°C for 16 h. Since the TS plasmid was unable to replicate autonomously at 41°C, mostly cells possessing integrated plasmid by homologous recombination survived antibiotic selection. Several single-crossover mutants were transferred to 5 ml Columbia broth and incubated at 30°C for 16 h. This step was repeated twice more to promote plasmid resolution from the chromosome. The third-pass broth culture growth was streaked onto Columbia blood agar plates and grown at 37°C for isolation. After 16 h, colonies were replica plated onto Columbia blood agar plates with or without 50 μg/ml kanamycin and grown at 37°C. Kanamycin-sensitive colonies were analyzed by PCR using the M. hemolytica aroA primer pair. The absence of the Kanr element and TS origin were confirmed by PCR using Tn903 Kanr and TS origin primer pairs. The positive-control template was M. hemolytica containing replacement plasmid.
(ii) P. multocida.
P. multocida strain NADC-TT94 was grown for 4 h in 100 ml Columbia broth containing 2,500 U hyaluronidase to digest the capsule and facilitate pelleting of cells. Cells were electroporated with 200 ng of plasmid DNA as previously described (7). After 2 h at 30°C, cells were spread onto Columbia agar plates containing 50 μg/ml kanamycin and then incubated for 24 h at 30°C. Six colonies were passed separately into 5 ml Columbia broth containing 50 μg/ml kanamycin and then incubated for 18 h at 30°C. Cells were spread (10 μl/plate) onto Columbia agar plates containing kanamycin and incubated for 16 h at 40°C to generate single-crossover mutants. Representative colonies were passed into 5 ml Columbia broth lacking antibiotic and grown at 30°C for 16 h to allow resolution. As described above, three 16-h passes in 5-ml Columbia broth cultures incubated at 30°C were done to promote plasmid resolution. Ten microliters of culture was spread onto Columbia plates and incubated at 37°C for 16 h for colony isolation. Kanamycin-sensitive colonies were analyzed by PCR as described above except that the P. multocida aroA primer pair was used.
(iii) H. somnus.
Purified replacement plasmid pHsΔaroA was in vitro methylated with HhaI methyltransferase as specified by the supplier (New England Biolabs, Beverly, MA). Plasmid DNA protection was verified by resistance to digestion with HsoI. H. somnus strain 2336 was grown in Levinthal's broth in 5% CO2 to late logarithmic phase. The cells were washed, and conditions for electroporation were identical to those described above. Medium for cultivation of H. somnus consisted of Chocolate agar plates and Levinthal's broth. Chocolate agar plates contained Columbia blood agar base (Difco Laboratories) supplemented with 5% defibrinated bovine blood at 90°C. Levinthal's broth was 3.7% brain heart infusion broth to which 7% (vol/vol) defibrinated bovine blood was added at 90°C which was supplemented by 6 ml of 3% sterile yeast autolysate. The protocol for mutant generation and analysis was identical to that described above except that the H. somnus aroA primer pair was used. All cultivation of H. somnus was done in 5% CO2.
Growth of the aroA mutants on chemically defined media.
The defined media for M. hemolytica and P. multocida cultivation were developed by Wessman (36) and Watko (35), respectively. These defined media contained specific amino acids which included phenylalanine and tyrosine but not tryptophan. Clones unable to grow on the chemically defined media for M. hemolytica and P. multocida cultivation were presumed to be aroA mutants. Media were made complete with respect to aromatic amino acids by the addition of 40 μg/ml tryptophan.
Nucleotide sequence accession number.
The H. somnus aroA sequence data were deposited in GenBank under accession number L47538.
RESULTS
Construction of the TS replacement plasmids.
The parent plasmid, pD70, is an endogenous 4.25-kb plasmid of M. hemolytica that encodes streptomycin resistance (Fig. 1). The Tn903 kanamycin resistance cassette was inserted into pD70 because of its broad host range and excellent selection achieved with kanamycin-containing media. Plasmid pD70KanR was subjected to mutagenesis with hydroxylamine (33), and after extensive screening and genetic characterization, a TS plasmid, pGA301, was shown to replicate in M. hemolytica at 30°C but was inactive at high temperature. A single base change of G to A in the plasmid origin caused TS replication in M. hemolytica. Moreover, pGA301 was shown to replicate in a temperature-dependent manner in P. multocida and in H. somnus. The derivative plasmid pGA301ori, which was made smaller to simplify engineering of Pasteurellaceae, was equally capable as the parent plasmid, pGA301, at temperature-dependent replication in the three species examined. The addition of the ColE1 origin to produce pGA301oriC allowed many routine steps for replacement plasmid construction to be carried out in E. coli (Fig. 1).
Cloning of aroA genes.
H. somnus and P. multocida aroA fragments of approximately 900 bp were obtained by PCR using degenerate primers (Table 1) complementary to highly conserved regions of bacterial aroA genes. Sequencing verified their authenticity. Full-length aroA clones were identified from P. multocida and H. somnus cosmid libraries from which 4-kb and 8-kb aroA fragments, respectively, were subcloned and sequenced. The aroA of M. hemolytica (GenBank accession number UO3068) used in this work was cloned and sequenced previously (32). When compared, the nucleotide sequences of the three aroA genes showed approximately 75% sequence similarity. Each subcloned aroA gene was capable of complementing E. coli AB2829 (aroA mutant) growth on minimal medium plates.
Construction and characterization of in-frame aroA deletion mutants of M. hemolytica, P. multocida, and H. somnus.
A generalized protocol for mutant generation using the TS replacement plasmid is shown in Fig. 3. In-frame deletions were engineered into each aroA gene, and the gene fragments were cloned into pGA301ori or pGA301oriC to produce replacement plasmids (Fig. 2). Sequencing of the respective replacement plasmids with custom-designed primers confirmed that the intended deletions were in frame. When required, specific methyl protection of replacement plasmid was carried out prior to electroporation into the appropriate hosts. Electroporation efficiencies into M. hemolytica, P. multocida, and H. somnus were approximately 104, 103, and 102 CFU/μg, respectively. Passage of transformants to the nonpermissive temperature for plasmid maintenance resulted in variable numbers of colonies ranging from 10 to >1,000, varying more between clones than between host strains.
FIG. 3.
Protocol for mutant production with the TS replacement plasmid. Cells transformed with the TS replacement plasmid were transferred onto selective plates and grown at the nonpermissive temperature for plasmid replication. Integration is necessary for cell growth and colony formation. The presumed single-crossover mutants were then transferred to broth culture without selection and grown at a permissive temperature for an active plasmid origin which greatly reduces the growth rate of single-crossover mutants. Cells having excised plasmid from their chromosome at one of two potential crossover points returned to a normal growth rate, and either mutant or wild-type progeny result. Cells were transferred from broth onto plates and then grown without antibiotic at a nonpermissive temperature to eliminate plasmid. Colonies were screened for resistance to kanamycin, and Kans colonies were analyzed by PCR to confirm their genetic identity.
Selected colonies which arose at the nonpermissive temperature for plasmid maintenance were passed successively in broth without selection at 30°C and at 40°C and then streaked for isolation. The ensuing colonies were replicated onto plates with and without antibiotic. The majority of the progeny from most cultures of the three species had lost antibiotic resistance after low-temperature passage but had not lost antibiotic resistance after high-temperature passage. Occasionally, selected isolates never resolved after repeated low-temperature passage and remained Kanr.
With M. hemolytica, more than 80% of the progeny of nine clones passed at low temperature had resolved plasmid and were Kans, while all progeny of the remaining clone passed at low temperature retained Kanr. The latter clone was found to contain plasmid which was no longer thermoregulated. Kanamycin-sensitive colonies were screened for the desired aroA mutation by PCR using primers flanking the deletion sites. Most of these colonies were shown to possess intact aroA, indicating that the second crossover occurred on the same arm of the replacement plasmid as integration (Fig. 2). Approximately 4% of the progeny were shown by PCR to possess a deletion in aroA (Fig. 4A). Amplification of clones possessing a deleted aroA by PCR yielded no product for kanamycin or TS origin using the respective primer pairs. Amplification of positive-control samples yielded the expected products.
FIG. 4.
A. Agarose gel of PCR samples. Lane 1, wt M. hemolytica; lane 2, M. hemolytica aroA mutant (templates were amplified using the MharoA primer pair). Lane 3, M. hemolytica transformed with replacement plasmid; lane 4, M. hemolytica aroA mutant (templates were amplified with the Kanr primer set). Lane 5, M. hemolytica transformed with replacement plasmid; lane 6, M. hemolytica aroA mutant (templates were amplified with the pD70ori primer set). B. Lane 1, wt P. multocida; lane 2, P. multocida aroA mutant (templates were amplified with the PmaroA primer set). Lane 3, P. multocida transformed with replacement plasmid; lane 4, P. multocida aroA mutant (templates were amplified with the Kanr primer set). Lane 5, P. multocida transformed with replacement plasmid; lane 6, P. multocida aroA mutant (templates were amplified with the pD70ori primer set). C. Lane 1, wt H. somnus; lane 2, H. somnus aroA mutant (templates were amplified with the HsaroA primer set). Lane 3, H. somnus transformed with replacement plasmid; lane 4, H. somnus aroA mutant (templates were amplified with the Kanr primer set). Lane 5, H. somnus transformed with replacement plasmid; lane 6, H. somnus aroA mutant (templates were amplified with the pD70ori primer set).
Identical steps were used to produce aroA mutants of P. multocida and H. somnus using the TS replacement plasmid. The rates of plasmid resolution and frequencies of mutant generation for H. somnus and P. multocida approximated that determined for M. hemolytica.
Characterization of aroA mutants on defined media.
Both M. hemolytica and P. multocida parent strains grew on the respective defined medium but neither of the mutants did so. However, upon the addition of tryptophan to the defined media, growth of the aroA mutants was restored. Since there is no known defined medium for H. somnus, inactivation of its aroA was not assessed for the mutant.
DISCUSSION
The temperature-sensitive plasmids pGA301ori and pGA301oriC were shown to be broadly effective for constructing unmarked mutants in M. hemolytica, P. multocida, and H. somnus. The minimum requirements for this system are presumed to be that the host be recombination proficient and able to support replication of the TS plasmid. Inclusion of the ColE1 origin on pGA301oriC was possible since this origin is inactive in the three species of Pasteurellaceae studied here. A clear advantage to this system is that the replicative TS plasmid does not require high transformation efficiencies to produce results. The relatively low transformation efficiency we experienced with H. somnus, even after specific methylation of plasmid, would have been problematic if aroA inactivation were attempted using a nonreplicative suicide vector.
This system was developed, in part, to produce attenuated vaccine candidates. It is thought that aroA mutants are attenuated because they are unable to synthesize chorismic acid, which is an intermediate product of p-aminobenzoic acid, aromatic amino acids, and folate biosynthesis (18). More broadly, however, application of this system would enable the construction of in-frame deletion mutants capable of expressing and transporting inactive antigenic protein(s) or toxoids, which may be important virulence factors as well as components for induction of protective immunity, such as leukotoxin of M. hemolytica. Furthermore, an ability to generate in-frame mutants is essential for unambiguously deciphering the role of virulence genes in pathogenesis. It is well known that insertion mutations have the potential for unintended consequences such as inactivating nontarget genes by polar termination (4) or altering the expression of the adjacent gene(s) because of the insertion marker's promoter (34). Attribution of altered virulence to the inactivation of a target gene through insertion mutagenesis may be faulty since the effect may arise through altered expression of nontargeted genes.
A possible limitation to this system is the requirement that the replacement plasmid be introduced into the host by electroporation. The inclusion of a recognition site for mobilization such as mob (31) onto the plasmid might allow plasmid transfer by conjugation in species which are difficult to transform by other means.
In early work with this system, a derivative of pGA301oriC containing Bacillus subtilis sacB (14) was constructed which was intended for counterselection of cells harboring plasmid sequences to eliminate single-crossover mutants from cultures containing the final products. Counterselection with varying sucrose concentrations and several medium formulations was inconsistent and typically allowed large numbers of M. hemolytica or P. multocida cells to continue proliferation despite the confirmed presence of sacB (data not shown). The use of sacB for counterselection was abandoned without discovering the cause of its failure. However, because resolution of integrated plasmid from single-crossover mutants during log-phase growth at 30°C was rapid for the species engineered in this study, the potential benefit gained by the application of sacB selection was modest.
Active plasmid origins of replication integrated into the chromosome are known to dramatically reduce the growth rate of E. coli (16). The rapid loss of plasmid from most single-crossover mutants examined in this study is likely due to a similar phenomenon. The occasional products that did not give rise to Kans progeny after repeated passage at low temperature were often found to possess free plasmids whose replication was no longer thermoregulated, presumably by random mutation of the plasmid ori. For this reason, we routinely use multiple parallel cultures to produce independent single-crossover products for mutant generation.
Although the corresponding aroA arms contained on the replacement plasmids were comparable in length, the frequencies of progeny bearing deleted alleles after resolution of plasmid were less than 5%. It was determined by PCR that crossover sites on the replacement plasmids in host chromosomes are preferential, occurring predominately on a particular arm of the mutated sequences (Fig. 3). We speculate that plasmid resolution also initiated most frequently on that same arm, resulting in a preponderance of wild-type progeny.
While not attempted, the gene replacement method described here could be applied to create strains possessing multiple mutations devoid of exogenous DNA. Furthermore, this system could be adapted for the delivery of transposon systems or to insert recombinant genes or DNA segments into the chromosome of intended hosts.
This study demonstrated the feasibility of using the TS plasmid pGA301oriC as a mutagenic vector in three species of bacteria. The TS plasmid-based system described here for genetically engineering members of Pasteurellaceae represents a valuable tool for investigators to perform genetic studies and create new vaccine strains in this group of economically important bacterial pathogens. Further confirmation of the utility of this system is provided by the subsequent construction of additional in-frame deletion mutants of Pasteurellaceae in our laboratory.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. Greene Publishing Associates and Wiley Interscience, New York, N.Y.
- 2.Azad, A. K., J. G. Coote, and R. Parton. 1994. Construction of conjugative shuttle and suicide vectors for Pasteurella haemolytica and P. multocida. Gene 145:81-85. [DOI] [PubMed] [Google Scholar]
- 3.Bates, P. F., and R. A. Swift. 1983. Double cos site vectors: simplified cosmid cloning. Gene 26:137-146. [DOI] [PubMed] [Google Scholar]
- 4.Berg, C. M., and D. E. Berg. 1996. Transposable element tools for microbial genetics, p. 2588-2612. In F. C. Neidhardt, R. Curtis III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Resnikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 5.Bills, M. M., J. M. Medd, R. J. Chappel, and B. Adler. 1993. Construction of a shuttle vector for use between Pasteurella multocida and Escherichia coli. Plasmid 30:268-273. [DOI] [PubMed] [Google Scholar]
- 6.Bochner, B. R., H. C. Huang, G. L. Schieven, and B. N. Ames. 1980. Positive selection for loss of tetracycline resistance. J. Bacteriol. 143:926-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Briggs, R. E., F. M. Tatum, T. A. Casey, and G. H. Frank. 1994. Characterization of a restriction endonuclease, PhaI, from Pasteurella haemolytica serotype A1 and protection of heterologous DNA by a cloned PhaI methyltransferase gene. Appl. Environ. Microbiol. 60:2006-2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7a.Briggs, R. E., and F. M. Tatum. 2005. Generation and molecular characterization of new temperature-sensitive plasmids intended for genetic engineering of Pasteurellaceae. Appl. Environ. Microbiol. 71:7187-7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carter, G. R. 1967. Pasteurellosis: Pasteurella multocida and Pasteurella haemolytica. Adv. Vet. Sci. 11:321. [PubMed] [Google Scholar]
- 9.Chung, J. Y., I. Wilkie, J. D. Boyce, K. M. Townsend, A. J. Frost, M. Ghoddusi, and B. Adler. 2001. Role of capsule in the pathogenesis of fowl cholera caused by Pasteurella multocida serogroup A. Infect. Immun. 69:2487-2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fedorova, N. D., and S. K. Highlander. 1997. Plasmids for heterologous expression in Pasteurella haemolytica. Gene 186:207-211. [DOI] [PubMed] [Google Scholar]
- 11.Fernandez de Henestrosa, A. R., I. Badiola, M. Saco, A. M. Perez de Rozas, S. Campoy, and J. Barbe. 1997. Importance of the galE gene on the virulence of Pasteurella multocida. FEMS Microbiol. Lett. 154:311-316. [DOI] [PubMed] [Google Scholar]
- 12.Frank, G. H. 1989. Pasteurellosis of cattle, p. 197-222. In C. Adlam and J. M. Rutter (ed.), Pasteurella and pasteurellosis. Academic Press Ltd., London, United Kingdom.
- 13.Fry, B. N., S. Feng, Y. Y. Chen, D. G. Newell, P. J. Coloe, and V. Korolik. 2000. The galE gene of Campylobacter jejuni is involved in lipopolysaccharide synthesis and virulence. Infect. Immun. 68:2594-2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gay, P., D. LeCoq, M. Steinmetz, T. Berkelman, and C. I. Kado. 1985. Positive selection procedure for entrapment of insertion sequence elements in gram-negative bacteria. J. Bacteriol. 164:918-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Griffen, D. 1997. Economical impact associated with respiratory disease in beef cattle. Vet. Clin. N. Am. 13:367-377. [DOI] [PubMed] [Google Scholar]
- 16.Hamilton, C. H., M. Aldea, B. K. Washburn, P. Babitzke, and S. R. Kushner. 1989. New method for generating deletions and gene replacements in Escherichia coli. J. Bacteriol. 171:4617-4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Highlander, S. K., N. D. Fedorova, D. M. Dusek, R. Panciera, L. E. Alvarez, and C. Rinehart. 2000. Inactivation of Pasteurella (Mannheimia) haemolytica leukotoxin causes partial attenuation of virulence in a calf challenge model. Infect. Immun. 68:3916-3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoiseth, S. K., and B. A. Stocker. 1981. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature 291:238-239. [DOI] [PubMed] [Google Scholar]
- 19.Homchampa, P., R. A. Strugnell, and B. Adler. 1992. Molecular analysis of the aroA gene of Pasteurella multocida and vaccine potential of a constructed aroA mutant. Mol. Microbiol. 23:3585-3593. [DOI] [PubMed] [Google Scholar]
- 20.Homchampa, P., R. A. Strugnell, and B. Adler. 1994. Construction and vaccine potential of an aroA mutant of Pasteurella haemolytica. Vet. Microbiol. 42:35-44. [DOI] [PubMed] [Google Scholar]
- 21.Homchampa, P., R. A. Strugnell, and B. Adler. 1997. Cross protective immunity conferred by a marker free aroA mutant of Pasteurella multocida. Vaccine 15:203-208. [DOI] [PubMed] [Google Scholar]
- 22.Hormaeche, C. E., H. S. Joysey, L. Desilva, M. Izhar, and B. A. Stocker. 1991. Immunity conferred by Aro− Salmonella live vaccines. Microb. Pathog. 10:149-158. [DOI] [PubMed] [Google Scholar]
- 23.Hoskins, I. C., and A. J. Lax. 1997. Identification of restriction barriers in Pasteurella multocida. FEMS Microbiol. Lett. 156:223-226. [DOI] [PubMed] [Google Scholar]
- 24.Humphrey, J. D., and L. R. Stephens. 1983. Haemophilus somnus: a review. Vet. Bull. 53:987-1004. [Google Scholar]
- 25.Ivins, B. E., J. W. Ezzell, Jr., J. Jemski, K. W. Hedlund, J. D. Ristroph, and S. H. Leppla. 1986. Immunization studies with attenuated strains of Bacillus anthracis. Infect. Immun. 52:454-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jackson, M., S. W. Phalen, M. Lagranderie, D. Ensergueix, P. Chavarot, G. Marchal, D. N. McMurray, B. Gicquel, and C. Guilhot. 1999. Persistence and protective efficacy of a Mycobacterium tuberculosis auxotroph vaccine. Infect. Immun. 67:2867-2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Noriega, F. R., G. Losonsky, C. Lauderbaugh, F. M. Liao, J. Y. Wang, and M. M. Levine. 1996. Engineered ΔguaB-A ΔvirG Shigella flexneri 2a strain CVD 1205: construction, safety, immunogenicity, and potential efficacy as a mucosal vaccine. Infect. Immun. 64:3055-3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pittard, J., and B. J. Wallace. 1966. Distribution and function of genes concerned with aromatic biosynthesis in Escherichia coli. J. Bacteriol. 91:1494-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 30.Sanders, J. D., Y. Tagawa, R. E. Briggs, and L. B. Corbeil. 1997. Transformation of a virulence associated gene of Haemophilus somnus into a strain lacking the gene. FEMS Microbiol. Lett. 154:251-258. [DOI] [PubMed] [Google Scholar]
- 31.Simon, R., V. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. BioTechnology 1:784-791. [Google Scholar]
- 32.Tatum, F. M., R. E. Briggs, and S. M. Halling. 1994. Molecular cloning and nucleotide sequencing and construction of an aroA mutant of Pasteurella haemolytica serotype A1. Appl. Environ. Microbiol. 60:2011-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thomas, C. M. 1987. Plasmid replication, p. 7-37. In K. G. Hardy (ed.), Plasmids: a practical approach. IRL Press, Oxford, United Kingdom.
- 34.Wang, A., and J. R. Roth. 1988. Activation of silent genes by transposons Tn5 and Tn10. Genetics 120:875-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Watko, L. P. 1966. A chemically defined medium for growth of Pasteurella multocida. Can. J. Microbiol. 12:933-937. [DOI] [PubMed] [Google Scholar]
- 36.Wessman, G. E. 1966. Cultivation of Pasteurella haemolytica in a chemically defined medium. Appl. Microbiol. 14:597-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wright, C. L., R. A. Strugnell, and A. L. Hodgson. 1997. Characterization of a Pasteurella multocida plasmid and its use to express recombinant proteins in P. multocida. Plasmid 37:65-79. [DOI] [PubMed] [Google Scholar]
- 38.Yanping, W. U., J. H. McQuiston, A. Cox, T. D. Pack, and T. J. Inzana. 2000. Molecular cloning and mutagenesis of a DNA locus involved in lipooligosaccharide biosynthesis in Haemophilus somnus. Infect. Immun. 68:310-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang, X., S. M. Kelly, W. S. Bollen, and R. Curtiss III. 1997. Characterization and immunogenicity of Salmonella typhimurium SL1344 and UK-1 Δcrp and Δcdt deletion mutants. Infect. Immun. 65:5381-5387. [DOI] [PMC free article] [PubMed] [Google Scholar]