Abstract
The growth of Bifidobacterium animalis subsp. lactis IPLA 4549 and its derivative with acquired resistance to bile, B. animalis subsp. lactis 4549dOx, was evaluated in batch cultures with glucose or the glucose disaccharide maltose as the main carbon source. The acquisition of bile salt resistance caused a change in growth pattern for both sugars, which mainly resulted in a preferential use of maltose compared to glucose, whereas the mother strain used both carbohydrates in a similar way. High-performance liquid chromatography and gas chromatography-mass spectrometry analyses were performed to determine the amounts of glucose consumption and organic acid and ethanol formation from glucose by buffered resting cells taken at different points during growth. Resting cells of the bile-adapted strain generally consumed less glucose than those of the nonadapted one but showed an enhanced production of ethanol and higher acetic acid-to-lactic acid as well as formic acid-to-lactic acid ratios. These findings suggest a shift in the catabolism of carbohydrates promoted by the acquisition of bile resistance that may cause changes in the redox potential and improvements in the cellular ATP yield.
Some strains of the genus Bifidobacterium are considered probiotics and are used as active ingredients in functional dairy-based products. Strains of Bifidobacterium animalis subsp. lactis have been largely applied in fermented dairy products instead of other species due to their better technological properties (7). Microorganisms of the genus Bifidobacterium can colonize the colon and are important components of the human intestinal microbiota, in which they may be present at concentrations of 109 to 1010 cells per gram of feces (36). To achieve this colonization, these bacteria must overcome biological barriers that include acid in the stomach and bile in the intestine (5, 28, 34).
Bile salts are synthesized in the liver from cholesterol and secreted as amino acid conjugates into the duodenum, where they facilitate fat absorption. Since these compounds are toxic for microbial cells, the autochthonous gastrointestinal microbiota must have developed strategies to defend themselves against the toxic action of bile, although these mechanisms still remain poorly understood.
Some dietary carbohydrates not digested in the upper gastrointestinal tract can act as substrates for bacterial fermentation in the colon (8). Among these carbohydrates, those that are selectively fermented by beneficial microorganisms are generally known as prebiotics (12). The ability to degrade oligo- or polysaccharides has been related to the presence in the cell of the corresponding glycoside-hydrolyzing activities (13, 17, 29, 42). In bifidobacteria, the fermentation of hexoses released from oligo- or polysaccharides occurs by the d-fructose 6-phosphate shunt, whose characteristic key enzyme, fructose 6-phosphate phosphoketolase (6, 24), is not present in other gram-positive intestinal bacteria. It has been reported that bifidobacteria may alter the preferential direction of their metabolic pathways, and subsequently the ratio of organic acids produced during fermentation, based upon the carbohydrates available for their use (29, 38, 42). Some of the end products of carbohydrate metabolism in bifidobacteria (lactic, acetic, and formic acids) can exert beneficial effects on human health (37).
A relationship seems to exist between the assimilation of oligosaccharides and the level of bile salt resistance in bifidobacteria. In general, the presence of a metabolizable carbon source could alleviate the toxic effect of bile salts (11, 30). It has been demonstrated that resistance to bile was increased in medium containing fructo-oligosaccharides compared to medium containing their monomeric components (30). Since the cellular detoxification of bile salts is an energy-dependent mechanism (30), the predicted higher yields of energy on oligosaccharides than that on monosaccharides might contribute towards increased bile salt resistance (11, 29, 35, 38). Therefore, it could be hypothesized that a bile-adapted microorganism should have optimized the cellular ways to obtain energy from available carbon sources. In a related study, we recently indicated that fructose 6-phosphate phosphoketolase and glycosidic activity levels could be modified in some cases as a consequence of the acquisition of bile salt resistance (26, 32). At the same time, preliminary evidence obtained by us indicated that the acquisition of bile salt resistance in bifidobacteria can modify growth in medium containing glucose or maltose as the carbon source (10).
Several studies of amylolytic Bifidobacterium strains have been conducted in past years (16, 21, 22). Starch is one of the main components of some tubers, legumes, and cereal grains (3). In spite of the importance of these vegetal foods in the human diet throughout the world, fermentation patterns of gluco-oligosaccharides have remained considerably less studied than those of fructo- and galacto-oligosaccharides (13, 15, 17, 30, 33, 38, 42).
Maltose ([1,4]-α-d linkages) is a glucose disaccharide that can result from the partial hydrolysis of starch by intestinal bacteria. In the present work, we analyze differences in growth and fermentation patterns of the strain B. animalis subsp. lactis IPLA 4549 and its bile-resistant derivative B. animalis subsp. lactis 4549dOx (14, 26) in medium containing glucose or maltose as the carbon source.
MATERIALS AND METHODS
Bacterial strains and culture media.
The microorganisms used in this study were the mother strain B. animalis IPLA 4549 and its bile-resistant derivative B. animalis 4549dOx, obtained previously by adaptation of the mother strain to progressively increasing concentrations of bovine bile (ox gall) (26). The induced bile resistance phenotype was stable after several subcultures in medium with glucose in the absence of bile (data not shown), and the adapted microorganism retained the resistance level until stationary phase. The mother and derivative strains, previously grown at 37°C in MRS broth (Biokar Diagnostics, Beauvais, France) with 0.05% (wt/vol) l-cysteine (Sigma Chemical Co., St. Louis, MO) (MRSC) and in MRSC supplemented with 0.8% (wt/vol) ox gall (Sigma), respectively, were maintained as frozen stocks at −80°C in MRSC with 20% (vol/vol) glycerol. Prior to the experiments, cultures were revived from frozen stocks by incubation overnight in 10 ml MRSC in an anaerobic chamber (Mac500; Down Whitley Scientific, West Yorkshire, United Kingdom) under a 10% (vol/vol) H2, 10% (vol/vol) CO2, and 80% (vol/vol) N2 atmosphere.
A basal fermentation medium (FM) (tryptone, 10 g/liter; bacteriological meat extract, 8 g/liter; yeast extract, 4 g/liter; dipotassium phosphate, 2 g/liter; diammonium citrate, 2 g/liter; sodium acetate, 5 g/liter; magnesium sulfate, 0.2 g/liter; manganese sulfate, 0.04 g/liter; and l-cysteine, 0.5 g/liter), added with 20 g/liter of the corresponding sugar, was used for fermentation experiments with glucose and maltose as the carbon source.
Identification and identity of mother and bile-resistant derivative strains.
Identification to the species level of the mother strain and its bile-resistant derivative as well as the identity between both strains was corroborated by partially sequencing the 16S rRNA gene. Isolated colonies of microorganisms were washed and resuspended in 500 μl deionized water. Cell suspensions were heated for 10 min at 98°C and then centrifuged (16,000 × g, 10 min). PCR mixtures were composed of 5 μl of the supernatant, a 0.25 μM concentration of each primer, a 200 μM concentration of each deoxynucleoside triphosphate (Amersham Biosciences, Uppsala, Sweden), and 2.5 U of Taq DNA polymerase (Roche Diagnostics, S.L., Barcelona, Spain) in a final volume of 50 μl. Amplification of the DNA was performed using the following two pairs of primers: Y1 and Y2 (43), which flanked a portion of approximately 350 bp, including the variable regions V1 and V2 of the 16S rRNA coding gene (25), and Bif164 and Bif662, which allowed the amplification of a fragment of ∼523 bp between variable regions V2 and V4 of the 16S rRNA gene (20). The thermal cycle program used consisted of the following time and temperature profile: an initial cycle of 95°C for 5 min, 30 cycles of 94°C for 45 s, 56°C for 1 min, and 72°C for 45 s, and a final extension step of 10 min at 72°C. DNA amplification was performed in a PCR Sprint thermal cycler (Hybaid Ltd., Ashford, Middlesex, United Kingdom). Amplified products were subjected to gel electrophoresis in 1% agarose gels and were visualized by ethidium bromide staining.
PCR products were purified using a GenElute PCR clean-up kit (Sigma) to remove unincorporated primers and nucleotides. Automated sequencing of one strand of the PCR products was done at the Servicio de Secuenciación (University of Oviedo, Spain) with primer Y1 or Bif164 in a 373A automated gene sequencer (Applied Biosystems, Foster City, Calif.). The Basic Local Alignment Search Tool (BLAST) (1) program was used to compare the sequences obtained with those of reference strains held in GenBank.
The identification of B. animalis strains to the subspecies level was achieved as described by Ventura and Zink (40, 41). DNAs were obtained as indicated above. A fragment of approximately 770 bp corresponding to the 16S-23S internal transcribed spacer rRNA gene region was amplified with primers 16S-for and 23Si. The PCR products were digested with the restriction enzyme Sau3AI and visualized by agarose gel electrophoresis and ethidium bromide staining.
Growth in medium containing glucose or maltose as a carbon source.
Overnight cultures in MRSC were washed twice with the same volume of FM at room temperature. Afterwards, 150-μl aliquots of washed cultures were resuspended in tubes containing 15 ml of FM with glucose or maltose. Batch cultures were incubated at 37°C in an anaerobic chamber, and the optical density at 600 nm (OD600) and the pH were measured throughout incubation. Two independent fermentations for each sugar were carried out simultaneously.
Preparation and incubation of resting cells.
Bifidobacterium cells were collected by centrifugation (10,000 × g, 15 min) from cultures in FM plus glucose or maltose at three defined sampling points through growth (exponential phase, beginning of stationary phase, and late stationary phase) (Table 1). The pellet of each tube was washed twice with 33 mM potassium phosphate buffer, pH 6.5, and then resuspended in 10 ml of 33 mM potassium phosphate buffer, pH 5.6, containing 25 mM glucose. The resting cell suspension was incubated with constant mild stirring at 37°C for 4 h in an anaerobic jar with the Anaerocult A system (Merck, Darmstadt, Germany).
TABLE 1.
Metabolic activity and related parameters of buffered resting cells of the mother strain B. animalis subsp. lactis IPLA4549 and its bile-resistant derivative B. animalis subsp. lactis 4549dOx collected at several sampling points throughout fermentation in FM supplemented with glucose or maltose as the carbon source
| Sampling pointa | Sugar added to FMb | Strain | Time of incubation (h) (OD) | Value for indicated parameter (mean ± SD)h
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Glucose consumed (mM/OD600) | Total organic acidsc (mM/OD600) | A/L ratiod | F/L ratiod | Formic acid carbon balancee | Ethanol carbon balancef | Theoretical ATP yieldg | ||||
| 1 | G | Mother | 8 (1.32 ± 0.32) | 3.70 ± 0.59 | 6.16 ± 1.30 | 3.23 ± 0.34 | 0 | 0 | 0.002 ± 0.001 | 1.05 ± 0.08 |
| Derivative | 24 (1.72 ± 0.22) | 2.30 ± 0.31* | 3.90 ± 0.21* | 3.11 ± 0.13 | 0.42 ± 0.03*** | 0.027 ± 0.003*** | 0.016 ± 0.004** | 1.25 ± 0.18 | ||
| M | Mother | 8 (1.98 ± 0.08) | 2.80 ± 0.16 | 4.94 ± 0.39 | 2.63 ± 0.12 | 0 | 0 | 0.002 ± 0.000 | 1.25 ± 0.03 | |
| Derivative | 10 (1.90 ± 0.07) | 1.45 ± 0.14*** | 3.08 ± 0.30** | 11.59 ± 1.48*** | 3.03 ± 0.41*** | 0.068 ± 0.001*** | 0.037 ± 0.004*** | 1.67 ± 0.01*** | ||
| 2 | G | Mother | 24 (4.84 ± 0.07) | 1.48 ± 0.13 | 2.45 ± 0.06 | 2.37 ± 0.29 | 0.23 ± 0.02 | 0.018 ± 0.001 | 0.007 ± 0.002 | 1.24 ± 0.18 |
| Derivative | 36 (3.42 ± 0.05) | 1.61 ± 0.24 | 3.21 ± 0.07*** | 3.00 ± 0.10* | 0.36 ± 0.01** | 0.027 ± 0.001*** | 0.011 ± 0.001* | 1.63 ± 0.08* | ||
| M | Mother | 24 (5.46 ± 0.71) | 1.58 ± 0.20 | 2.57 ± 0.92 | 2.03 ± 0.25 | 0.04 ± 0.00 | 0.004 ± 0.002 | 0.002 ± 0.002 | 1.26 ± 1.04 | |
| Derivative | 24 (6.04 ± 0.42) | 1.04 ± 0.01** | 1.62 ± 0.02 | 3.51 ± 0.14** | 0.73 ± 0.01*** | 0.036 ± 0.001*** | 0.017 ± 0.001*** | 1.08 ± 0.01 | ||
| 3 | G | Mother | 48 (5.07 ± 0.03) | 1.63 ± 0.22 | 1.90 ± 0.50 | 1.77 ± 0.41 | 0.11 ± 0.00 | 0.008 ± 0.002 | 0.003 ± 0.001 | 0.67 ± 0.59 |
| Derivative | 48 (4.13 ± 0.47) | 1.55 ± 0.13 | 2.97 ± 0.36* | 3.35 ± 0.24** | 0.42 ± 0.09** | 0.028 ± 0.003** | 0.012 ± 0.000*** | 1.49 ± 0.10 | ||
| M | Mother | 48 (5.92 ± 0.04) | 1.05 ± 0.07 | 2.05 ± 0.07 | 2.12 ± 0.37 | 0.10 ± 0.00 | 0.010 ± 0.001 | 0.002 ± 0.000 | 1.64 ± 0.19 | |
| Derivative | 48 (6.47 ± 0.36) | 0.72 ± 0.03** | 1.41 ± 0.15** | 4.41 ± 0.10*** | 0.55 ± 0.00*** | 0.030 ± 0.002*** | 0.007 ± 0.001** | 1.48 ± 0.16 | ||
Point 1, exponential phase; point 2, beginning of stationary phase; point 3, late stationary phase.
G, glucose; M, maltose.
Total acids, acetic acid plus formic acid plus lactic acid.
A, acetic acid; L, lactic acid; F, formic acid.
Formic acid carbon balance = 1 × mmol of formic acid formed/6 × mmol of glucose consumed.
Ethanol carbon balance = 2 × mmol of ethanol formed/6 × mmol of glucose consumed.
Theoretical ATP yield = mmol of ATP formed/mmol of glucose consumed.
One-way ANOVA was used to determine statistical significance. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Determination of organic acids and glucose by HPLC analysis.
Cells were removed from the suspension of resting cells by centrifugation, and the supernatant was analyzed by high-performance liquid chromatography (HPLC) to determine the amounts of glucose consumption and acetic, lactic, and formic acid formation. A chromatographic system composed of an Alliance 2690 module injector, a Photodiode Array PDA 996 detector, a 410 differential refractometer detector, and Millennium 32 software (Waters, Milford, MA) was used. Resting cell supernatants (50 μl) were separated isocratically at a flow rate of 0.7 ml/min at 65°C on a 300- by 7.8-mm HPX-87H Aminex ion-exchange column (Hewlett Packard, Palo Alto, CA) protected by a H+ cation Microguard cartridge (Bio-Rad Laboratories, Richmond, CA), using 3 mM sulfuric acid as the mobile phase. The PDA 996 detector was used for the identification and quantification of organic acids at 210 nm, whereas the amount of glucose was analyzed with the 410 refractometer. Standard solutions of lactic, acetic, and formic acids as well as glucose were run separately for the identification and quantification of each compound. Results at each sampling point were expressed as concentrations per cell OD600 (mM/OD600 unit).
For mother and derivative strains grown in glucose and maltose, we calculated for the previously defined sampling points the acetic acid-to-lactic acid (A/L) and formic acid-to-lactic acid (F/L) molar ratios as well as the carbon balance of formic acid formation, using the following formula:
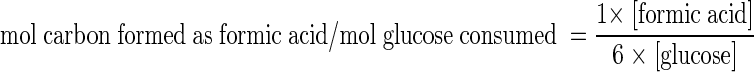 |
where amounts within brackets indicate concentrations (mM) of the corresponding products.
Considering that for the fructose 6-phosphate shunt (Fig. 1) the production of 1 mol of acetic acid gives rise to the formation of 1 mol of ATP, 1 mol of lactic or formic acid generates 2 mol of ATP, and the phosphorylation of glucose at the beginning of glucolysis consumes 1 mol of ATP (2), we estimated the theoretical energetic ATP yield (theoretical mol of ATP formed per mol of glucose consumed) for the same sampling points indicated above, using the following formula:
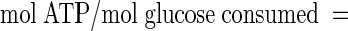 |
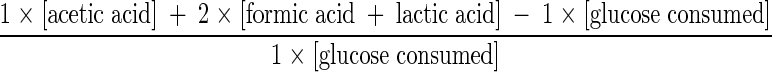 |
where amounts within brackets indicate concentrations (mM) of the corresponding products.
FIG. 1.
Short schematic representation of the main steps of the fructose 6-phosphate shunt in Bifidobacterium (2, 6). Shaded boxes indicate molecules of ADP, ATP, NAD+, and NADH formed.
Determination of ethanol level by dynamic headspace extraction and GC-MS analysis.
Dynamic headspace extraction and gas chromatography-mass spectrometry (GC-MS) analysis were used to determine the amount of ethanol in supernatants of resting cells from cultures with glucose or maltose as the carbon source. A system composed of a 3100 purge and trap concentrator (Tekmar and Dohrmann, Cincinnati, OH) fitted with a Vocarb 3000 trap (Supelco/Sigma-Aldrich Química S.A., Madrid, Spain) was used. The concentrator was coupled to a gas chromatograph (GC 6890N; Agilent Technologies Inc., Palo Alto, CA) connected to a mass spectrometer detector (MS 5973N; Agilent). Data were collected with Enhanced ChemStation G1701DA software (Agilent).
Resting cell supernatants (1.5 ml), obtained as previously described, were mixed with 10 μl of an internal standard (IS) aqueous solution containing 0.156 mg/ml of propyl acetate (Chem Service Inc., West Chester, PA). Samples were placed in a 25-ml needle sparger tube and purged with helium at a flow rate of 45 ml/min for 20 min at 50°C. The volatile compounds were trapped by adsorption to the trap, which was maintained at 35°C with 4.5 lb/in2 back pressure. Water was removed by flushing the trap with helium for 2 min (dry purge). The volatile compounds were desorbed from the trap (by heating at 250°C for 2 min) directly into the injection port at 220°C, with a split ratio of 1:50 and a pressure at the column head of 30 lb/in2. Separation was carried out in an HP-Innowax capillary column (60-m length by 0.25-mm internal diameter, with a 0.25-μm film thickness; Agilent). Chromatographic conditions were as follows: initial oven temperature of 40°C, 1°C/min up to 45°C, 2 min at 45°C, 5°C/min up to 55°C, 20°C/min up to 210°C, and 2 min at 210°C. The column was directly connected to an MS detector, and the electron impact energy was set to 70 eV. The data collected were in the range of 25 to 250 atomic mass units (at 3.25 scans/s). Ethanol was identified by comparison of its mass spectrum with that in the HP-Wiley 138 library (Agilent) and by comparison of its retention time with that of an ethanol standard (Fluka/Sigma-Aldrich Química S.A.). The peak for ethanol was quantified as the relative abundance with respect to the IS (area of total ionic counts of the sample/area of total ionic counts of the IS). The concentration (mM) of ethanol was calculated using the linear regression equation (R2 ≥ 0.99) from a standard curve obtained with at least three replicate measures. Results at each sampling point were expressed as concentrations of ethanol per cell OD600 (mM/OD600 unit).
For mother and derivative strains grown in glucose and maltose, we calculated for the previously defined sampling points the carbon balance of ethanol formation, using the following formula:
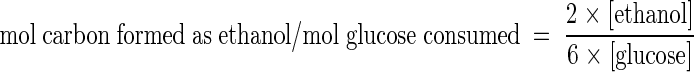 |
where amounts within brackets indicate concentrations (mM) of the corresponding products.
Statistical analysis.
Data for the amounts of organic acids and ethanol formed as well as the amount of sugar consumed by resting cells were subjected to independent one-way analysis of variance using SPSS 11.0 software for Windows (SPSS Inc., Chicago, IL). Tests were performed within each sugar category, using the factor “type of strain” with two categories, “mother” and “derivative.”
Nucleotide sequence accession numbers.
The partial nucleotide sequences of the 16S rRNA V1-V2 genes flanked by primers Y1 and Y2 of B. animalis IPLA 4549 and B. animalis 4549dOx have been deposited in the GenBank database under accession numbers AY835936 and AY835937, respectively. The nucleotide sequences of the V2-V4 region flanked by primers Bif164 and Bif662 of B. animalis IPLA 4549 and B. animalis 4549dOx were given the accession numbers AY835938 and AY835939, respectively, in the GenBank database.
RESULTS AND DISCUSSION
Identification and identity of strains.
An alignment of the partial sequences of the 16S rRNA gene covering regions V1 and V2 and the fragment between regions V2 and V4 of the bile-resistant derivative and its mother strain showed, in both cases, 100% identity between both microorganisms. These results indicated that no contamination or selection of a strain different from the mother one occurred during the experiments involving adaptation to bile salts (data not shown). The mother and bile-resistant derivative strains were referred to in previous works as Bifidobacterium bifidum CECT (Spanish Type Culture Collection) 4549 (26, 32) and B. bifidum 4549dOx, respectively. However, the partial analysis of the 16S rRNA gene carried out in the present work identified them as B. animalis (100% similarity with sequences held in GenBank for regions V1 and V2 and 99% similarity for V2-V4 sequences). According to Ventura and Zink (41), the restriction patterns obtained from PCR-amplified fragments corresponding to the internal transcribed spacer region of the 16S-23S rRNA gene which showed restriction fragment length polymorphism assigned the strains to B. animalis subsp. lactis (data not shown). In addition, the sequence of our mother/derivative pair differed from that of the collection strain B. bifidum CECT 4549, which was identified in its turn as Bifidobacterium infantis/Bifidobacterium longum (99% similarity with sequences from GenBank for regions V1 and V2). These results suggested that our mother strain was probably isolated as a coculture with the strain B. bifidum CECT 4549. A pure culture of this mother strain was then stored in the IPLA collection and was used in the present work. The microorganism was thus renamed B. animalis subsp. lactis IPLA 4549 for this and future works.
Growth in medium containing glucose or maltose as the carbon source.
Figure 2 shows the growth and pH changes of cultures containing glucose and maltose. No differences in pH values were found at late stationary phase between the mother strain and the derivative growing in medium with either sugar. However, differences in growth were evident for each microorganism between glucose- and maltose-containing medium as well as between the mother strain and its derivative on each carbohydrate. On glucose, IPLA 4549 displayed higher cell densities throughout growth than 4549dOx, showing that the latter microorganism had a longer lag phase and slower growth in exponential phase than the mother strain. When maltose was used as the carbon source, the growth of both strains in exponential phase was similar, although the derivative reached slightly higher optical densities in stationary phase than those reached by the mother strain. It is notable that considerably higher cell densities were obtained for the bile-resistant derivative on medium with maltose than on medium with glucose, whereas differences between both sugars were less pronounced for the mother strain. Our results indicated that the acquisition of bile resistance caused a shift in the growth pattern of the 4549dOx strain on glucose and maltose. The changes mainly resulted in the adaptation of the bile-resistant strain to a preferential use of maltose, to the detriment of its ability to grow in glucose.
FIG. 2.
Microbial growth of the original strain B. animalis IPLA 4549 (black symbols) and its bile-resistant derivative B. animalis 4549dOx (white symbols) in FM supplemented with glucose (a) or maltose (b). Squares, OD600; circles, pH values.
We demonstrated in a previous work that the acquisition of bile salt resistance in Bifidobacterium also promotes an increase in the tolerance to low pH (26), which suggests that the mechanisms of cellular response to these two environmental stimuli could be related. Interestingly, van der Meulen and coworkers (38) recently reported a commercial B. animalis dairy strain, probably adapted to acidic environments, which was unable to grow in a medium with glucose as the sole energy source. Since dairy microorganisms are subjected to acid stress, the adaptation to acidic environments of that strain might have promoted, as occurred with our bile-resistant derivative, a decrease in the ability to ferment glucose or even an inability to use this monosaccharide.
Several authors have previously found differences in Bifidobacterium growth depending on the species and the carbohydrate used (15, 27, 29, 42). In general, short oligosaccharides are better substrates for the growth of bifidobacteria than are monosaccharides or polysaccharides (15, 31, 38, 39). The fact that our bile-resistant derivative showed abrupt changes in the growth pattern on glucose but not on maltose could suggest the presence of different transport systems into the cell for each carbohydrate and probably a deep modification of the glucose uptake system. Although specific transport systems for oligosaccharides have been suggested for lactobacilli and bifidobacteria (13, 17, 29), very little is known about carbohydrate transport in Bifidobacterium. The uptake of glucose in Bifidobacterium breve is dependent on sugar phosphorylation (9), and two proton symport systems have been suggested to exist in B. bifidum for the uptake of glucose and lactose (18, 19). Another possibility to explain the changes in growth pattern between the mother and derivative strains is the presence of different expression levels of enzymatic activities involved in the catabolism of these carbohydrates (26, 32).
Changes occurring in the pattern of carbohydrate utilization by the bile-resistant derivative could also affect the end products obtained from the d-fructose 6-phosphate shunt. Therefore, studies on glucose consumption and end-product formation (acetic, lactic, and formic acids and ethanol) were achieved with buffered suspensions of cells (resting cells) from the mother and derivative strains taken throughout growth in medium containing each of the two carbohydrates studied.
Glucose consumption by resting cells.
Microbial samples were taken from FM plus glucose or maltose at defined sampling points throughout the incubation (Table 1). At exponential phase, cells of the mother strain were withdrawn after 8 h of incubation, and cells of the derivative were taken after 24 and 10 h of incubation for cultures in glucose and maltose, respectively. Cells at the beginning of stationary phase were taken at 24 and 36 h for the original and derivative strains, respectively, in medium with glucose and at 24 h for both strains grown in maltose. Microbial samples at late stationary phase were taken after 48 h of incubation.
A comparison of the amounts of glucose consumed by resting cells, as corrected according to the OD600 of cultures, was favorable to the mother strain throughout growth in maltose and at the exponential and late stationary phases when grown in glucose (Table 1). In a previous work, we found higher glucose consumption by resting cells of the derivative than by cells of the mother strain during the late stationary phase of cultures in glucose (32). The differences between both works could be attributed to the different culture medium used in each case. In fact, the MRS broth (Biokar) that was formerly used (32) promoted similar growth patterns (slightly slower for the derivative) in exponential phase for both the mother and derivative strains, whereas the FM-plus-glucose medium used for the present work readily retarded the growth of the bile-resistant derivative. Therefore, the composition of the medium seems to exert a strong influence on the ability of the derivative to use glucose as a carbon source, which in its turn results in sharp modifications of the growth pattern.
Metabolite production by resting cells throughout fermentation.
The production of organic acids from glucose by the original and derivative strains was determined in buffered resting cells previously grown on medium containing glucose or maltose at the same sampling points previously chosen for the determination of glucose consumption (Table 1; Fig. 3). As expected from the results of glucose consumption, the amount of organic acids produced (acetic plus lactic plus formic acids /OD600) was higher in the mother strain than in the bile-resistant derivative, with the exception of cultures at stationary phase in the presence of glucose. As well established for bifidobacteria (4), acetic acid was the major metabolite produced, followed successively by lactic and formic acids. With respect to resistance to bile salts and according to the results for total acids produced, resting cells of the mother strain produced more acetic and lactic acids than those of the derivative throughout growth in maltose and only at exponential phase when grown in glucose. However, in stationary phase the amount of acetic acid formed by the derivative grown in glucose surpassed that produced by the original strain (P < 0.05; statistical analysis not shown). In contrast, formic acid was formed in significantly larger amounts by the derivative than by the mother strain throughout growth in the presence of both sugars (P < 0.01; statistical analysis not shown). Moreover, formic acid was not detected at the beginning of fermentation in the mother strain but appeared at the same point in the derivative. With respect to ethanol, as expected it was detected at considerably lower levels than acetic and lactic acids (Fig. 3). In agreement with the results obtained with formic acid, ethanol was produced at higher levels by the derivative than by the mother strain throughout growth in glucose and maltose (P < 0.01 in most cases; statistical analysis not shown). The data presented here indicate that the preferential direction of the pathways followed for the production of organic acids and ethanol by our B. animalis strains is dependent not only on the growth substrate, as indicated previously by other authors (23, 29, 38), but also on the adaptation of cells to high bile salt concentrations.
FIG. 3.
HPLC analysis of acetic, lactic, and formic acids and GC-MS analysis of ethanol produced by resting cells of the original strain B. animalis IPLA 4549 (black symbols) and its bile-resistant derivative B. animalis 4549dOx (white symbols) collected at several points throughout fermentation in FM supplemented with glucose (a) or maltose (b). Results were corrected to the optical densities at 600 nm. The sampling points throughout growth in each medium are indicated in Table 1.
In the genus Bifidobacterium, lactic and formic acids are produced from pyruvate through the fructose 6-phosphate shunt (Fig. 1). The formation of lactic acid allows the equilibration of the redox balance, whereas the production of formic acid at the expense of lactic acid formation could yield an extra ATP or, alternatively, could allow the cell to regenerate NAD+ by means of the formation of ethanol (2, 6). Therefore, shifts in the production of organic acids and ethanol can influence the molar ratios among these compounds and hence the redox balance and ATP yield of the cell. In order to elucidate whether changes in carbohydrate fermentation patterns related to the acquisition of bile salt resistance could have affected the parameters indicated above, we compared the A/L and F/L molar ratios, formic acid and ethanol carbon balances, and theoretical ATP yields in resting cells of the mother and derivative strains (Table 1). In most cases, the derivative displayed higher A/L and F/L ratios than the mother strain (P < 0.05) throughout the fermentation. It is noteworthy that the carbon balances of the derivative for formic acid and ethanol were higher than the carbon balances of the mother strain (P < 0.01). Interestingly, the theoretical ATP yields were favorable to the bile-resistant strain throughout growth in glucose and at exponential phase when grown in maltose. It has been suggested by several authors (2, 23, 29, 38) that at high intracellular sugar concentrations, bifidobacteria preferentially convert pyruvate to lactic acid through the conventional catabolic route, whereas under conditions of carbon limitation this intermediate metabolite can be converted to formic acid and acetyl-coenzyme A, which finally renders acetate or ethanol (Fig. 1). Our results indicated that the acquisition of bile salt resistance by the 4549dOx strain promoted a metabolic adaptation in this microorganism that enhanced the production of ethanol and formic acid and increased the A/L ratio. In spite of the generally lower glucose consumption rates of the bile-adapted microorganism than those of its mother strain, this adaptation theoretically could improve the ATP yield in exponential phase of bile-resistant cells grown in maltose and could promote a general energetic improvement in medium with glucose. The enhanced production of ethanol could also allow the bile-resistant cells to regenerate NAD+ more efficiently through the fructose 6-phosphate shunt.
From a physiological and ecological point of view, it can be supposed that when undigested carbohydrates reach the colon at the end of the digestive process (under nonlimiting nutrient conditions), the bile-resistant derivative may use glucose disaccharides, which are abundant in this situation, with preference. However, the microorganism retains the ability to slowly use glucose under nutrient-starving conditions, when only small amounts of carbohydrates are available from the diet, obtaining in this situation a better energetic balance from glucose than the mother strain. Finally, the modification of the redox balance of cells could improve the survival of the bile-resistant microorganism in the reducing environment of the colon.
This study will support biochemical and molecular analyses that could help us to understand the interrelationships between bile salt adaptation and modifications of fluxes and fates of intermediate compounds through the fructose 6-phosphate shunt in bifidobacteria.
Acknowledgments
This work was financed by European Union FEDER funds and the Spanish Plan Nacional de I + D (projects AGL2001-2296 and AGL2004-06088-C02-01). P. Ruas-Madiedo was supported by an I3P postdoctoral research contract granted by CSIC and FEDER funds.
We thank Isabel Cuevas for excellent technical assistance.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Shäffer, J. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ballongue, J. 1998. Bifidobacteria and probiotic action, p. 519-587. In S. Salminen and A. von Wright (ed.), Lactic acid bacteria: microbiology and functional aspects. Marcel Dekker Inc., New York, N.Y.
- 3.Belitz, H. D., and W. Grosch. 1999. Food chemistry. Springer-Verlag, Berlin, Germany.
- 4.Bezkorovainy, A. 1989. Nutrition and metabolism of bifidobacteria, p. 93-130. In A. Bezkorovainy and R. Miller-Catchpole (ed.), Biochemistry and physiology of bifidobacteria. CRC Press, Boca Raton, Fla.
- 5.Bezkorovainy, A. 2001. Probiotics: determinants of survival and growth in the gut. Am. J. Clin. Nutr. 73:399S-405S. [DOI] [PubMed] [Google Scholar]
- 6.Biavati, B., B. Sgorbati, and V. Scardovi. 1991. The genus Bifidobacterium, p. 816-833. In A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K. H. Schleifer (ed.), The prokaryotes, 2nd ed. Springer-Verlag, New York, N.Y.
- 7.Biavati, B., P. Mattarelli, and F. Crociani. 1992. Identification of bifidobacteria from fermented milk products. Microbiologica 15:7-14. [PubMed] [Google Scholar]
- 8.Cummings, J. H., and H. Englyst. 1995. Gastrointestinal effects of food carbohydrates. Am. J. Clin. Nutr. 61:938S-945S. [DOI] [PubMed] [Google Scholar]
- 9.Degnan, B. A., and G. T. Macfarlane. 1993. Transport and metabolism of glucose and arabinose in Bifidobacterium breve. Arch. Microbiol. 160:144-151. [DOI] [PubMed] [Google Scholar]
- 10.De los Reyes-Gavilán, C. G., P. Ruas-Madiedo, L. Noriega, I. Cuevas, B. Sánchez, and A. Margolles. 2005. Effect of the acquired resistance to bile salts on enzymatic activities involved in the utilisation of carbohydrates by bifidobacteria. Lait 85:113-123. [Google Scholar]
- 11.De Smet, I., L. van Hoorde, M. van de Woestyne, H. Christiaens, and W. Verstraete. 1995. Significance of bile salt hydrolytic activities of lactobacilli. J. Appl. Bacteriol. 79:292-301. [DOI] [PubMed] [Google Scholar]
- 12.Gibson, G. R., and M. B. Roberfroid. 1995. Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J. Nutr. 125:1401-1412. [DOI] [PubMed] [Google Scholar]
- 13.Gopal, P. K., P. A. Sullivan, and J. B. Smart. 2001. Utilisation of galacto-oligosaccharides as selective substrates for growth by lactic acid bacteria including Bifidobacterium lactis DR10 and Lactobacillus rhamnosus DR20. Int. Dairy J. 11:19-25. [Google Scholar]
- 14.Gueimonde, M., L. Noriega, A. Margolles, C. G. de los Reyes-Gavilán, and S. Salminen. 2005. Ability of Bifidobacterium strains with acquired resistance to bile to adhere to human intestinal mucus. Int. J. Food Microbiol. 101:341-346. [DOI] [PubMed] [Google Scholar]
- 15.Hopkins, M. J., J. H. Cumming, and G. T. Macfarlane. 1998. Inter-species differences in maximum specific growth rates and cell yields of bifidobacteria culture on oligosaccharides and other simple carbohydrate sources. J. Appl. Microbiol. 85:381-386. [Google Scholar]
- 16.Ji, G. E., K. K. Han, S. W. Yun, and S. L. Rhim. 1992. Isolation of amylolytic Bifidobacterium sp. Int57 and characterization of amylase. J. Microbiol. Biotechnol. 2:85-91. [Google Scholar]
- 17.Kaplan, H., and R. W. Hutkins. 2003. Metabolism of fructooligosaccharides by Lactobacillus paracasei 1195. Appl. Environ. Microbiol. 69:2217-2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krzewinski, F., C. Brassart, F. Gavini, and S. Bouquelet. 1996. Characterization of the lactose transport system in the strain Bifidobacterium bifidum DSM 20082. Curr. Microbiol. 32:301-307. [DOI] [PubMed] [Google Scholar]
- 19.Krzewinski, F., C. Brassart, F. Gavini, and S. Bouquelet. 1997. Glucose and galactose transport in Bifidobacterium bifidum DSM 20082. Curr. Microbiol. 35:175-179. [DOI] [PubMed] [Google Scholar]
- 20.Lanjendijk, P. S., F. Schut, G. J. Jansen, G. C. Raangs, G. R. Kamphuis, M. H. F. Wilkinson, and G. W. Welling. 1995. Quantitative fluorescence in situ hybridisation of Bifidobacterium spp. with genus-specific 16S rRNA-targeted probes and its application in fecal samples. Appl. Environ. Microbiol. 61:3069-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee, J. H., S. K. Lee, K. H. Park, I. K. Hwang, and G. E. Ji. 1999. Fermentation of rice using amylolytic Bifidobacterium. Int. J. Food Microbiol. 50:155-161. [Google Scholar]
- 22.Lee, S. K., Y. B. Kim, and G. E. Ji. 1997. Purification of amylase secreted from Bifidobacterium adolescentis. J. Appl. Microbiol. 83:267-272. [DOI] [PubMed] [Google Scholar]
- 23.Marx, S. P., S. Winkler, and W. Hartmeier. 2000. Metabolization of β-(2-6)-linked fructose-oligosaccharides by different bifidobacteria. FEMS Microbiol. Lett. 182:163-169. [DOI] [PubMed] [Google Scholar]
- 24.Meile, L., L. M. Rohr, T. A. Geissmann, M. Herensperger, and M. Teuber. 2001. Characterisation of the d-xylulose-5-phosphate/d-fructose-6-phosphate phosphoketolase gene (xfp) from Bifidobacterium lactis. J. Bacteriol. 183:2929-2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neefs, J. M., Y. van de Peer, P. de Rijk, S. Chapelle, and R. de Wachter. 1993. Compilation of small ribosomal subunit RNA structures. Nucleic Acids Res. 21:3025-3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Noriega, L., M. Gueimonde, B. Sánchez, A. Margolles, and C. G. de los Reyes-Gavilán. 2004. Effect of the adaptation to high bile salt concentrations on glycosidic activity, survival at low pH and cross-resistance to bile salts in Bifidobacterium. Int. J. Food Microbiol. 94:79-86. [DOI] [PubMed] [Google Scholar]
- 27.Olano-Martín, E., G. R. Gibson, and R. A. Rastell. 2002. Comparison of the in vitro bifidogenic properties of pectins and pectic-oligosaccharides. J. Appl. Microbiol. 93:505-511. [DOI] [PubMed] [Google Scholar]
- 28.Ouwehand, A. C., S. Salminen, and E. Isolauri. 2002. Probiotics: an overview of beneficial effects. Antonie Leeuwenhoek 82:279-289. [PubMed] [Google Scholar]
- 29.Palframan, R. J., G. R. Gibson, and R. A. Rastall. 2003. Carbohydrate preferences of Bifidobacterium species isolated from the human gut. Curr. Issues Intest. Microbiol. 4:71-75. [PubMed] [Google Scholar]
- 30.Perrin, S., J. P. Grill, and F. Schneider. 2000. Effects of fructooligosaccharides and their monomeric components on bile salt resistance on three species of bifidobacteria. J. Appl. Microbiol. 88:968-974. [DOI] [PubMed] [Google Scholar]
- 31.Perrin, S., M. Warchol, J. P. Grill, and F. Schneider. 2001. Fermentations of fructo-oligosaccharides and their components by Bifidobacterium infantis ATCC 15697 on batch culture in semi-synthetic medium. J. Appl. Microbiol. 90:859-865. [DOI] [PubMed] [Google Scholar]
- 32.Sánchez, B., L. Noriega, P. Ruas-Madiedo, C. G. de los Reyes-Gavilán, and A. Margolles. 2004. Acquired resistance to bile increases fructose-6-phosphate phosphoketolase activity in Bifidobacterium. FEMS Microbiol. Lett. 235:35-41. [DOI] [PubMed] [Google Scholar]
- 33.Sghir, A., J. M. Chow, and R. I. Mackie. 1998. Continuous culture selection of bifidobacteria and lactobacilli from human faecal samples using fructo-oligosaccharide as selective substrate. J. Appl. Microbiol. 85:769-777. [DOI] [PubMed] [Google Scholar]
- 34.Shah, N. P. 2000. Probiotic bacteria: selective enumeration and survival in dairy foods. J. Dairy Sci. 83:894-907. [DOI] [PubMed] [Google Scholar]
- 35.Strobel, H. J., F. C. Caldwell, and K. A. Dawson. 1995. Carbohydrate transport by the anaerobic thermophile Clostridium thermocellum LQRI. Appl. Environ. Microbiol. 61:4012-4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tannock, G. W. 1995. Microecology of the gastrointestinal tract in relation to lactic acid bacteria. Int. Dairy J. 5:1059-1070. [Google Scholar]
- 37.Topping, D. L., and P. M. Clifton. 2001. Short-chain fatty acids and human colonic function: roles of resistant starch and non-starch polysaccharides. Physiol. Rev. 81:1031-1064. [DOI] [PubMed] [Google Scholar]
- 38.Van der Meulen, R., L. Avonts, and L. de Vuyst. 2004. Short fractions of oligofructose are preferentially metabolized by Bifidobacterium animalis DN-173 010. Appl. Environ. Microbiol. 70:1923-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van Laere, K. M. J., T. Abee, H. A. Schols, G. Beldman, and A. G. J. Voragen. 2000. Characterization of a novel β-galactosidase from Bifidobacterium adolescentis DSM 20083 active towards transgalactooligosaccharides. Appl. Environ. Microbiol. 66:1379-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ventura, M., and R. Zink. 2002. Rapid identification, differentiation, and proposed new taxonomic classification of Bifidobacterium lactis. Appl. Environ. Microbiol. 68:6429-6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ventura, M., and R. Zink. 2003. Comparative sequence analysis of the tuf and recA genes and restriction fragment length polymorphism of the internal transcribed spacer region sequences supply additional tools for discriminating Bifidobacterium lactis from Bifidobacterium animalis. Appl. Environ. Microbiol. 69:7517-7522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang, X., and R. Gibson. 1993. Effects of the in vitro fermentation of oligofructose and inulin by bacteria growing in the human large intestine. J. Appl. Bacteriol. 75:373-380. [DOI] [PubMed] [Google Scholar]
- 43.Ward, L., J. Brown, and D. Graham. 1998. Two methods for the genetic differentiation of Lactococcus lactis and cremoris based on differences in the 16S rRNA gene sequence. FEMS Microbiol. Lett. 166:15-21. [DOI] [PubMed] [Google Scholar]





