Abstract
Acidithiobacillus ferrooxidans is one of the main acidophilic chemolithotrophic bacteria involved in the bioleaching of metal sulfide ores. The bacterium-mineral interaction requires the development of biofilms, whose formation is regulated in many microorganisms by type AI-1 quorum sensing. Here, we report the existence and characterization of a functional type AI-1 quorum-sensing system in A. ferrooxidans. This microorganism produced mainly acyl-homoserine lactones (AHL) with medium and large acyl chains and different C-3 substitutions, including 3-hydroxy-C8-AHL, 3-hydroxy-C10-AHL, C12-AHL, 3-oxo-C12-AHL, 3-hydroxy-C12-AHL, C14-AHL, 3-oxo-C14-AHL, 3-hydroxy-C14-AHL, and 3-hydroxy-C16-AHL. A quorum-sensing genetic locus that includes two open reading frames, afeI and afeR, which have opposite orientations and code for proteins with high levels of similarity to members of the acyl synthase (I) and transcriptional regulator (R) protein families, respectively, was identified. Overexpression of AfeI in Escherichia coli and the associated synthesis of AHLs confirmed that AfeI is an AHL synthase. As determined by reverse transcription-PCR, the afeI and afeR genes were transcribed in A. ferrooxidans. The transcription levels of the afeI gene were higher in cells grown in sulfur and thiosulfate media than in iron-grown cells. Phosphate starvation induced an increase in the transcription levels of afeI which correlated with an increase in AHL levels. Two afe boxes which could correspond to the AfeR binding sites were identified upstream of the afeI gene. This is the first report of a functional type AI-1 quorum-sensing system in an acidophilic chemolithotrophic microorganism, and our results provide a very interesting opportunity to explore the control and regulation of biofilm formation during the bioleaching process.
Quorum sensing (QS) is a widespread phenomenon that enables bacterial cells to establish cell-cell communication and to regulate the expression of specific genes in response to local changes in cell density (6, 48, 49). QS provides the means to coordinate the activities of cells so that they function as a multicellular unit and communicate with eukaryotic hosts (6, 20, 48, 49). In gram-negative bacteria, depending on the autoinductor (AI) molecule, two QS processes have been described: type AI-1, which is involved mainly in intraspecies communication, and type AI-2, which is related to interspecies communication (20, 49).
The type AI-1 QS regulatory system is composed of four elements: (i) a transcriptional regulator (protein family R); (ii) a cis-acting DNA palindromic sequence; (iii) an acyl-homoserine lactone (AHL), which is the signaling molecule or autoinducer (AI-1); and (iv) the AHL synthase protein (protein family I), which synthesizes the AI (6, 20, 48, 49). It is currently accepted that AI-1 diffuses freely between the cellular and external environments and complexes with the R protein only at a high cell density. The AHL-R complex binds through the R carboxyl domain to the specific site which corresponds to a palindromic sequence centered at about position −40 with respect to the transcriptional start sites of the target genes (15, 48, 49).
Acidithiobacillus ferrooxidans is an acidophilic gram-negative bacterium that is capable of oxidizing ferrous iron or reduced sulfur compounds to obtain energy for growth. Its energetic metabolism is directly involved in biomining processes. For this reason, there is great interest in understanding the molecular mechanisms of this peculiar physiological kind of life (34). During bioleaching A. ferrooxidans adheres to solid substrates by means of extracellular polymeric substances, such as exo- or lipopolysaccharides (36). A. ferrooxidans is also able to develop biofilm structures and exhibits morphological modifications during the cellular adhesion process (7, 13, 22, 36). AHL-mediated gene regulation has been shown to influence exopolysaccharide production and biofilm formation in many proteobacteria (12, 16, 21, 24, 49).
The purpose of the present work was to determine whether A. ferrooxidans possesses a functional type AI-1 quorum-sensing system. Different types of AHLs were found in the growth medium of A. ferrooxidans cultures. An A. ferrooxidans quorum-sensing locus was identified and designated afeIR. The afeI and afeR genes were expressed in A. ferrooxidans, and it was demonstrated that AfeI is an AHL synthase. Our results suggest that there is functional type AI-1 quorum sensing in A. ferrooxidans which could be part of a regulon controlling some physiological functions, such as exopolysaccharide synthesis and biofilm formation.
MATERIALS AND METHODS
Bacterial strains and growth media.
A. ferrooxidans ATCC 23270 was grown in ferrous iron-containing modified 9K medium at pH 1.5 as described previously (4), and organisms were grown on elemental sulfur at pH 2.5 with 5% (wt/vol) sulfur prills (3, 4). A. ferrooxidans was grown on thiosulfate at pH 4.6 in DSMZ medium 71 containing 20 mM thiosulfate as described previously (32). Escherichia coli strain BL21(DE3) and derivatives of this strain were grown in Luria-Bertani (LB) medium (37).
Preparation of crude AHL extracts and LC-MS-MS analysis.
The AHLs and oxo-AHLs were extracted and characterized as described previously (29). Briefly, after the bacterial cells were removed from the media of grown cultures by centrifugation, the supernatants were extracted twice with 1 volume of high-performance liquid chromatography (HPLC)-grade dichloromethane. The dichloromethane extracts were dried over anhydrous magnesium sulfate, filtered, and evaporated to dryness. Residues were dissolved in 1 ml of HPLC-grade acetonitrile and analyzed by using reverse-phase liquid chromatography coupled with positive-ion electrospray ionization and ion trap mass spectrometry (LC-MS-MS) (29). The 3-hydroxy-AHLs were analyzed using the same protocol. These compounds were identified by comparison with synthetic 3-hydroxy-AHLs based on three criteria: the MS-MS fragmentation product ions ([M+H-H2O]+ and m/z 102), their relative intensities, and the HPLC retention times.
AHL bioassays.
The different bioassay steps were performed as described previously (39). A 250-μl portion of an overnight culture of the Agrobacterium tumefaciens NTL4(pZLR4) AHL reporter strain grown in LB medium with gentamicin was inoculated into AB minimal glucose medium (ABm) and grown for 8 h at 30°C with shaking. The culture was then mixed with an equal volume of 1.5% TOP agar containing 80 μg/ml 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal), and the preparation was used as an overlay on ABm agar plates. The dichloromethane extracts obtained from the A. ferrooxidans media were spotted at the center of the overlaid ABm agar plates and incubated overnight at 30°C.
Bioinformatic tools.
The BLAST program (2) was used to search for open reading frames (ORFs) encoding LuxR and LuxI homologs in the genome sequence of A. ferrooxidans ATCC 23270. The search for afe boxes was done at the Genomic and Bioinformatic Center, Catholic University of Chile (www.cgb.cl). Palindromic sequences were detected in the intergenic region of the afeIR locus by using the algorithm bl2seq from the BLAST software (41). Hidden Markov models (HMM) were constructed with the results obtained by using the HMMER v2.3.2 software (14). Hidden Markov models were compared with the genome sequence to identify afe boxes.
For homology modeling and evaluation of AfeR and AfeI protein structures, 130 models were constructed for each protein with the MODELLER program (28). The templates were chain C of the TraR structure of A. tumefaciens (PDB ID no. 1L3L), determined by X-ray diffraction at 1.66-Å resolution, and the LasI structure of Pseudomonas aeruginosa (PDB ID no. 1RO5), determined by X-ray diffraction at 2.3-Å resolution (17, 44, 50). All models were evaluated using the Verify-3D program (27). The models with the highest three-dimensional profile scores were finally selected.
Cloning the afeI gene.
The afeI gene was amplified from A. ferrooxidans chromosomal DNA by PCR using the following primers: 5′-CATATGCAGGTTATAACCGGGCCA-3′ (5′ end) and 5′-CGGTTAGTCCAGATCTATCCAGC-3′ (3′ end). The 5′ end primer included an NdeI restriction site (underlined). The PCR was performed by using 3 min at 95°C, followed by 30 cycles of 30 s at 95°C, 30 s at 56°C, and 40 s at 72°C and finally 3 min at 72°C. The PCR fragment was purified from agarose gels with Wizard PCR Preps from Promega and cloned in pCRT7-NT-TOPO (Invitrogen) according to the manufacturer's recommendations. The different plasmid constructions were checked by automatic DNA sequencing at the Biotechnology Center of the Faculty of Sciences, University of Chile.
RNA manipulations.
A ferrooxidans ATCC 23270 total RNA was prepared from thiosulfate-, iron- or sulfur-grown cells by a modified hot-phenol method as described previously (19, 45). Primer extension was performed with the Superscript II RNase H reverse transcriptase (Invitrogen) by using 15 μg of total RNA and the AfeIRT2 primer (5′-GGAAAGATCTCGCCCAACAG-3′) labeled with [γ- 32P]ATP. The sequencing reaction was performed with the fmoI DNA cycle sequencing system as described by Promega.
The following synthetic oligonucleotides were used in reverse transcription (RT)-PCR assays for the reverse transcription step: 5′-CGGTTAGTCCAGATCTATCCAGC-3′, 5′-CGATCACGACAGCAACCCGAGCA-3′, and 5′-GATGTTGCTTCGTGGGAATC-3′ for the afeI, afeR, and orf3 genes, respectively. For PCRs, the following primers were used: for afeI, 5′-CATATGCAGGTTATAACCGGGCCA-3′ and 5′-CGGTTAGTCCAGATCTATCCAGC-3′; for afeR, 5′-CATATGGCGTCCGAAATGGCGCGT-3′ and 5′-AGGTCAACATGCCGCCCATC-3′; and for orf3, 5′-CGGACAAAAGATGCACCAGA-3′ and 5′-GGTAGCCTGTTCTTATCCGA-3′. The RT step was carried out with 1 μg of DNase I-treated total RNA. PCRs were done using 3 μl and 5 μl of the afeI and afeR RT reaction mixtures, respectively. The following program was used to perform the PCRs: an initial denaturation step of 95°C for 3 min; 40 cycles of 30 s at 95°C, 30 s at 60°C, and 30 s at 72°C; and a final extension step of 72°C for 3 min. For each RT-PCR experiment, a control RT reaction without reverse transcriptase was carried out to check for the absence of genomic DNA contamination in the RNA preparations used. RT-PCR products were checked by electrophoresis in a 1% agarose gel in 0.5× Tris-acetate-EDTA buffer.
Macroarray analysis.
Macroarray production was performed manually by using the colony copier VP381 (V&P Scientific). The PCR products were printed onto Inmobilon-NY+ membranes (Millipore). The different steps for expression analysis were performed as described previously (1). The afeI and afeR genes were amplified with the following oligonucleotide pairs: 5′-CATATGCAGGTTATAACCGGGCCA-3′ and 5′-CGGTTAGTCCAGATCTATCCAGC-3′ for the afeI gene and 5′-CATATGGCGTCCGAAATGGCGCGT-3′ and 5′-CGATCACGACAGCAACCCGAGCA-3′ for the afeR gene. Exposed PhosphorzImager screens were scanned with a PhosphorImager (Molecular Imager FX Systems, Bio-Rad) at a resolution of 50 μm/pixel. To normalize and quantify the results, we used a “spiked” RNA (exp-1 of Prunus persica) as an internal control for the labeling reaction and hybridization steps.
Nucleotide sequence accession number.
The nucleotide sequence of the afeIR locus has been deposited in the EMBL database under accession number no. AJ879454.
RESULTS AND DISCUSSION
Characterization of AHLs produced by A. ferrooxidans.
Extracts obtained from the media of grown cultures were analyzed with a bioassay in solid medium using the following two reporter strains: A. tumefaciens NTL4(pZLR4), which is specific for AHLs having medium and long acyl chains; and Chromobacterium violaceum CV026, which is specific for AHLs having short acyl chains and is inhibited by long-chain AHLs. No positive results were obtained with C. violaceum, while the bioassays with NTL4(pZLR4) revealed the characteristic formation of a blue halo (not shown). This screening procedure showed that A. ferrooxidans produces AHLs with medium and/or long acyl chains. To determine precisely the chemical structure of the AHLs synthesized, the extracts were analyzed by LC-MS-MS.
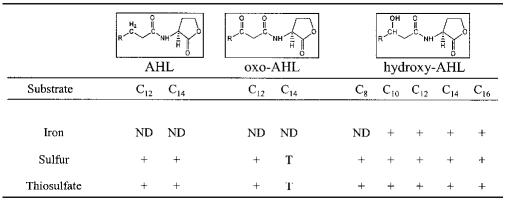
The dichloromethane extracts obtained from A. ferrooxidans grown in different media contained AHLs with diverse C-3 substitutions and only even numbers of carbons in the acyl chain. 3-Hydroxy-C10-, 3-hydroxy-C12-, 3-hydroxy-C14-, and 3-hydroxy-C16-AHLs were present in iron-grown cells; C12-, C14-, 3-oxo-C12-, 3-oxo-C14-, 3-hydroxy-C8, 3-hydroxy-C10-, 3-hydroxy-C12-, 3-hydroxy-C14-, and 3-hydroxy-C16-AHLs were present in sulfur-grown cells; and C12-, C14-, 3-oxo-C12, 3-oxo-C14-, 3-hydroxy-C8-, 3-hydroxy-C10-, 3-hydroxy-C12-, 3-hydroxy-C14-, and 3-hydroxy-C16-AHLs were present in thiosulfate-grown cells (Table 1). In our experimental conditions, A. ferrooxidans was able to produce nine different kinds of AHLs which included all the known types of C-3 substitutions (oxo and hydroxyl), and all of them had medium or long acyl chains with an even number of carbons. For the five types of 3-hydroxy-AHL and independent of the medium, the large acyl chains with 12 or 14 carbons were the predominant forms.
TABLE 1.
Identification of the different AHLs produced by A. ferrooxidansa
C8, R—CH3(CH2)4; C10, R—CH3(CH2)6; C12, R—CH3(2)8; C14, R—CH3(CH2)12; ND, not detected; T, trace.
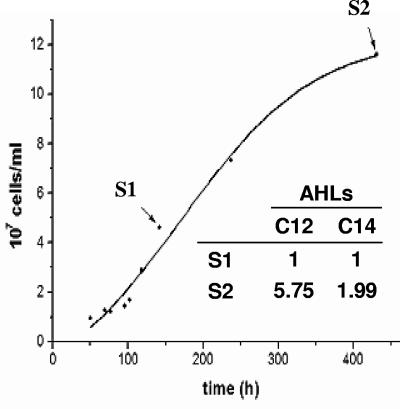
To determine whether some of these AHLs were produced in a cell density-dependent manner, as the quorum sensing paradigm requires, the presence of these compounds was analyzed in the three media obtained from A. ferrooxidans cultures in the early exponential and early stationary growth phases. Of the nine AHLs synthesized in A. ferrooxidans grown in sulfur medium, only two (C12-AHL and C14-AHL) showed a detectable increase in the early stationary phase (Fig. 1, inset). Similar results were obtained with C12-AHL and 3-hydroxy-C8-, C10-, and C12-AHLs in thiosulfate medium (results not shown). On the other hand, in iron medium, the 3-hydroxy-AHL levels appeared to be low and constant (result not shown).
FIG. 1.
Production of acyl-AHLs by A. ferrooxidans cells grown in sulfur. The correlation between growth phase and AHL amount was analyzed. AHLs were extracted from the culture medium with dichloromethane during the early exponential (S1) and stationary (S2) phases. The relative quantities of the large-acyl-chain AHLs produced are shown in the inset.
It is commonly accepted that AHL synthases can synthesize more than one type of AHL. Some Rhizobium spp. produce high numbers of AHLs by using AHL synthase-encoding genes located in the bacterial chromosome and/or plasmids (16). On the other hand, the AHL synthase SinI from Sinorhizobium meliloti Rm1021 is able to synthesize five different kinds of AHLs which involve different types of C-3 substitutions, a monounsaturated acyl chain, and the largest characterized acyl chain (C18) (16).
Family I proteins catalyze the synthesis of AHLs from both substrates, S-adenosylmethionine (SAM) and acylated acyl carrier protein (acyl-ACP). In addition to the putative structural specificity of each AHL synthase, the capacity to synthesize different kinds of AHLs has been associated with the available pool of acyl-ACP substrates in each microorganism (47). Recently, for Erwinia sp., it has been postulated that growth conditions could also affect the acyl-ACP availability (8). This could explain why C12- and C14-AHLs and oxo-C12- and oxo-C14-AHLs were produced only in sulfur- and thiosulfate-grown cells. However, the way in which the energy source (iron versus sulfur or thiosulfate) could affect the nature of the AHLs is still unknown, since no information relating external acidic pH, energy metabolism, and cell wall metabolism is currently available for A. ferrooxidans. Therefore, the nature of the pool of acyl-ACPs in A. ferrooxidans is unknown. Our results are the first results to suggest that hydroxy-acyl-ACPs with medium and large acyl chains are present in A. ferrooxidans irrespective of the energy source. With regard to the second precursor, little is known about sulfur metabolism and the related pool of SAM in bacteria living under extreme conditions, such as acid pH. Recently, it was suggested that A. ferrooxidans could regulate sulfur assimilation in a manner comparable to the manner described for other bacteria (43). Since the first step in sulfur assimilation corresponds to sulfate uptake, the pool of SAM in A. ferrooxidans should depend on sulfate availability. Sulfate is present at high concentrations in all the different media used to grow A. ferrooxidans, and it cannot be assumed that sulfate is a limiting factor. Therefore, AHL synthesis could not be affected under the three growth conditions that we employed.
Identification and characterization of AfeI, a LuxI homolog.
Some bacteria possess various loci involved in AHL synthesis (16, 20, 49). On the other hand, three AHL synthase families have been characterized (16). To determine how many loci for AHL production were present in the genomic sequence of A. ferrooxidans ATCC 23270, a search for ORFs encoding AHL synthases was performed. The amino acid sequences of LuxI (accession no. AAA27552) and AinS (AAP33508) from Vibrio fischeri and HdtS (AAG30826) from Pseudomonas fluorescens, which belong to the known AHL synthase families 1, 2, and 3, respectively (16), were used as queries in the tblastn search.
Orthologs were found only for AHL synthases belonging to families 1 and 3. The first gene coded for a putative protein with 53% similarity to HdtS, the first identified member of the third AHL synthase family. In P. fluorescens, HdtS directs the synthesis of three AHLs, C6-AHL, C10-AHL, and N-(3-hydroxy-7-cis-C14)-AHL (25).
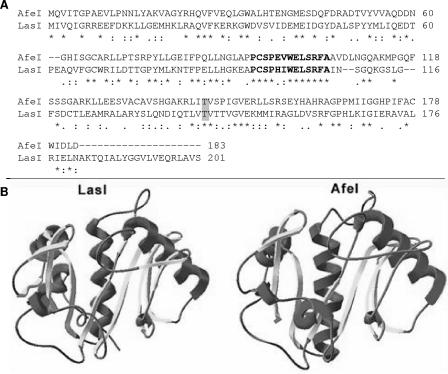
The second gene coded for a protein that is 64% similar to protein BveI from Burkholderia cepacia, a member of the LuxI family. The deduced protein of A. ferrooxidans was designated AfeI, and it had 183 amino acids, a molecular mass of 19.9 kDa, and a theoretical isoelectric point of 5.77. Based on the recently solved structure of the LasI protein of P. aeruginosa (17), we constructed an AfeI model structure (Fig. 2). Our modeling approach took advantage of the higher level of similarity between AfeI and LasI (56%) (Fig. 2A). The amino acid sequence of AfeI revealed the presence of a threonine residue (Fig. 2A) which is involved in the oxo-C3 substitution in the AHLs synthesized by LuxI, EsaI, and LasI of V. fischeri, Pantonea stewwartii subsp. stewartii, and P. aeruginosa, respectively (47). In agreement with this, we also characterized oxo-AHLs in the growth media of A. ferrooxidans. The highest three-dimensional profile score yielded an AfeI structure model with high structural similarity to the LasI structure (Fig. 2B).
FIG. 2.
Structure modeling of AfeI. (A) LasI and AfeI amino acid sequence alignment used for homology modeling. Boldface type indicates the most conserved region in the two proteins. Identical residues are indicated by asterisks, and similar residues are indicated by colons. A residue involved in oxo substitution (47) is indicated by a gray box. (B) AfeI modeled structure (right) based on LasI solved structure (left).
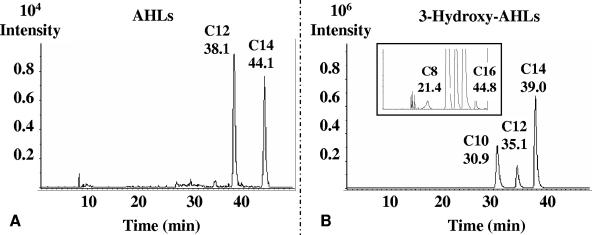
To confirm that the afeI gene product was an AHL synthase, afeI was overexpressed in an E. coli strain which does not produce AHLs. The afeI gene was cloned in plasmid pCRT7-NT-TOPO and transformed into the E. coli BL21(DE3) strain. The recombinant strain was induced with 0.4 M isopropyl-β-d-thiogalactopyranoside (IPTG) for 1 h, and the media of the induced bacteria were extracted with dichloromethane and analyzed by LS-MS-MS (Fig. 3). C12-AHL and C14-AHL (Fig. 3A) and five different hydroxy-AHLs (C8, C10, C12, C14, and C16) (Fig. 3B) were characterized, while in the control E. coli strain carrying the same vector without the insert no AHLs were found (results not shown). The more abundant hydroxy-AHLs were C10, C12, and C14, in agreement with the results obtained for A. ferrooxidans cells. This result definitively confirmed that AfeI is an AHL synthase able to synthesize AHLs and hydroxy-AHLs with large acyl chains (including C16), depending on the available pool of acyl-ACPs. No other homoserine lactone synthase-encoding gene (except the htdS-like gene, whose function in the quorum-sensing pathway is still unknown) was identified in the available genome sequence of A. ferrooxidans. In S. meliloti, the AHL synthase SinI is responsible for the synthesis of a series of long-chain AHLs ranging in size from C12-AHL to C18-AHL, including some oxo-AHLs and a monounsaturated AHL (16). Therefore, we concluded that AfeI could be responsible for the synthesis of various long-chain AHLs in A. ferrooxidans.
FIG. 3.
Characterization of the AHLs produced by AfeI in E. coli carrying the afeI gene. E. coli transformed with plasmid pCRT7-NT-TOPO with the afeI gene was grown in LB medium. After the cells were removed, the growth medium was extracted with dichloromethane, and the presence of AHLs was analyzed by LS-MS-MS. The different MS-MS spectra of the selected ion m/z 102, which is specific for the lactone ring of the homoserine lactone, are shown. AHLs (A) and hydroxy-AHLs (B) were characterized. The large amounts of hydroxy-C10, hyrodxy-C12-, and hydroxy-C14-AHLs (B) made it necessary to present this spectrum with two different intensity scales (106 for the common size and 104 for the inset). The designation of each AHL and the different elution times are indicated in each spectrum.
Organization and transcription of the afeI-afeR locus.
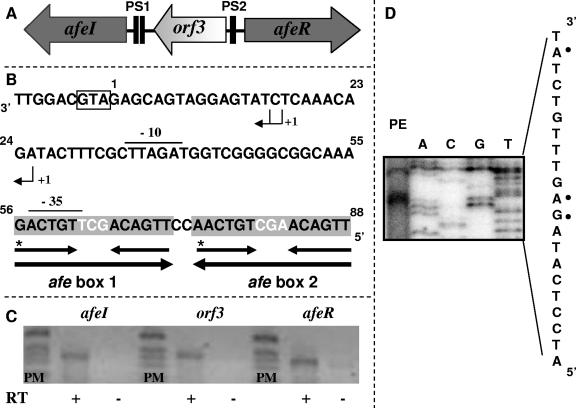
At a position 746 bp downstream of the afeI gene and in the opposite orientation, we found an ORF which exhibited 59% similarity to BviR, an R protein family member from B. cepacia. The deduced protein was designated AfeR and had 214 amino acids, a molecular mass of 23.7 kDa, and an isoelectric point of 9.89. Despite the low level of amino acid identity with TraR (less than 25%), we constructed a protein structure model for AfeR, which was very similar to the TraR structure (results not shown). The high level of structural similarity between the two proteins strongly suggests that AfeR is a transcriptional regulator of the R protein family, and, most likely, it acts in a dimeric form by recognizing lux-type boxes. A third ORF designated orf3 was identified in the afeIR intergenic region. orf3 had the same orientation as afeI, and it coded for a protein with 51% similarity to a hypothetical protein from B. cepacia R18194. The corresponding genes (afeI, orf3, and afeR) formed the A. ferrooxidans quorum-sensing locus (afeIR) shown in Fig. 4A. Despite the fact that A. ferrooxidans is a γ-proteobacterium, higher similarity scores for AfeI and AfeR (69% and 66%, respectively) were obtained with the genomic data for other members of the Burkholderiaceae belonging to the β-proteobacterial subdivision. This is in agreement with the idea of coevolution and acquisition of the IR regulatory cassettes by horizontal transmission (18).
FIG. 4.
Quorum-sensing genetic locus of A. ferrooxidans. (A) Schematic map of the quorum-sensing locus of A. ferrooxidans composed of three genes: afeR encoding the transcriptional regulator, afeI encoding the AHLs synthase, and orf3 having an unknown function. Two palindromic sequences (PS1 and PS2) were located (solid boxes). (B) Nucleotide sequence of the putative afe boxes. The large palindromic sequence (PS1) was conformed by a 32-bp palindromic sequence (large arrows) which was built over internal, hierarchical and smaller palindromic sequences (small arrows) called afe box 1 and afe box 2. The box at the 3′ end indicates the translational start codon for afeI. +1 indicates the transcriptional initiation sites identified; overlining indicates the E. coli σ70-type promoter; and asterisks indicate the purine base transition at the 3′ ends of the afe boxes. (C) Transcription analysis of the quorum-sensing genetic locus by RT-PCR analysis. RT reactions were carried out with 1 μg of total RNA from thiosulfate-grown A. ferrooxidans cells and were performed with (+) and without (−) the Moloney murine leukemia virus reverse transcriptase in order to exclude amplification due to genomic DNA contamination. RT-PCR products were analyzed by 1% agarose gel electrophoresis. (D) Determination of transcriptional initiation sites for the afeI gene by a primer extension experiment. The relevant DNA sequence (complementary to the sequence shown in panel B between nucleotides 8 and 27) is shown on the right, and the positions of the possible start sites are indicated by solid dots.
Intergenic sequence analysis of the afeIR locus revealed two palindromic sequences, PS1 and PS2 (Fig. 4A). PS2 was located 36 bp upstream of orf3 and 168 bp from afeR, and its function is unknown. The analysis of the PS1 sequence revealed a hierarchical organization (Fig. 4B). Each arm of the 30-bp palindromic sequence could be subdivided into two identical palindromic sequences whose sizes (15 bp) were similar to that of the known lux box. The nucleotide sequences of the two afe boxes were identical except for the three central bases (Fig. 4B), suggesting that these bases are not essential for AfeR binding but could play a role as a physical spacer for the binding of the dimeric form of AfeR. In the 3′ end of each afe box, a purine base was conserved. In addition, each afe box could be sufficient to bind a dimeric form of AfeR, as suggested by the modeling of the (AfeR)2-afe box complex based on the (TraR-AHL)2-DNA structure data (not shown). We designated these boxes afe box 1 and afe box 2 (Fig. 4B) since they could correspond to the typical type AI-1 QS regulator binding sites located close to a putative −35 transcriptional site and could play roles similar to those of lux and tra boxes (15, 48).
To study if the afeIR locus was functional in A. ferrooxidans, gene expression was analyzed by RT-PCR. Total RNA was prepared from planktonic cells of A. ferrooxidans grown in thiosulfate medium. The results clearly showed that afeI, orf3, and afeR were expressed in A. ferrooxidans (Fig. 4C). Nevertheless, in our experimental conditions orf3 and afeI were not cotranscribed.
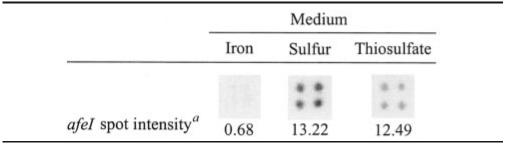
As the AHLs produced in the presence of iron, sulfur, or thiosulfate were different (Table 1), we analyzed the transcription levels of the afeI gene in the different media in DNA macroarray experiments. The afeI transcription levels in thiosulfate and sulfur media were similar. Compared to iron-grown cells, transcription of the afeI gene was increased 19- and 18-fold in sulfur- and thiosulfate-grown cells, respectively (Table 2). In iron medium, the transcription level was very low. This could explain the lack of detection of C12- and C14-AHLs and oxo-C12- and oxo-C14-AHLs (Table 1). Interestingly, the increase in the transcription levels of the afeI gene was not related to the transcription levels of afeR since the expression of this gene did not change when iron- and sulfur-grown cells were compared (results not shown).
TABLE 2.
Effect of the energy source on the transcription levels of the afeI gene
Relative intensity.
To highlight the afe box function, the 5′ end of the afeI gene transcript was determined by primer extension analysis with RNA samples prepared from A. ferrooxidans cells grown in thiosulfate as the energy source. Two transcriptional initiation sites were determined with oligonucleotide AfeIRT2 (Fig. 4D). The first site had a double transcription initiation site (G and A), while the second site had a single transcription initiation site (A) (Fig. 4B). The two transcription initiation sites were located 17 or 18 bp and 28 bp, respectively, upstream of the translational start codon of afeI (Fig. 4B). An E. coli σ70-type promoter (5′-TTGTCA-16 bp-TAGATT-3′) was identified and correctly positioned upstream of the second transcription initiation sites (Fig. 2B). As observed for the lux and tra boxes, afe box 1 overlapped the −35 transcriptional region determined (Fig. 4B).
Based on the lux box-tra box models, we decided to investigate the presence of the afe boxes (Fig. 4B) in the entire genome sequence of the ATCC 23270 strain. By using hidden Markov models constructed with the HMMER v2.3.2 software (14), we characterized various putative afe boxes (not shown). However, our results are preliminary results which only suggest the existence of a quorum-sensing regulon in A. ferrooxidans. Proteomic and transcriptomic studies like those performed with other bacteria (P. aeruginosa, B. cepacia H111, and S. meliloti) should help demonstrate the existence of a QS regulon in A. ferrooxidans (5, 10, 30, 35, 38, 46).
Phosphate starvation activates transcription of afeI.
Different results have revealed that type AI-1 QS influences exopolysaccharide production and biofilm formation in many proteobacteria (16, 21, 24, 49), and phosphate and polyphosphate metabolism has been linked to biofilm formation and the quorum-sensing regulatory pathway (26, 33, 40). On the other hand, in Serratia sp. strain ATCC 390006, mutation of the pstS gene, which belongs to the Pho regulon and whose product is part of a high-affinity phosphate transporter, mimicked phosphate limitation and caused a three- to fourfold increase in transcription of the AHL synthase-encoding gene (smaI) of Serratia type AI-I QS (40).
During bioleaching A. ferrooxidans adheres to solid substrates by means of extracellular polymeric substances, such as exo- or lipopolysaccharides, and develops biofilm structures during the cellular adhesion process (7, 13, 22, 36). Previous immunological results have demonstrated that the amounts of lipopolysaccharides which are part of the polysaccharide matrix involved in ore colonization are increased in phosphate-starved A. ferrooxidans cells (3), and a pho regulon has been described in A. ferrooxidans (45). Therefore, the effect of phosphate starvation on the transcription of the afeI gene was investigated by using a DNA macroarray analysis.
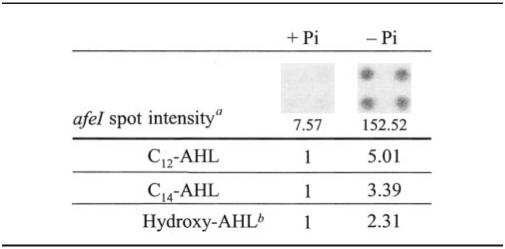
Transcription of the afeI gene was increased 20-fold when A. ferrooxidans cells were cultured in a low-phosphate medium (−Pi medium; Pi concentration, 0.22 mM) compared to a high-phosphate medium (+Pi medium; Pi concentration, 22 mM) (Table 3). The overexpression of the afeI gene in phosphate-starved cells was directly correlated with increases in the C12-AHL, C14-AHL, and hydroxy-AHL levels (Table 3). These results are in agreement with those obtained with the smaI gene of Serratia sp. (40). Therefore, the AfeIR quorum-sensing system appears to be modulated by Pi availability. Slater et al. (40) suggested that the expression of smaI could be enhanced through the two-component regulatory system PhoR-PhoB. In A. tumefaciens phosphate-starved cells, biofilm formation is positively affected through the PhoR-PhoB regulatory pathway (11). Nevertheless, how these different regulatory levels are related and affect biofilm formation in A. ferrooxidans is still an open question.
TABLE 3.
Effect of the phosphate starvation on afeI expression and AHL synthesis in thiosulfate-grown cells
Relative intensity.
Average values including hydroxy-C10, hydroxy-C14, and hydroxy-C16-AHLs.
Recently, preliminary evidence for the occurrence of homoserine lactone signal production in archaea has been reported (31). Some biomining microorganisms, such as Ferroplasma type II and Lepotspirillum ferrooxidans type II, are also able to form a natural and mixed acidophilic biofilm (42). Therefore, the existence of an AHL communication system in A. ferrooxidans and its impact on the biofilm structure and abundance of bacteria and archaea in biomining microbial communities are very relevant.
Obviously, further studies with A. ferrooxidans are necessary (i) to determine if AfeR is a positive transcriptional regulator or negative transcriptional regulator or both, as in the case of AhyR of Aeromonas hydrophila (23), (ii) to understand the biological significance of both afe boxes upstream of the afeI gene, (iii) to understand the role of the product of the orf3 gene, and (iv) to identify all the genes which form the quorum-sensing regulon. Since no genetic transfer techniques are currently available for A. ferrooxidans, strategies using afeI mutants or any kind of gene cloning are not possible. However, the use of proteomic and transcriptomic analyses of mimicked null mutants obtained with quorum-sensing inhibitors (9) should be a successful global approach for identification of all the components of the quorum-sensing regulon in A. ferrooxidans.
Acknowledgments
This work was supported by grants from Universidad de Chile (DID I-02/4-2), Fundación Andes, and FONDECYT (1040676).
We thank Juan Gonzalez, who kindly provided the reporter strains. We acknowledge Bruce Cassel's laboratory for assistance with acyl-AHL extract preparation. We thank Francisco Chavez and Simon Beard for critical reading of the manuscript and experimental support, respectively. We acknowledge Juan-Carlos Mobarec for structure modeling. Sequence data for A. ferrooxidans strain ATCC 23270 were obtained from the NCBI microbial genome site.
Footnotes
Dedicated to the memory of Jean-Noël Guiliani.
REFERENCES
- 1.Acosta, M., S. Beard, J. Ponce, M. Vera, J. C. Mobarec, and C. A. Jerez. 2005. Identification of putative sulfurtransferase genes in the extremophilic Acidithiobacillus ferrooxidans ATCC 23270 genome: structural and functional characterization of the proteins. OMICS 9:13-29. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amaro, A. M., M. Seeger, R. Arredondo, M. Moreno, and C. A. Jerez. 1993. The growth conditions affect Thiobacillus ferrooxidans attachment to solids, p. 577-585. In A. E. Torma, M. L. Apel, and C. L. Brierley (ed.), Biohydrometallurgical technologies. The Minerals, Metals and Material Society, Warrendale, Pa.
- 4.Amaro, A. M., D. Chamorro, M. Seeger, R. Arredondo, I. Peirano, and C. A. Jerez. 1991. Effect of external pH perturbations on in vivo protein synthesis by the acidophilic bacterium Thiobacillus ferrooxidans. J. Bacteriol. 173:910-915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arevalo-Ferro, C., M. Hentzer, G. Reil, A. Gorg, S. Kjelleberg, M. Givskov, K. Riedel, and L. Eberl. 2003. Identification of quorum-sensing regulated proteins in the opportunistic pathogen Pseudomonas aeruginosa by proteomics. Environ. Microbiol. 5:1350-1369. [DOI] [PubMed] [Google Scholar]
- 6.Bassler, B. 2002. Small talk: cell-to-cell communication in bacteria. Cell 109:421-424. [DOI] [PubMed] [Google Scholar]
- 7.Blais, J., R. Tyagi, N. Meunier, and J. Auclair. 1994. The production of extracellular appendages during bacterial colonization of elemental sulphur. Process Biochem. 29:475-482. [Google Scholar]
- 8.Brader, G., S. Sjoblom, H. Hyytiainen, K. Sims-Huopaniemi, and E. T. Palva. 2005. Altering substrate chain length specificity of an acylhomoserinelactone synthase in bacterial communication. J. Biol. Chem. 280:10403-10409. [DOI] [PubMed] [Google Scholar]
- 9.Castang, S., B. Chantegrel, C. Deshayes, R. Dolmazon, P. Gouet, R. Haser, S. Reverchon, W. Nasser, N. Hugouvieux-Cotte-Pattat, and A. Doutheau. 2004. N-Sulfonyl homoserine lactones as antagonists of bacterial quorum sensing. Bioorg. Med. Chem. Lett. 14:5145-5149. [DOI] [PubMed] [Google Scholar]
- 10.Chen, H., M. Teplitski, J. B. Robinson, B. G. Rolfe, and W. D. Bauer. 2003. Proteomic analysis of wild-type Sinorhizobium meliloti responses to N-acyl homoserine lactone quorum-sensing signals and the transition to stationary phase. J. Bacteriol. 185:5029-5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Danhorn, T., M. Hentzer, M. Givskov, M. R. Parsek, and C. Fuqua. 2004. Phosphorus limitation enhances biofilm formation of the plant pathogen Agrobacterium tumefaciens through the PhoR-PhoB regulatory system. J. Bacteriol. 186:4492-4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davies, D. G., M. R. Parsek, J. P. Pearson, B. H. Iglewski, J. W Costerton, and E. P. Greenberg. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280:295-298. [DOI] [PubMed] [Google Scholar]
- 13.DiSpirito, A., M. Silver, L. Voss and, O. Tuovinen. 1982. Flagella and pili of iron-oxidizing thiobacilli isolated from uranium mine in northern Ontario, Canada. Appl. Environ. Microbiol. 43:1196-1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eddy, S. R. 1998. Profile hidden Markov models. Bioinformatics 14:755-763. [DOI] [PubMed] [Google Scholar]
- 15.Fuqua, W. C., and S. Winans. 1996. Conserved cis-acting promoter elements are required for density-dependent transcription of Agrobacterium tumefaciens conjugal transfer genes J. Bacteriol. 178:435-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gonzalez, J. E., and M. M. Marketon. 2003. Quorum sensing in nitrogen-fixing rhizobia. Microbiol. Mol. Biol. Rev. 67:574-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gould, T. A., H. P. Schweizer, and M. E. Churchill. 2004. Structure of the Pseudomonas aeruginosa acyl-homoserinelactone synthase LasI. Mol. Microbiol. 53:1135-1146. [DOI] [PubMed] [Google Scholar]
- 18.Gray, K., and J. Garey. 2001. The evolution of bacterial LuxI and LuxR quorum sensing regulators. Microbiology 147:2379-2387. [DOI] [PubMed] [Google Scholar]
- 19.Guiliani, N., A. Bengrine, F. Borne, M. Chippaux, and V. Bonnefoy. 1997. Alanyl-tRNA synthetase gene of the extreme acidophilic chemolithoautotrophic Thiobacillus ferrooxidans is highly homologous to alaS genes from all living kingdoms but cannot be transcribed from its promoter in Escherichia coli. Microbiology 143:2179-2187. [DOI] [PubMed] [Google Scholar]
- 20.Henke, J. M., and B. L. Bassler. 2004. Bacterial social engagements. Trends Cell Biol. 14:648-656. [DOI] [PubMed] [Google Scholar]
- 21.Huber, B., K. Riedel, M. Hentzer, A. Heydorn, A. Gotschlich, M. Givskov, S. Molin, and L. Eberl. 2001. The cep quorum-sensing system of Burkholderia cepacia H111 controls biofilm formation and swarming motility. Microbiology 147:2517-2528. [DOI] [PubMed] [Google Scholar]
- 22.Karamanev, D. 1991. Model of the biofilm structure of Thiobacillus ferrooxidans. J. Biotechnol. 20:51-64. [Google Scholar]
- 23.Kirke, D. F., S. Swift, M. J. Lynch, and P. Williams. 2004. The Aeromonas hydrophila LuxR homologue AhyR regulates the N-acyl homoserine lactone synthase, AhyI, positively and negatively in a growth phase-dependent manner. FEMS Microbiol. Lett. 241:109-117. [DOI] [PubMed] [Google Scholar]
- 24.Labbate, M., S. Y. Queck, K. S. Koh, S. A. Rice, M. Givskov, and S. Kjelleberg. 2004. Quorum sensing-controlled biofilm development in Serratia liquefaciens MG1. J. Bacteriol. 186:692-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laue, B. E., Y. Jiang, S. R. Chhabra, S. Jacob, G. S. Stewart, A. Hardman, J. A. Downie, F. O'Gara, and P. Williams. 2000. The biocontrol strain Pseudomonas fluorescens F113 produces the Rhizobium small bacteriocin, N-(3-hydroxy-7-cis-tetradecenoyl)homoserine lactone, via HdtS, a putative novel N-acylhomoserine lactone synthase. Microbiology 146:2469-2480. [DOI] [PubMed] [Google Scholar]
- 26.Ledgham, F., C. Soscia, A. Chakrabarty, A. Lazdunski, and M. Foglino. 2003. Global regulation in Pseudomonas aeruginosa: the regulatory protein AlgR2 (AlgQ) acts as a modulator of quorum sensing. Res. Microbiol. 154:207-213. [DOI] [PubMed] [Google Scholar]
- 27.Lüthy, R., J. U. Bowie, and D. Eisenberg. 1992. Assessment of protein models with three-dimensional profiles Nature 356:83-85. [DOI] [PubMed] [Google Scholar]
- 28.Marti-Renom, M. A., A. C. Stuart, A. Fiser, R. Sánchez, F. Melo, and A. Sali. 2000. Comparative protein structure modeling of genes and genomes Annu. Rev. Biophys. Biomol. Struct. 29:291-325. [DOI] [PubMed] [Google Scholar]
- 29.Morin, D., B. Grasland, K. Vallee-Rehel, C. Dufau, and D. Haras. 2003. On-line high-performance liquid chromatography-mass spectrometric detection and quantification of N-acylhomoserine lactones, quorum sensing signal molecules, in the presence of biological matrices. J. Chromatogr. A 1002:79-92. [DOI] [PubMed] [Google Scholar]
- 30.Nouwens, A. S., S. A. Beatson, C. B. Whitchurch, B. J. Walsh, H. P. Schweizer, J. S. Mattick, and S. J. Cordwell. 2003. Proteome analysis of extracellular proteins regulated by the las and rhl quorum sensing systems in Pseudomonas aeruginosa PAO1. Microbiology 149:1311-1322. [DOI] [PubMed] [Google Scholar]
- 31.Paggi, R. A., C. B. Martone, C. Fuqua, and R. E. De Castro. 2003. Detection of quorum sensing signals in the haloalkaliphilic archaeon Natronococcus occultus. FEMS Microbiol. Lett. 221:49-52. [DOI] [PubMed] [Google Scholar]
- 32.Ramirez, P., N. Guiliani, L. Valenzuela, S. Beard, and C. A. Jerez. 2004. Differential protein expression during growth of Acidithiobacillus ferrooxidans on ferrous iron, sulfur compounds, or metal sulfides. Appl. Environ. Microbiol. 70:4491-4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rashid, M. H., K. Rumbaugh, L. Passador, D. G. Davies, A. N. Hamood, B. H. Iglewski, and A. Kornberg. 2000. Polyphosphate kinase is essential for biofilm development, quorum sensing, and virulence of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 97:9636-9641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rawlings, D. E. 2002. Heavy metal mining using microbes. Annu. Rev. Microbiol. 56:65-91. [DOI] [PubMed] [Google Scholar]
- 35.Riedel, K., C. Arevalo-Ferro, G. Reil, A. Gorg, F. Lottspeich, and L. Eberl. 2003. Analysis of the quorum-sensing regulon of the opportunistic pathogen Burkholderia cepacia H111 by proteomics. Electrophoresis 24:740-750. [DOI] [PubMed] [Google Scholar]
- 36.Rohwerder, T., T. Gehrke, K. Kinzler, and W. Sand. 2003. Bioleaching review part A: progress in bioleaching: fundamentals and mechanisms of bacterial metal sulfide oxidation. Appl. Microbiol. Biotechnol. 63:239-248. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook, J., and D. W. Russell. 1989. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 38.Schuster, M., C. P. Lostroh, T. Ogi, and E. P. Greenberg. 2003. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J. Bacteriol. 185:2066-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shaw, P. D., G. Ping, S. L. Daly, C. Cha, J. E. Cronan, Jr., K. L. Rinehart, and S. K. Farrand. 1997. Detecting and characterizing N-acyl-homoserine lactone signal molecules by thin-layer chromatography. Proc. Natl. Acad. Sci. USA 94:6036-6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Slater, H., M. Crow, L. Everson, and G. P. Salmond. 2003. Phosphate availability regulates biosynthesis of two antibiotics, prodigiosin and carbapenem, in Serratia via both quorum-sensing-dependent and -independent pathways. Mol. Microbiol. 47:303-320. [DOI] [PubMed] [Google Scholar]
- 41.Tatusova, T. A., and T. L. Madden. 1999. BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol. Lett. 174:247-250. [DOI] [PubMed] [Google Scholar]
- 42.Tyson, G. W., J. Chapman, P. Hugenholtz, E. E. Allen, R. J. Ram, P. M. Richardson, V. V. Solovyev, E. M. Rubin, D. S. Rokhsar, and J. F. Banfield. 2004. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428:37-43. [DOI] [PubMed] [Google Scholar]
- 43.Valdes, J., F. Veloso, E. Jedlicki, and D. Holmes. 2003. Metabolic reconstruction of sulfur assimilation in the extremophile Acidithiobacillus ferrooxidans based on genome analysis. BMC Genomics 4:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vannini, A., C. Volpari, C. Gargioli, E. Muraglia, R. Cortese, R. De Francesco, P. Neddermann, and S. Marco. 2002. The crystal structure of the quorum sensing protein TraR bound to its autoinducer and target DNA. EMBO J. 21:4393-4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vera, M., N. Guiliani, and C. A. Jerez. 2003. Proteomic and genomic analysis of the phosphate starvation response of Acidithiobacillus ferrooxidans. Hydrometallurgy 71:125-132. [Google Scholar]
- 46.Wagner, V. E., D. Bushnell, L. Passador, A. I. Brooks, and B. H. Iglewski. 2003. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J. Bacteriol. 185:2080-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Watson, W. T., T. D. Minogue, D. L. Val, S. B. von Bodman, and M. E. Churchill. 2002. Structural basis and specificity of acyl-homoserine lactone signal production in bacterial quorum sensing. Mol. Cell 9:685-694. [DOI] [PubMed] [Google Scholar]
- 48.Whitehead, N., A. Barnard, H. Slater, N. Simpson, and G. Salmond. 2001. Quorum sensing in Gram-negative bacteria. FEMS Microbiol. Rev. 25:365-404. [DOI] [PubMed] [Google Scholar]
- 49.Winans, S., and B. Bassler. 2002. Mob psychology. J. Bacteriol. 184:873-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang, R., T. Pappas, J. Brace, P. Miller, T. Oulmassov, J. Molyneaux, J. Anderson, J. Bashkin, S. Winans, and A. Joachimiak. 2002. Structure of a bacterial quorum-sensing transcription factor complexed with pheromone and DNA. Nature 417:971-974. [DOI] [PubMed] [Google Scholar]