Abstract
A nucleic acid sequence-based amplification (NASBA) assay in combination with a molecular beacon was developed for the real-time detection and quantification of hepatitis A virus (HAV). A 202-bp, highly conserved 5′ noncoding region of HAV was targeted. The sensitivity of the real-time NASBA assay was tested with 10-fold dilutions of viral RNA, and a detection limit of 1 PFU was obtained. The specificity of the assay was demonstrated by testing with other environmental pathogens and indicator microorganisms, with only HAV positively identified. When combined with immunomagnetic separation, the NASBA assay successfully detected as few as 10 PFU from seeded lake water samples. Due to its isothermal nature, its speed, and its similar sensitivity compared to the real-time RT-PCR assay, this newly reported real-time NASBA method will have broad applications for the rapid detection of HAV in contaminated food or water.
Outbreaks of acute gastroenteritis due to hepatitis A virus (HAV) have been attributed to consumption of drinking water and foods considered safe on the basis of bacterial standards (23). These outbreaks indicate that meeting bacterial standards does not always ensure the absence of infectious viruses, and more reliable approaches are needed to detect HAV and other enteroviruses in environmental samples.
Conventional methods for the detection of HAV are based on cell culture propagation, which is often difficult to perform and can take several weeks and continuous propagation in one or more cell lines before a sufficient amount of viral antigen or nucleic acid is produced to allow detection (5, 6). In addition, no single cell line is currently recommended for the detection of HAV (19, 22).
To date, molecular methods, such as the reverse transcription (RT)-PCR technique, are the most commonly studied, offering improved sensitivity, specificity, and the possibility of direct detection of HAV in environmental samples (11, 16). In RT-PCR, the viral RNA is first converted to a single-stranded complementary DNA in a reverse transcription step, followed by PCR amplification of the target complementary DNA sequences to a detectable level. However, RT-PCR procedures have the disadvantages of requiring a two-step amplification process and relying on the use of expensive thermal-cycling equipment, which add to the complexity and the cost of their implementation for routine testing programs.
Unlike RT-PCR, nucleic acid sequence-based amplification (NASBA) is a homogeneous, isothermal nucleic acid amplification method (17) that is particularly suited to RNA targets in a double-stranded DNA background (12). A cocktail of three enzymes (reverse transcriptase, T7 RNA polymerase, and RNase H) acting in concert allows the rapid amplification of target sequences by more than 108-fold without the use of expensive thermal-cycling equipment. NASBA has proved successful in the detection of various mRNAs (3, 9) and in the detection of both viral (17) and bacterial (21) RNAs. Diagnostic procedures based on NASBA methodology have been described for several viruses, including human immunodeficiency virus type 1 (10), cytomegalovirus (26), enterovirus (13), West Nile and St. Louis encephalitis viruses (17), parainfluenza virus (14), and hepatitis C virus (8).
Molecular beacons (MBs) are single-stranded nucleic acid sequences that possess a stem-loop structure that is double labeled with a fluorescent dye and a universal quencher at the 5′ and 3′ ends, respectively (25). By combining the standard NASBA technology with MB, a real-time detection system can be generated. During the NASBA reaction, the MB hybridizes to the target RNA, separating the reporter dye and the quencher and yielding a measurable fluorescence emission directly proportional to the concentration of the target sequence (20).
In the present study, the development of an MB-based real-time NASBA assay for the detection of hepatitis A virus was investigated. The sensitivity of the assay was evaluated using serial dilutions of HAV. The specificity of the procedure was tested against closely related viruses from the Picornaviridae. The possibility of combining such an assay with immunomagnetic separation (IMS) for the detection of the virus in surface water samples was also demonstrated.
MATERIALS AND METHODS
Virus strain and propagation.
A cytopathic HAV strain, HM175, was obtained from the ATCC (Manassas, Va.). Fetal rhesus monkey kidney (FRhK-4) cells were used for the propagation and titration of HAV. HAV titers were measured by performing plaque assays. Briefly, cell monolayers were grown overnight in 12-well culture plates at 37°C in the presence of 5% CO2. A 100-μl portion of the virus dilution was inoculated into each of three wells. The virus was allowed to be in contact with the cells for 90 min at 37°C in the presence of 5% CO2, and then 2 ml of semisolid agarose was overlaid upon each well. The plates were incubated at 37°C in the presence of 5% CO2 for 8 days. This was followed by fixation and staining for plaque counting as described previously (7). Other viruses used in this study (e.g., coxsackievirus B6, echovirus 11, bacteriophage φX174, poliovirus 1 LscAb [ATCC], rotavirus SA11, and echovirus 19) were grown separately on their specific mammalian or bacterial host cells.
RNA purification.
Total viral RNA was extracted by the phenol-chloroform method (22, 24). Briefly, 100 μl of hepatitis A virus-infected cell culture was mixed with 100 μl of phenol-chloroform (1:1) and vortexed vigorously before centrifugation. The RNA-containing aqueous top layer was mixed with a mixture of 4.0 M LiCl and ice-cold 100% ethanol and placed at −20°C overnight. Following the recovery of RNA pellets by centrifugation, the pellets were washed with 70% ethanol and dried under a vacuum. The dried RNA pellets were dissolved in 100 μl RNase-free water and kept at −20°C.
Primers and molecular beacon.
The 5′ noncoding region (5′ NCR) of HAV, which is conserved among different HAV isolates (1), was chosen as the target region for amplification. The primer pair UC1 (5′-AATGGATCCGTAGGAGTCTAAATTGGGGA-3′) and T7KH2 (5′-AATTCTAATACGACTCACTATAGGGAGACGGCGTTGAATGGTTTTT-3′) was used to amplify a 202-bp region of the 5′ NCR of the HAV genome. The 3′ antisense NASBA primer was elongated with the preferred transcription initiation sequence at the 5′ end (underlined) and indicated with the prefix T7.
A molecular beacon (HAV MB: 6-FAM-5′-CTTGCGGGATAGGGTAACAGCGGCGGCGCAAG-3′-DABCYL [the stem sequence of the MB is underlined, and the target sequence of HAV is in bold]) (Midland Certified Reagent Co., Midland, TX), which is perfectly complementary to a 20-bp region of the amplicon, was designed based on aligning the sequences of 15 different HAV strains obtained from GenBank as described previously (1). The molecular beacon was labeled at the 5′-end with the fluorophore FAM (6-carboxyfluorescein), and DABCYL [4-(4′-dimethylaminophenylazo) benzoic acid] was used as the quencher at the 3′ end.
Real-time NASBA reaction.
A Bio-Rad (Hercules, CA) icycler iQ Real-Time PCR Detection System was used to perform the real-time NASBA assay. The NASBA reactions were performed as described previously (2) with some modifications. The final reaction mixture volume was 25 μl. An 18-μl pre-reaction mixture was prepared to give a final concentration in 25 μl of 40 mM Tris-HCl (pH 8.5), 50 mM KCl, 12 mM MgCl2, 1.0 mM (each) deoxyribonucleoside triphosphate, 2.0 mM (each) ribonucleotide-5′-triphosphate, 2.0 mM dithiothreitol, 15% (vol/vol) dimethylsulfoxide, 0.2 μM of each cartridge-purified primer, and 100 nM of HAV MB. Five microliters of purified viral RNA was added to the 18 μl of pre-reaction mixture in a 0.5-ml Bio-Rad PCR plate, which was incubated for 5 min at 65°C in order to disrupt any secondary structure in the target RNA. The plate was then placed at 40°C. After 5 min, 2 μl of an enzyme mixture containing 2.6 μg of bovine serum albumin (Promega Co.), 40 international units (IU) of T7 RNA polymerase (Novagen Inc., Madison, WI), 8 IU of avian myeloblastosis virus reverse transcriptase (Seikagaku, Ijamsville, Md.), 0.2 IU of RNase H (New England Biolabs, Beverly, MA), and 12.5 IU of RNasin (Amersham Biotech) were added to each well, followed by incubation at 40° ± 1°C for 150 min. Fluorescence intensity data were recorded every minute of the NASBA reaction. The threshold cycle of each amplification reaction was calculated based on the first cycle at which the fluorescence was 10-fold higher than the standard deviation of the mean baseline emission.
To estimate the minimal detection limit, 10-fold serial dilutions of viral-RNA samples were made in autoclaved double-deionized water. The viral loading was represented in terms of PFU. For each experiment, a control containing no HAV template was used. The amplification products were analyzed by quick gel electrophoresis using a 2.0% ethidium bromide-stained agarose gel. To confirm that the increase in the fluorescence was due to the formation of the amplification product, the thermal denaturation profiles of the NASBA products were determined.
Specificity of the real-time NASBA assay.
Several viruses and phages obtained from various sources were used to determine the selectivity of the real-time NASBA reaction for the detection of HAV strain HM175. The selectivity of our assay was investigated by using either 103 PFU of HAV or 104 PFU of phages and other members of the Picornaviridae.
Combined IMS-real-time NASBA assay for the detection of HAV in surface water.
Different dilutions of hepatitis A virus were added to surface water samples obtained from Lake Elsinore in southern California. An IMS technique was used to concentrate the virus particles from the water sample. The conditions of IMS were first optimized using rabbit polyclonal anti-HAV antibodies and protein A-modified magnetic Dynabeads (Dynal Biotech, WI). One hundred micrograms of polyclonal rabbit anti-HAV antibodies and protein A Dynabeads were used in the IMS step, which was carried out according to the manufacturer's instructions. Briefly, protein A beads were washed two times with sodium phosphate buffer (pH 8.1), mixed with anti-HAV immunoglobulin G (IgG), and rotated for 20 min at 4°C to allow antibody-protein A complexation. After coating was completed, unbound IgG was removed and the beads were washed three times with 500 μl sodium phosphate buffer. The IgG-functionalized beads were then mixed with different HAV dilutions for 2 h. After incubation, the beads were trapped against the wall of the microcentrifuge tube by using a strong magnetic particle separator stand (Novagen Inc., Madison, WI), and the supernatant was then removed. Finally, the beads were washed three times with 1 ml phosphate-buffered saline buffer (pH 7.4).
The concentrated virus was then extracted by heating the coated beads in NASBA buffer at 95°C for 5 min, cooling them immediately on ice, and removing the beads by centrifugation at 10,000 rpm for 5 min. This was followed by carrying out the real-time NASBA assay with the extracted RNA sample under the same conditions described before. The experiment was repeated twice for each dilution.
RESULTS AND DISCUSSION
Molecular-beacon-based real-time NASBA assay for HAV.
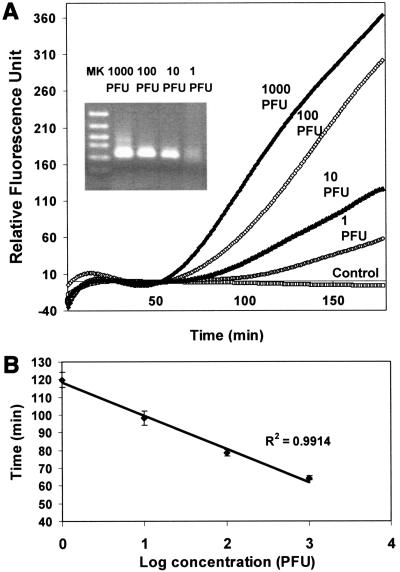
The ability of the MB-based real-time NASBA assay to detect HAV was investigated. The sensitivity of the assay was assessed using 10-fold serial dilutions of HAV samples. Any fluorescent signal that was 10-fold higher than the standard deviation of the mean baseline emission was indicative of a positive detection. A 202-bp fragment was correctly amplified by the primers, and the amplicon was detected by the MB as a significant increase in fluorescence (Fig. 1A). As little as 1 PFU was detected within 100 min using the real-time NASBA assay. This is substantially shorter than the 210 min reported recently for HAV detection using a conventional NASBA assay (14), which is based on end point detection of amplification products by electrophoresis. The substantial improvement in analysis time is primarily due to the early detection of fluorescent signal afforded by the use of MB.
FIG. 1.
Sensitivity of the real-time NASBA assay. (A) Detection of serial dilutions of HAV viral RNA at 1,000, 100, 0, and 1 PFU per reaction. A negative control containing only water was used for comparison. (B) Standard curve generated by plotting the time required for signal detection versus PFU. Fluorescence intensity data were recorded every minute of the NASBA reaction. The threshold cycle of each amplification reaction was calculated based on the first cycle at which the fluorescence was 10-fold higher than the standard deviation of the mean baseline emission. The data represent the results from four independent experiments.
The reproducibility of the assay was evaluated by comparing the detection times from four different NASBA assays. The results showed less than 5% variability among the assays (Table 1). A linear standard curve from 1 to 1,000 PFU was obtained by plotting the time required for the signal detection versus PFU (Fig. 1B).
TABLE 1.
Reproducibility of real-time NASBA reactions for different viral RNA loads
| Run no. | Time (min.) at:
|
|||
|---|---|---|---|---|
| 1,000 PFU | 100 PFU | 10 PFU | 1.0 PFU | |
| 1 | 64.25 | 78.25 | 88.61 | 119.83 |
| 2 | 66.40 | 75.06 | 94.23 | 121.10 |
| 3 | 66.80 | 77.40 | 96.05 | 129.03 |
| 4 | 65.83 | 79.23 | 98.11 | 120.90 |
| Mean ± SD | 65.82 ± 1.12 | 77.485 ± 1.78 | 94.25 ± 4.08 | 122.72 ± 4.247 |
Specificity of the real-time NASBA assay.
The specificity of the molecular-beacon-based NASBA assay depends on the selected sequence of the primer set (UC1 and T7KH2) and the probe moiety of the beacon, both of which were based on the 5′ NCR of HAV strain HM-175. The presence of other microorganisms or viruses in food and water samples could potentially affect the specificity of the assay. To test this, NASBA assays were conducted with other potential pathogens (e.g., coxsackievirus B6, echovirus 11, and echovirus 19) and indicator microorganisms (e.g., bacteriophage φX174, Escherichia coli, and rotavirus SA11) commonly found in contaminated food or water. None of these species produced significant fluorescence that could be detected (data not shown), demonstrating the specificity of the assay. To investigate whether the presence of other pathogens could affect the sensitivity of HAV detection, similar assays were conducted by mixing 103 PFU of HAV with 104 PFU of other viruses or phages. No noticeable differences in the fluorescent signal were detected, again indicating the highly specific nature of the primers and the molecular beacon (data not shown).
Combined IMS-real-time NASBA reaction for the detection of HAV particles in surface water.
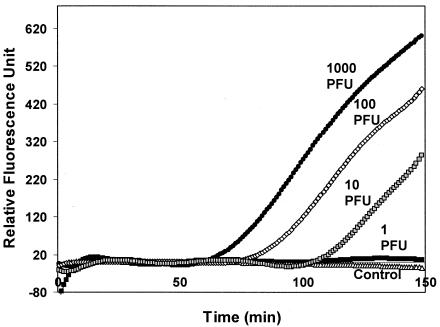
The utility of the real-time RT-PCR assay for environmental samples was tested using surface water samples from Lake Elsinore in southern California seeded with HAV. When different dilutions of HAV were added to the lake water, no visible increase in fluorescence was observed from the real-time NASBA assay, indicating severe interference from the inhibitors present in the samples. Although many pretreatment methods are available for the concentration of virus from other inhibitory substrates presented in water samples, IMS is particularly attractive because of the potential for detecting intact and infectious viruses. As little as 10 PFU was detected in the seeded lake water samples (Fig. 2). These results demonstrate the potential of the combined IMS-real-time NASBA assay for rapid and quantitative detection of HAV in contaminated water and food.
FIG. 2.
Detection of HAV in seeded lake water samples by a combined IMS-MB NASBA assay. Different dilutions of hepatitis A virus were added to surface water samples obtained from Lake Elsinore in southern California. An IMS technique was used to concentrate the virus particles from the water sample. Results for 1 to 1,000 PFU are shown. A control using lake water alone was also used.
Conclusions.
A rapid and quantitative technique to detect the presence of HAV in contaminated food and water samples is essential to provide the required rapid diagnostic capability and to allow an evaluation of the possible health risk. Although traditional methods based on multiplication of HAV in cell culture have been replaced by modern molecular techniques, such as RT-PCR and NASBA, quantification is labor-intensive and typically requires hybridization with a radiolabeled probe.
In this study, we have developed a molecular-beacon-based real-time NASBA assay for the detection of HAV. Introduction of a target-specific molecular beacon to the NASBA assay enables simultaneous amplification and real-time detection of RNA molecules in a closed-tube format (20). This is substantially better than other methods of detecting NASBA products, such as Northern blotting and the electrochemiluminescence technique, which require opening of the tubes after amplification and carry the risk of contamination. In addition, NASBA has a much higher inherent amplification capability than RT-PCR, resulting in faster detection (∼100 min) than TaqMan RT-PCR (∼180 min) (4) and molecular-beacon-based RT-PCR (1) assays. These benefits, combined with the isothermal nature (at 40° ± 1°C) of NASBA, allow high-throughput detection of HAV in a real-time manner.
Acknowledgments
This work was funded in part by grants from the UC Water Resources Center and the U.S. Environmental Protection Agency (R828040). Khaled H. Abd El Galil was supported by the Egyptian Ministry of High Education.
REFERENCES
- 1.Abd El Galil, K. H., M. A. El Sokkary, S. M. Kheira, A. M. Salazar, M. V. Yates, W. Chen, and A. Mulchandani. 2004. A combined IMS-molecular beacon RT-PCR assay for detection of hepatitis A from environmental samples. Appl. Environ. Microbiol. 70:4371-4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blais, B. W., G. Turner, R. Sooknanan, and L. T. Malek. 1997. A nucleic acid sequence-based amplification system for detection of Listeria monocytogens hlyA sequences. Appl. Environ. Microbiol. 63:310-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blok, M. J., V. J. Goosens, S. J. V. Vanherle, B. Top, N. Tacken, J. M. Middeldorp, M. H. L. Christiaans, J. P. Van Hooff, and C. A. Bruggeman. 1998. Diagnostic value of monitoring human cytomegalovirus late pp67 mRNA expression in renal-allograft recipients by nucleic acid sequence-based amplification. J. Clin. Microbiol. 36:1341-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Costa-Mattioli, M., S. Monpeoho, E. Nicand, M. H. Aleman, S. Billaudel, and V. Ferré. 2002. Quantification and duration of viraemia during hepatitis A infection as determined by real-time RT-PCR. J. Virol. Methods 9:101-109. [DOI] [PubMed] [Google Scholar]
- 5.Cromeans, T., M. D. Sobsey, and H. A. Fields. 1987. Development of a plaque assay for a cytopathic, rapidly replicating isolate of hepatitis A virus. J. Med. Virol. 22:45-56. [DOI] [PubMed] [Google Scholar]
- 6.Cromeans, T., H. A. Fields, and M. D. Sobsey. 1989. Replication kinetics and cytopathic effect of hepatitis A virus. J. Gen. Virol. 70:2051-2062. [DOI] [PubMed] [Google Scholar]
- 7.Cromeans, T. L., A. S. Foust, R. Baric, B. Robertson, and M. D. Sobsey. 2002. Replication of wild-type HAV and detection of replicative forms (RF), p. 472. Abstr. 102nd Annu. Meet. Am. Soc. Microbiol. American Society for Microbiology, Washington, D.C.
- 8.Damen, M., P. Sillekins, H. T. M. Cuypers, I. Frantzen, and R. Melsert. 1999. Characterization of the quantitative HCV NASBA assay. J. Virol. Methods 82:45-54. [DOI] [PubMed] [Google Scholar]
- 9.Darke, B. M., S. K. Jackson, S. M. Hanna, and J. D. Fox. 1998. Detection of human TNF-alpha mRNA by NASBA™. J. Immunol. Methods 212:19-28. [DOI] [PubMed] [Google Scholar]
- 10.De Baar, M. P., A. M. Van der Schoot, J. Goudsmit, F. Jacobs, R. Ehren, K. H. M. van der Horn, P. Oudshoorn, F. De Wolf, A. De Ronde, et al. 1999. Design and evaluation of human immunodeficiency virus type 1 RNA assay using nucleic acid sequence-based amplification technology able to quantify both group M and O viruses by using the long terminal repeat as target. J. Clin. Microbiol. 37:1813-1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graff, J., J., Ticehurst, and B. Flehmig. 1993. Detection of hepatitis A virus in sewage sludge by antigen capture polymerase chain reaction. Appl. Environ. Microbiol. 59:3165-3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heim, A., I. M. Grumbach, S. Zeuke, and B. Top. 1998. Highly sensitive detection of gene expression of an intronless gene: amplification of mRNA, but not genomic DNA by nucleic acid sequence based amplification. Nucleic Acid Res. 26:2250-2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heim, A., and J. Schumann. 2002. Development and evaluation of a nucleic acid sequence based amplification protocol for the detection of enterovirus RNA in cerebrospinal fluid samples. J. Virol. Methods 103:101-107. [DOI] [PubMed] [Google Scholar]
- 14.Hibbitis, S., A. Rahman, R. John, D. Westmoreland, and J. D. Fox. 2003. Development and evaluation of Nuclisens basic kit NASBA for diagnosis of parainfluenza virus infection with end point and real-time detection. J. Virol. Methods 108:145-155. [DOI] [PubMed] [Google Scholar]
- 15.Jean, J., B. Blais, A. Darveau, and I. Fliss. 2001. Detection of hepatitis A virus by the nucleic acid sequence-based amplification technique and comparison with reverse transcription-PCR. Appl. Environ. Microbiol. 67:5593-5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jothikumar, N., K. Aparna, S. Kamatchiammal, R. Paulmurugan, S. Saravanadevi, and P. Khanna. 1993. Detection of hepatitis A virus in raw and treated wastewater with the polymerase chain reaction. Appl. Environ. Microbiol. 59:2558-2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kievits, T., B. Van Gemen, D. Van Strijp, R. Schukkink, M. Dircks, H. Adriaanse, L. Malek, R. Sooknanan, and P. Lens. 1991. NASBA™ isothermal enzymatic in vitro nucleic acid amplification optimized for the diagnosis of HIV-1 infection. J. Virol. Methods 35:273-286. [DOI] [PubMed] [Google Scholar]
- 18.Lanciotti, R. S., and A. J. Kerst. 2001. Nucleic acid sequence-based amplification assays for rapid detection of West Nile and St. Louis encephalitis. J. Clin. Microbiol. 39:4506-4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lemon, S. M., L. N. Binn, and R. H. Marchwicki. 1983. Radioimmunoassay for quntitation of hepatitis A virus in cell cultures. J. Clin. Microbiol. 17:834-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leone, G., H. Van Schijndel, B. Van Gemen, F. R. Kramer, and C. D. Schoen. 1998. Molecular beacon probes combined with amplification by NASBA enable homogeneous, real-time detection of RNA. Nucleic Acids Res. 26:2150-2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morre, S. A., P. Sillekens, M. V. Jacobs, P. Van Aarle, S. de Blok, B. Van Gemen, J. M. Walboomers, C. J. Meijer, and A. J. Van denBrule. 1996. RNA amplification by nucleic acid sequence-based amplification with an internal standard enables reliable detection of Chlamydia trachomatis in cervical scrapings and urine samples. J. Clin. Microbiol. 34:3108-3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nasser, A. M., and T. G. Metcalf. 1987. Production of cytopathology in FRhK-4 cells by Bs-C-1 passaged hepatitis A virus. Appl. Environ. Microbiol. 53:2967-2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Portnoy, B. L., P. A. Mackowiak, C. T. Caraway. J. A. Walker, T. W. Mckinley, and C. A. Klein. 1975. Oyster-associated hepatitis: failure of certification programs to prevent outbreaks. JAMA 233:1065-1068. [DOI] [PubMed] [Google Scholar]
- 24.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Tyagi, S., and F. R. Kramer. 1996. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 14:303-308. [DOI] [PubMed] [Google Scholar]
- 26.Witt, D. J., M. Kemper, A. Stead, P. Sillekins, C. C. Ginocchio, A. M. Caliendo, et al. 2000. Analytical performance and clinical utility of a nucleic acid sequence-based amplification assay for detection of Cytomegalovirus. J. Clin. Microbiol. 38:3994-3999. [DOI] [PMC free article] [PubMed] [Google Scholar]