Abstract
Dilution culture experiments were conducted in western North Pacific coastal regions to determine growth and grazing mortality rates of bacterial phylogenetic groups (α-, β-, and γ-proteobacteria and the Cytophaga-Flavobacter cluster) detected by fluorescent in situ hybridization. Growth rates varied greatly (1.2- to 4.0-fold) among different groups, and they were related to environmental variables (chlorophyll a concentrations and temperature) in a group-specific fashion. Growth rates of α-proteobacteria, the most abundant group in all the samples examined, were generally lower than those of less abundant groups, including the Cytophaga-Flavobacter cluster and γ-proteobacteria. Grazing mortality rates and mean cell volumes varied little among different groups. These results provide insights into factors that affect distributions of different groups, but growth and grazing mortality alone did not fully explain bacterial community compositions at a broad phylogenetic level.
Recent evidence has indicated that bacterial communities in marine surface waters are generally dominated by α-proteobacteria (19, 30, 35) and the Cytophaga-Flavobacter cluster (6, 14, 23), although γ-proteobacteria may also account for a significant fraction of total communities, depending on environments (11, 18). Bacterial community compositions might be accounted for by differences in growth rates among different groups (bottom-up control). Factors that may affect bacterial growth include temperature and substrate supply. For bulk bacterial communities, interactive effects of temperature and substrate supply in enhancing bacterial growth have been documented (24, 31). Recent work has suggested that even broad phylogenetic groups of bacteria differ in the uptake of dissolved organic matter (7, 26) and growth (9, 41). These results suggest that responses of bacterial growth to environmental variables may differ among different bacterial groups. However, data regarding relationships between environmental variables and the growth of individual groups are scarce.
Other factors that may control bacterial community composition include bacterial mortality due to grazing and lysis (top-down control). Grazing by protists on planktonic bacteria is known to be selective. Critical determinants of the selectivity include the size, morphology, motility, and surface characteristics of prey cells (20). Studies have suggested that actively growing, large bacteria are more rapidly eliminated by grazers than slow-growing, small bacteria (17, 37). This probably explains the persistence of slow-growing (or dormant) cells in coastal marine systems, although the relationship between these attributes (cell size and grazing mortality) and phylogenetic affiliations of bacteria has yet to be clarified fully (3, 29).
Our objective here was to test the hypothesis that growth (bottom-up control) and grazing mortality (top-down control) explain bacterial community compositions at a broad phylogenetic level in coastal marine waters. Our approach was to use the dilution culture method in combination with fluorescent in situ hybridization (FISH) (2, 41) in order to determine cell size distributions and growth rates of bacteria belonging to four phylogenetic groups (α-, β-, and γ-proteobacteria and the Cytophaga-Flavobacter cluster). Grazing mortality rates were determined by the use of the serial dilution technique (25). Our data show high variability in growth rates among different groups, but growth and grazing mortality alone did not explain bacterial community compositions at a broad phylogenetic level.
MATERIALS AND METHODS
Water samples were collected at three sampling stations located at the upper (station A) (39o20′N, 141o55′E; water depth, 25 m), middle (station B) (39o20′N, 141o57′E; water depth, 45 m), and lower (station C) (39o21′N, 141o59′E; water depth, 70 m) parts of the Otsuchi Bay, a semiclosed embayment (20.2 km2) on the northeast coast of Japan, on 22 May 2001 and 24 May 2002. A 10-liter Van Dorn water sampler was used to collect samples from surface and subsurface layers (20 m, 40 m, and 65 m for stations A, B, and C, respectively; these depths correspond to about 5 m above the bottom) in 2001, whereas only surface waters were collected in 2002. Water samples were also collected during a cruise aboard the R/V Tansei-Maru (KT-02-10 cruise). Sampling stations were located in the center of Sagami Bay (station 1) (35o00′N, 139o20′E; water depth, 1,500 m; 17 July 2002) and off the Kuroshio (station 4) (32o55′N, 138o39′E; water depth, 3,000 m; 18 July 2002). At each station, samples were collected with a 10-liter Niskin CTD rosette sampler at depths of 1 and 100 m.
Water samples were filtered through either 0.2-μm or 0.8-μm-pore-size polycarbonate filters (Millipore). One hundred milliliters of the 0.8-μm filtrate and 900 ml of the 0.2-μm filtrate were mixed in an acid-washed 1-liter polycarbonate bottle. Powder-free plastic gloves were worn, and care was taken to minimize contamination. The incubation was carried out in the dark at in situ temperature (9 to 28°C) for 12 to 24 h depending on the experiments. In order to determine the abundance and size distribution of individual bacterial phylogenetic groups (see below), subsamples were withdrawn at the beginning (from the 0.8-μm filtrate and the 0.2-μm filtrate) and at the end of the incubation. Growth rates (μ) (day−1) of individual groups were calculated with an assumption of exponential growth; i.e., μ = (1/t)ln(Nt/N0), where N0 and Nt are the abundances of cells (cells milliliter−1) at the beginning and at the end of the incubation, respectively, and t is the incubation period in days. Growth rates estimated by this method are conservative because viruses that pass through the 0.8-μm-pore-size filter would cause bacterial mortality during the incubations.
Serial dilution culture experiments were conducted to determine growth and grazing mortality rates of bacteria (25). This approach, originally designed for the determination of the growth and grazing mortality of phytoplankton (25), has been tested and applied for the measurement of bacterivory (see e.g., reference 39). Surface waters were collected at station B of the Otsuchi Bay on 28 and 30 May 2002 with a 10-liter Van Dorn water sampler. Sample waters were first filtered through a 200-μm-mesh screen, and a portion of this filtrate was further filtered through 0.2-μm-pore-size filters (Gelman SporCap filters). These filtrates were mixed to generate a dilution gradient, as follows: (volume of 200-μm filtrate)/[(volume of 200-μm filtrate) + (volume of 0.2-μm-pore-size filtrate)] × 100 = 22, 44, 67, and 100. Each mixture was contained in duplicate 5-liter-capacity polycarbonate bottles, amended with ammonium (10 μM) and phosphate (1 μM), and incubated under natural irradiance in an outside tank through which Otsuchi Bay water was circulated. Incubations were started at 1700 h and lasted for 24 h. Subsamples were withdrawn at the beginning and at the end of the incubation for analysis of the abundance and cell size of individual bacterial groups (see below).
For each phylogenetic group and each dilution series, we calculated the apparent growth rate (r) (day−1) as follows: r = (1/t)ln(Nt/N0), where N0 and Nt are the abundances of cells (cells milliliter−1) at the beginning and at the end of the incubation, respectively, and t is the period of incubation (days). The relationship between r (y variable) and the proportion of the 200-μm filtrate in each bottle (x variable) was analyzed by linear regression to obtain estimates of slope (grazing mortality rate) (day−1) and y intercept (growth rate) (day−1) (25).
The different phylogenetic groups and their cell abundances and biovolumes in each of the dilution culture samples were determined by FISH in combination with semiautomated image analysis (7). Samples were preserved with paraformaldehyde solution (final concentration, 2%). After fixation at 4°C for 24 h, bacteria were collected on 0.2-μm-pore-size polycarbonate filters (Millipore) and stored at −20°C until analysis. The hybridization solution contained 2.5 μg ml−1 of probe, 0.9 M NaCl, 20 mM Tris-HCl (pH 7.2), 5 mM EDTA, 0.01% sodium dodecyl sulfate, and the concentration of formamide determined to achieve specificity for the bacterial groups targeted by the different probes (26): Eub338 (5′-GCTGCCTCCCGTAGGAGT-3′; formamide, 30%) (1) for eubacteria, Alf968 (5′-GGTAAGGTTCTGCGCGTT-3′; formamide, 30%) (18) for α-proteobacteria, Bet42a (5′-GCCTTCCCACTTCGTTT-3′; formamide, 30%) (28) for β-proteobacteria, Gam42a (5′-GCCTTCCCACATCGTTT-3′; formamide, 30%) (27) for γ-proteobacteria, CF319a (5′-TGGTCCGTGTCTCAGTAC-3′; formamide, 35%) (27) for the Cytophaga-Flavobacter cluster, and a negative control probe (5′-TAGTGACGCCGTCGA-3′; formamide, 30% (22) for nonspecific probe bindings. To avoid misbinding between Bet42a and Gam42a, we used the same concentration for unlabeled competitor probes (i.e., unlabeled Gam42a with labeled Bet42a and unlabeled Bet42a with labeled Gam42a). All probes were commercially synthesized and labeled with Cy3 (Operon, Inc.). After hybridization, the samples were transferred to a wash solution containing 20 mM Tris-HCl (pH 7.2), 5 mM EDTA, 0.01% sodium dodecyl sulfate, and the concentration of NaCl appropriate for the probe (11, 42). Hybridized and DAPI (4′,6′-diamidino-2-phenylindole)-stained cells were detected by using an Olympus BX 50 microscope equipped with a charge-coupled-device camera (Hamamatsu C-4742-95-12NR). Semiautomated image analysis (Image-pro4.0; MediaCybernetics, Inc.) was conducted to determine cell abundance and biovolume as described elsewhere (8, 41).
For each phylogenetic group, 10 microscopic fields were examined. The number of DAPI-positive cells detected in the 10 fields was 1,050 ± 950 (average ± standard deviation). The counts of Cy3-positive cells varied depending on the relative contribution; for the dominant group, the average counts were 440 ± 560 per sample. The cell abundance of each group was calculated by multiplying the ratio of hybridized (Cy3-positive) cell counts to DAPI-positive counts by the estimate of the total abundance of DAPI-positive cells. Propagation errors were calculated as described by Bevington (4). Background Cy3 counts obtained with the negative control probe were <1% of DAPI counts. Because the distribution of cell volumes skewed to the right, the cell volume data were log transformed for the derivation of mean cell volume and statistical comparisons.
RESULTS
Environmental characteristics of the sampling sites and compositions of the bacterial communities.
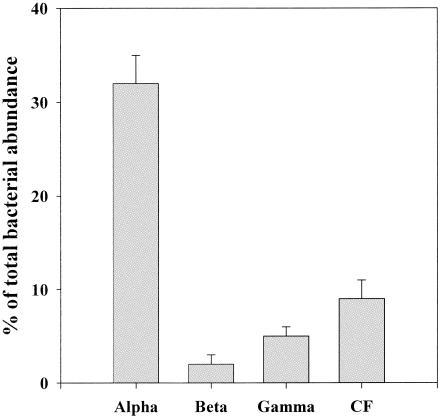
Dilution culture experiments were carried out using coastal waters (salinity, 25.7 to 34.9 practical salinity units) collected at different locations and depths in order to cover a range of water temperature (8.5 to 27.6°C) and chlorophyll a concentrations (0.09 to 3.43 μg liter−1) (Table 1). Along this environmental gradient, the abundance of total DAPI-positive cells varied in the range of 0.3 × 106 to 1.8 × 106 cells ml−1 (Table 1). In all the samples, the relative abundance of α-proteobacteria was highest among the four phylogenetic groups; α-proteobacteria accounted for 32% ± 4% (mean ± standard error; range, 17 to 61%) of total DAPI-positive cells (Table 1; Fig. 1). The second dominant group was the Cytophaga-Flavobacter cluster (average contribution to total DAPI-positive cells was 9% ± 2%), followed by γ-proteobacteria (5% ± 1%) (Table 1; Fig. 1). β-Proteobacteria were the least abundant group, whose contribution was detectable (detection limit, 1%) at only four locations (Table 1). The sum of the abundances of four phylogenetic bacterial groups accounted for 49% ± 6% (range, 26 to 95%) of total DAPI-positive cells, whereas the average detection rate with the general eubacterial probe (Eub338) was 40% (range, 14 to 57%) (Table 1). These detection rates are within the range obtained in previous studies, which have generally reported lower values in more oligotrophic environments (5). The detection by the sum of four probes on occasions exceeded that by Eub338 (Table 1). Similar results have been reported for some previous studies. For example, for their experiment conducted in Plymouth Sound, Fuchs et al. (14) reported that the summed detection by four specific probes exceeded that by Eub338 in eight out of nine cases (see Fig. 2 of reference 14). This phenomenon might be in part attributed to the submaximum accessibility of the Eub338 probe to the target rRNA. In support of this hypothesis, Fuchs et al. (13) have found that the relative probe fluorescence (accessibility) of Eub338 was lower than those of some specific probes (e.g., CF319a) in Escherichia coli, although probe-dependent variations in the detectability by FISH of marine communities has yet to be examined fully.
TABLE 1.
Environmental variables, total abundance of DAPI-positive cells, and bacterial community compositions at coastal sampling stations
| Location (date, day/mo/yr) | Station (depth, m) | Water temp (°C) | Chlorophyll aa (μg liter−1) | Total abundance of DAPI-positive cells (106 cells ml−1) | % of total bacterial abundanceb (avg ± SE, n = 10)
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| Eub | Alpha | Beta | Gamma | CF | Σ | |||||
| Otsuchi Bay (22/05/01) | A (0) | 12.9 | 1.77 | 1.4 | 57 ± 1 | 33 ± 1 | 4 ± 1 | 8 ± 1 | 13 ± 1 | 58 |
| A (20) | 9.6 | 1.65 | 1.6 | 57 ± 1 | 61 ± 1 | 6 ± 1 | 10 ± 1 | 18 ± 1 | 95 | |
| B (0) | 12.9 | 2.42 | 1.8 | 54 ± 2 | 17 ± 1 | 8 ± 1 | <1 | 6 ± 1 | 33 | |
| B (40) | 8.7 | 3.09 | 1.5 | 43 ± 2 | 17 ± 1 | 3 ± 1 | 7 ± 1 | 4 ± 1 | 31 | |
| C (0) | 10.4 | 1.18 | 1.5 | 51 ± 2 | 33 ± 1 | <1 | 7 ± 1 | 17 ± 1 | 57 | |
| C (65) | 8.5 | 1.68 | 1.1 | 22 ± 1 | 33 ± 1 | <1 | <1 | 8 ± 1 | 41 | |
| Otsuchi Bay (24/05/02) | A (0) | 13.4 | 1.92 | 1.5 | 54 ± 1 | 39 ± 1 | <1 | 6 ± 0 | 13 ± 1 | 58 |
| B (0) | 13.4 | 2.56 | 1.5 | 51 ± 2 | 46 ± 1 | <1 | 8 ± 1 | 12 ± 1 | 66 | |
| C (0) | 11.7 | 1.85 | 1.2 | 23 ± 2 | 22 ± 3 | <1 | <1 | <1 | 26 | |
| Sagami Bay (17/07/02) | 1 (0) | 21.9 | 3.43 | 1.5 | 49 ± 1 | 36 ± 1 | <1 | 6 ± 1 | 8 ± 1 | 50 |
| 1 (100) | 16.0 | 0.09 | 0.3 | 27 ± 2 | 35 ± 2 | <1 | 8 ± 1 | 7 ± 1 | 50 | |
| Off Kuroshio (18/07/02) | 4 (0) | 27.6 | 0.18 | 0.7 | 17 ± 1 | 27 ± 1 | <1 | <1 | <1 | 27 |
| 4 (100) | 22.4 | 0.12 | 0.3 | 14 ± 1 | 23 ± 1 | <1 | <1 | 5 ± 1 | 28 | |
| Total | 40 ± 5 | 32 ± 4 | 3 ± 2 | 5 ± 1 | 9 ± 2 | 49 ± 6 | ||||
Fluorometric measurement after the extraction of pigments with N,N-dimethylformamide (H. Ogawa and H. Fukuda, personal communication).
Bacteria were classified by using rRNA binding oligonucleotide probes specific for eubacteria (Eub), α-proteobacteria (Alpha), β-proteobacteria (Beta), γ-proteobacteria (Gamma), and the Cytophaga-Flavobacter cluster (CF). Σ, sum of four phylogenetic bacterial groups.
FIG. 1.
Percentages of α-, β-, and γ-proteobacteria and the Cytophaga-Flavobacter cluster (CF) with respect to total bacterial abundance at the beginning of incubation. Each bar represents the average (± standard error) of the data compiled for all the stations examined (n = 13).
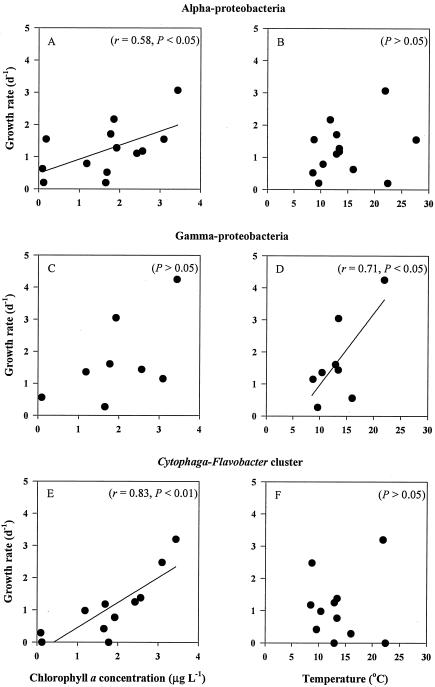
FIG. 2.
Relationships between specific growth rates of individual phylogenetic bacterial groups and the environmental variables chlorophyll a concentration (A, C, and E), and water temperature (B, D, and F). Pearson's correlation coefficient (r) and statistical significance (P) are shown in parentheses.
Growth rates of individual bacterial groups.
The growth rates of α-proteobacteria varied in the range of 0.2 to 3.1 day−1 (Table 2). These values were generally lower than the corresponding rates of less abundant groups, including γ-proteobacteria and Cytophaga-Flavobacter cluster; the growth rates of α-proteobacteria were less than those of γ-proteobacteria in 5 of 8 cases and less than those of the Cytophaga-Flavobacter cluster in 7 of 10 cases (Table 2). The low relative abundance (<1%) of β-proteobacteria hampered the accurate measurement of growth rates for this group; the rates were measurable for only three cases (Table 2). In these cases, the growth rates of β-proteobacteria were lower than those of α-proteobacteria.
TABLE 2.
Specific growth rates of bacteria belonging to different phylogenetic bacterial groups
| Location (date, day/mo/yr) | Station (depth, m) | Growth rate, day−1 (avg ± SE, n = 10)a
|
|||
|---|---|---|---|---|---|
| Alpha | Beta | Gamma | CF | ||
| Otsuchi Bay (22/05/01) | A (0) | 1.71 ± 0.22 | 0.60 ± 0.20 | 1.61 ± 0.39 | ND |
| A (20) | 0.20 ± 0.02 | ND | 0.27 ± 0.07 | 0.42 ± 0.05 | |
| B (0) | 1.11 ± 0.36 | 0.85 ± 0.57 | — | 1.25 ± 0.94 | |
| B (40) | 1.55 ± 0.44 | 1.39 ± 0.64 | 1.15 ± 0.25 | 2.48 ± 0.91 | |
| C (0) | 0.79 ± 0.16 | — | 1.36 ± 0.17 | 0.98 ± 0.29 | |
| C (65) | 0.52 ± 0.08 | — | — | 1.18 ± 0.24 | |
| A (0) | 1.28 ± 0.27 | — | 3.05 ± 0.43 | 0.77 ± 0.45 | |
| B (0) | 1.18 ± 0.20 | — | 1.44 ± 0.35 | 1.38 ± 0.46 | |
| C (0) | 2.17 ± 1.60 | — | — | — | |
| Sagami Bay (17/07/02) | 1 (0) | 3.07 ± 0.82 | — | 4.25 ± 1.19 | 3.20 ± 1.91 |
| 1 (100) | 0.63 ± 0.14 | — | 0.56 ± 0.34 | 0.29 ± 0.29 | |
| Off Kuroshio (18/07/02) | 4 (0) | 1.55 ± 0.70 | — | — | — |
| 4 (100) | 0.20 ± 0.14 | — | — | ND | |
For abbreviations, see Table 1, footnote b. When the increase in cell abundance during the incubation period was less than 20% of initial abundance, the growth rate was judged to be below the detection limit (ND). Dashes indicate that the growth rate could not be calculated because the contribution of the phylogenetic group to total DAPI-positive cells was below the detection limit (1%) for samples collected at the beginning and/or at the end of the incubation.
We conducted a Pearson's correlation analysis to examine if growth rates of three of the phylogenetic groups (α- and γ-proteobacteria and the Cytophaga-Flavobacter cluster) covaried with temperature and chlorophyll a concentrations. Significant positive correlations were found between chlorophyll a concentrations and the growth rates of α-proteobacteria (r = 0.58; P < 0.05) and the Cytophaga-Flavobacter cluster (r = 0.83; P < 0.01), whereas temperature was correlated with the growth rate of γ-proteobacteria (r = 0.71; P < 0.05) (Fig. 2). A forward stepwise regression analysis was conducted by using chlorophyll a concentration and temperature as two independent variables. Temperature and chlorophyll a concentration were not correlated with each other (P > 0.05); the colinearity was not significant. Results showed that 62% of the variations in growth rates of α-proteobacteria was accounted for by the combination of temperature and chlorophyll a concentration. The multiple regression equation to show this extent of variation is GRα = 0.612 (± 0.160) · CHL + 0.080 (± 0.029) · T − 0.975 (± 0.607) (r2 = 0.62; P < 0.01; n = 13), where GRα, CHL, and T are the growth rate of α-proteobacteria (day−1), the chlorophyll a concentration (μg liter−1), and temperature (oC), respectively. In the above equation, parameters are given with standard errors in parentheses. The standard partial regression coefficient of chlorophyll a (0.43) was greater than that of temperature (0.04), suggesting that chlorophyll a explained a larger amount of the variation of GRα.
The relationships between predictor variables (chlorophyll a concentration and temperature) and growth rates for the Cytophaga-Flavobacter cluster (GRCF) and γ-proteobacteria (GRγ) were different from that for the α-proteobacteria. A significant fraction (73%) of variation in GRCF was explained by chlorophyll a concentration (GRCF = 0.798 [± 0.182] · CHL − 0.271 [± 0.404] [r2 = 0.73; P < 0.01; n = 9]), although temperature did not significantly contribute to the variation of GRCF (P = 0.28). In contrast, temperature accounted for a significant fraction of the variation in growth rates of γ-proteobacteria (GRγ = 0.212 [± 0.087] · T − 1.005 [± 1.191] [r2 = 0.46; P = 0.045; n = 9]), whereas a forward stepwise regression analysis did not select chlorophyll as a significant predictor variable (P = 0.11).
Grazing mortality rates of individual bacterial groups.
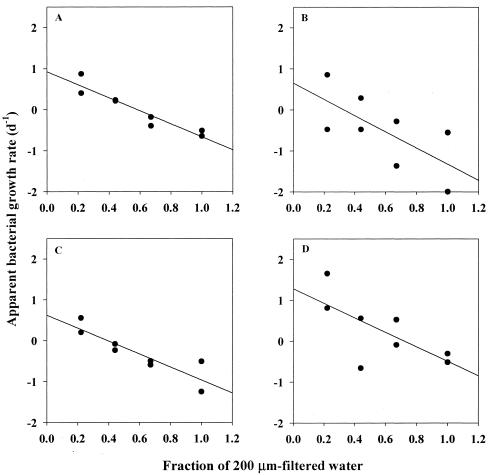
Two serial dilution culture experiments (25) were also conducted at Otsuchi Bay to test the hypothesis that grazing mortality rates differed among different phylogenetic groups of bacteria. For the three phylogenetic groups examined, apparent growth rates (r) linearly decreased with an increase of the fraction of <200-μm filtrate (f) (r2 = 0.47 to 0.89) (Table 3; Fig. 3), suggesting that grazing pressures were significant in both experiments 1 and 2. The grazing mortality rates of the bulk bacterial community and each phylogenetic group were estimated as slopes of the linear regression equations that relate r to f. The results showed that the grazing mortality rates of bulk bacterial communities were 1.59 ± 0.23 (average ± standard error) day−1 and 2.92 ± 0.14 day−1 for experiments 1 and 2, respectively. These values are comparable to those reported for other coastal waters (see, e.g., reference 16). The grazing mortality for individual groups varied in the range of 1.59 to 1.97 day−1 and 3.03 to 4.41 day−1 for experiments 1 and 2, respectively (Table 3). In each experiment, the analysis of covariance (ANCOVA) results indicated that these values did not differ significantly among different phylogenetic groups (P = 0.4 to 0.9). In contrast, growth rates estimated as y intercepts varied significantly among different phylogenetic groups for both experiments (P < 0.05). The Cytophaga-Flavobacter cluster and the γ-proteobacteria exhibited the highest growth rates in experiments 1 and 2, respectively (Table 3).
TABLE 3.
Summary of the regression parameters for estimating the growth (slope) and grazing mortality rate (y intercept) of the bulk bacterial community (total) and bacteria belonging to three different phylogenetic groups (α- and γ-proteobacteria and the Cytophaga-Flavobacter cluster [CF])a
| Expt | Group | r2 | P | Grazing rate, day−1 (avg ± SE) | Growth rate, day−1 (avg ± SE) |
|---|---|---|---|---|---|
| 1 | Alpha | 0.47 | <0.05 | 1.97 ± 0.85 | 0.65 ± 0.55 |
| Gamma | 0.80 | <0.01 | 1.59 ± 0.21 | 0.63 ± 0.33 | |
| CF | 0.49 | <0.05 | 1.77 ± 0.74 | 1.28 ± 0.46 | |
| Total | 0.89 | <0.01 | 1.59 ± 0.23 | 0.93 ± 0.15 | |
| 2 | Alpha | 0.74 | <0.01 | 4.41 ± 1.07 | 2.71 ± 0.69 |
| Gamma | 0.81 | <0.01 | 3.24 ± 0.65 | 2.84 ± 0.42 | |
| CF | 0.89 | <0.01 | 3.03 ± 0.44 | 1.90 ± 0.29 | |
| Total | 0.98 | <0.01 | 2.92 ± 0.14 | 2.23 ± 0.09 |
Duplicate bottles were used for each dilution series (n = 8). ANCOVA results to test the differences in slope and y intercept among different bacterial groups are as follows. For slope, df = 2, 18 (degree of freedom, residual degree of freedom), F=0.082, and P = 0.921 for experiment 1, and df = 2, 18, F = 0.944, and P = 0.407 for experiment 2. For y intercept, df = 2, 20, F = 4.313, and P < 0.05 for experiment 1, and df = 2, 20, F = 4.543, and P < 0.05 for experiment 2.
FIG. 3.
Relationships between apparent growth rates of bacterial groups (A, total bacteria detected by DAPI; B, α-proteobacteria; C, γ-proteobacteria; D, Cytophaga-Flavobacter cluster) and the degree of dilution of 200-μm-filtered seawater with 0.2-μm-filtered seawater (the data obtained in experiment 1 are shown as an example). Solid lines are linear fits of the data points. Statistical parameters for each regression are tabulated in Table 3.
Cell volumes of individual bacterial groups.
We measured the cell volumes of individual groups of bacteria for all the samples collected in the growth experiments. The results indicated that bacteria at the beginning of incubation varied in the range of 0.011 to 0.060, 0.014 to 0.020, 0.013 to 0.059, and 0.013 to 0.049 μm3 cell−1 for α, β-, and γ-proteobacteria and the Cytophaga-Flavobacter cluster, respectively (Table 4). For none of the phylogenetic groups did the mean cell volume correlate with either group-specific growth rates or environmental variables (temperature and chlorophyll a concentration) (Pearson's correlation, P > 0.05). On some occasions during incubation, mean cell volume increased significantly (Student's t test, P < 0.05) to a moderate extent (<50% increase except for γ-proteobacteria at the subsurface of station B and the surface of station C and for the Cytophaga-Flavobacter cluster at the subsurface of station C in the 2001 experiment) (Table 4). In order to determine if the mean cell volume differed among different phylogenetic groups, we conducted three-way analysis of variance by using experiment, incubation (the beginning and end of incubation), and phylogenetic groups (α- and γ-proteobacteria and the Cytophaga-Flavobacter cluster; β-proteobacteria was not used for this test because the data were insufficient) as factors. We used results only from experiments in which the cell size data for all three phylogenetic groups were available. We found that after eliminating the effect of experiment and incubation, there was no significant difference in mean cell volume among different phylogenetic groups (P = 0.14). Differences associated with experiment were highly significant (P < 0.001), but there were no significant differences between sizes at the beginning and at the end of the incubation (P = 0.35). Notably, the variation among experiments indicated that the mean cell volumes (measured at the beginning of incubation) of different phylogenetic groups were strongly positively correlated with each other (α- versus γ-proteobacteria, r = 0.98, P < 0.001, n = 9; α-proteobacteria versus Cytophaga-Flavobacter cluster, r = 0.92, P < 0.001, n = 9; γ-proteobacteria versus Cytophaga-Flavobacter cluster, r = 0.91, P < 0.01, n = 8).
TABLE 4.
Mean cell volume of bacterial cells for individual phylogenetic groups
| Location (date, day/mo/yr) | Station (depth, m) | Incubation | Mean cell vol (10−3 μm3)a
|
|||
|---|---|---|---|---|---|---|
| Alpha | Beta | Gamma | CF | |||
| Otsuchi Bay (22/05/01) | A (0) | Initial | 16 (15-18, 233) | 14 (11-19, 36) | 17 (15-20, 62) | 18 (16-21, 105) (−)** |
| End | 14 (14-15, 1,211) | 14 (12-17, 41) | 21 (19-22, 316) | 15 (13-17, 79) | ||
| A (20) | Initial | 11 (10-12, 464) (+)*** | — | 17 (14-19, 88) | 13 (12-14, 150) (+)*** | |
| End | 15 (14-15, 805) | — | 13 (12-15, 149) | 19 (17-20, 201) | ||
| B (0) | Initial | 18 (16-20, 190) | 17 (14-19, 69) | — | 18 (15-22, 59) (+)*** | |
| End | 18 (17-19, 517) | 16 (15-18, 194) | — | 26 (24-27, 385) | ||
| B (40) | Initial | 16 (14-17, 155) (−)*** | 20 (14-31, 21) | 13 (10-15, 52) (+)*** | 13 (10-17, 19) (+)*** | |
| End | 12 (11-13, 321) | 24 (21-28, 88) | 22 (20-24, 179) | 15 (14-16, 209) | ||
| C (0) | Initial | 15 (14-16, 278) | — | 14 (12-17, 60) (+)*** | 16 (15-17, 134) (+)*** | |
| End | 14 (13-15, 243) | — | 35 (31-39, 195) | 21 (19-23, 199) | ||
| C (65) | Initial | 11 (10-12, 131) (+)*** | — | — | 15 (13-17.50) (+)*** | |
| End | 30 (28-32, 470) | — | — | 37 (34-40, 167) | ||
| Otsuchi Bay (24/05/02) | A (0) | Initial | 38 (32-45, 67) (+)*** | — | 38 (22-64, 12) (+)* | 48 (33-72, 12) |
| End | 60 (55-64, 189) | — | 47 (44-49, 249) | 53 (45-61, 52) | ||
| B (0) | Initial | 60 (57-63, 382) (−)*** | — | 59 (51-67, 67) | 49 (43-55, 104) (−)*** | |
| End | 48 (46-49, 595) | — | 51 (47-56, 120) | 33 (30-37, 139) | ||
| C (0) | Initial | 55 (47-64, 27) | — | — | — | |
| End | 63 (60-66, 479) | — | — | — | ||
| Sagami Bay (17/07/02) | 1 (0) | Initial | 34 (33-35, 1,041) (+)*** | — | 31 (29-33, 226) (+)*** | 33 (30-37, 113) (−)*** |
| End | 40 (40-41, 2,752) | — | 38 (37-39, 1,597) | 26 (25-28, 514) | ||
| 1 (100) | Initial | 38 (36-41, 286) (−)*** | — | — | — | |
| End | 31 (25-37, 31) | — | — | — | ||
| Off Kuroshio (18/07/02) | 4 (0) | Initial | 41 (37-44, 234) | — | — | — |
| End | 34 (30-38, 101) | — | — | — | ||
| 4 (100) | Initial | 34 (32-36, 313) | — | — | — | |
| End | 39 (17-91, 10) | — | — | — | ||
For abbreviations, see Table 1, footnote b. Values in parentheses are the 95% confidence interval and the number of cells determined. If the mean cell volume at the end of incubation differs significantly (by Student's t test) from that at the beginning of incubation, symbols are added to indicate the direction of the change (increase [+] or decrease [−]) and the level of significance, as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001. Calculations of means and 95% confidence intervals as well as the statistical test were carried out by using log-transformed data (see Materials and Methods); back (log−1) transformed values are reported in this table. Dashes indicate that the contribution of the phylogenetic group to total DAPI-positive cells was below the detection limit (1%).
DISCUSSION
In the western North Pacific coastal regions that we examined, the dominant bacterial group was α-proteobacteria, accounting for about one-third of total DAPI counts, followed by the Cytophaga-Flavobacter cluster and γ-proteobacteria (Fig. 1). β-Proteobacteria accounted for only a minor fraction of bacterioplankton communities. We found that different phylogenetic groups exhibited different relationships with water temperature and chlorophyll a concentrations. These results provide insights into factors that affect broad patterns in distributions of different groups, although correlation results should not be regarded as the demonstration of causal relationships between the variables. Our multiple regression analysis revealed that a significant fraction of the variations in growth of α-proteobacteria is explained by combinations of temperature and chlorophyll a (a proxy for the substrate supply), a pattern consistent with previous propositions that bulk bacterial growth is largely regulated by temperature and substrate supply in surface waters (24, 31). The fact that chlorophyll a concentration alone explained a substantial fraction (73%) of variations in the growth rate of the Cytophaga-Flavobacter cluster appears to reflect adaptation of the Cytophaga-Flavobacter cluster to organic-rich environments. Previous work has found that the Cytophaga-Flavobacter cluster is abundant on organic aggregates (10) and during the decay of phytoplankton blooms (34), whereas it is less abundant in newly upwelled, less productive water masses (38). Cottrell and Kirchman (7) have found that the Cytophaga-Flavobacter cluster is the most abundant group using high-molecular-weight dissolved organic matter. These data suggest that labile, polymeric substrates (e.g., polysaccharide) released by phytoplankton largely support the growth of the Cytophaga-Flavobacter cluster. In contrast, the growth rate of γ-proteobacteria was correlated with temperature but not with chlorophyll a concentration. γ-Proteobacteria are known to be an “opportunistic group” that responds rapidly to the pulse of nutrient enrichment (11, 33). These characteristics probably make it difficult to predict the growth of γ-proteobacteria, with a large fraction (54%) of variations in growth rates left unexplained.
Our measurements of bacterial growth rate used a dilution culture method with incubation of water samples contained in bottles. Potential problems in the dilution culture experiment include selective growth enhancement of specific bacterial groups by wall effects, artificial enrichment of substrates during filtration and handling, and the disturbance of the organic matter regimen caused by the elimination of particles by filtration (12, 14). Fuchs et al. (14) have suggested that high growth rates of γ-proteobacteria in dilution culture are due to responses of this bacterial group to artificial enrichment of substrate. In our experiments, care was taken to minimize filtration pressure and contaminations, and the incubation period was kept relatively short (<24 h). Our previous study in the Delaware estuary revealed that bacterial production rates estimated by the dilution culture technique generally agreed with those determined by the thymidine method, suggesting that artificial enhancement of bacterial growth was minimal (41). Eub338-negative cells might include dormant or inactive cells with low ribosome contents. The increase in ribosome content, and thus enhanced detection by oligonucleotide probes, in dormant cells during the incubation (14) may account for some of the increase in cell abundance detected by specific probes. However, our result of three-way analysis of variance on cell size distributions of individual groups revealed that changes in cell size, which generally increases with increasing ribosome content (40), during the incubation were insignificant (P = 0.35). Future studies are required to examine to what extent the estimates of growth and grazing parameters derived by the approach that we used are biased by changes in the physiological state of bacterial cells.
One hypothesis to explain the prevalence of α-proteobacteria in coastal waters that we examined is that growth rates of this group exceeded those of the less abundant groups. However, our data showed that, in most cases, either γ-proteobacteria or the Cytophaga-Flavobacter cluster, which were much less abundant than α-proteobacteria, grew faster than α-proteobacteria, suggesting that growth rate alone did not account for the dominance of α-proteobacteria. Although we cannot totally exclude the possibility that the growth of γ-proteobacteria and the Cytophaga-Flavobacter cluster was selectively enhanced by bottle containment (see above), our results are consistent with other studies using radiotracers (9, 43) with minimal perturbation of in situ conditions; these studies have suggested that the most abundant group does not necessarily grow most rapidly in coastal waters.
An alternative hypothesis to explain the dominance of α-proteobacteria is that grazers selectively eliminate coexisting groups in preference to α-proteobacteria. Bacterivorous protists are known to discriminate prey cells on the basis of cell size, morphology, motility, and surface properties (20). If α-proteobacteria have a characteristic trait (or traits) that effectively reduces grazing pressure relative to other groups, α-proteobacteria could dominate even if their intrinsic growth rates are lower than those of other groups. However, in two experiments conducted in Otsuchi Bay, we found that grazing mortality rates varied little among different phylogenetic groups, a result consistent with the data indicating that the mean cell volume, an important determinant of grazing selectivity (20), differed little among different groups in coastal waters that we examined. Thus, it does not appear that the prevalence of slow-growing α-proteobacteria is accounted for simply by preferential grazing on fast-growing groups. Although it is premature to generalize this conclusion to other environments, our data suggest that further studies are needed to examine the role of other forces, such as viral infection (15), in determining bacterial community compositions in coastal systems.
Our analysis used broad phylogenetic probes (2) that have been used by many others in previous studies (5, 7, 9, 18, 21). Clearly, each broad group potentially contains many different species and phylotypes; i.e., the growth and grazing mortality that we measured represent “collective properties” of the groups, which probably reflect the traits of the dominant species or subgroups. At a narrow phylogenetic level or at the species level, different patterns in controls of community compositions might be detected, given large variations in ecological traits among subclusters of phylogenetic groups. For example, a recent study has shown that SAR11 and Roseobacter exhibit distinctive patterns in large-scale distributions in the oceans (35) even though both subgroups belong to the same broad phylogenetic group, i.e., α-proteobacteria. Experiments have also suggested that responses to substrate additions differ greatly between two γ-proteobacteria subgroups, Alteromonas and SAR86 (11). Nevertheless, growing bodies of evidence have suggested that different ecological traits, including the uptake of dissolved organic matter (6, 26), production and growth (8, 9, 41, 43), and grazer avoidance (21, 32, 36), can be detected by the use of broad phylogenetic probes, presumably because a trait of the broad group largely reflects the trait of the dominant subgroup (or species) in oceanic communities, a hypothesis to be tested by future studies. These differences in ecological traits at a broad phylogenetic level probably explain our results demonstrating that different groups of bacteria vary greatly not only in growth rates but also in relationships between growth rates and oceanographic variables (temperature and chlorophyll a concentrations). These results have important implications for better modeling of bacterial dynamics in marine systems, and support the assertion that broad phylogenetic level analysis is a useful approach in order to examine the ecological roles of specific bacterial groups in aquatic environments.
Acknowledgments
Matthew T. Cottrell provided helpful suggestions concerning the image analysis system. Serial dilution culture experiments were conducted with the assistance of Yumiko Obayashi and Tsuyoshi Shiotani. Hiroshi Ogawa and Hideki Fukuda kindly provided the chlorophyll data. We thank Yoko Nishimura and Chulgoo Kim for assistance and discussions. Thanks are also due to the staff of the International Coastal Research Center, ORI, University of Tokyo, and the captain, officers, and crews of R/V Tansei-Maru for logistic support. We thank DOBIS colleagues, especially Isao Koike and Kazuhiro Kogure, for encouragement throughout the study.
This study was conducted as a part of the DOBIS project (Dynamics of the Ocean Biosystems, MEXT Grant-in-Aid for Creative Basic Research 12NP0201). Financial support was also provided by the 21st Century COE program (grant A14) of Kyoto University and by JSPS grants (12800016 and 13308029) awarded to T.N. T.Y. was supported by a Research Fellowship of the JSPS for Young Scientists.
REFERENCES
- 1.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Deverux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann, R. I., W. Ludwig, and K. Schleiffer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beardsley, C., J. Pernthaler, W. Wosniok, and R. Amann. 2003. Are readily culturable bacteria in coastal North Sea waters suppressed by selective grazing mortality? Appl. Environ. Microbiol. 69:2624-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bevington, P. R. 2003. Data reduction and error analysis for the physical science, 3rd ed. McGraw-Hill, New York, N.Y.
- 5.Bouvier, T. C., and P. A. del Giorgio. 2003. Factors influencing the detection of bacterial cells using fluorescence in situ hybridization (FISH): a quantitative review of published reports. FEMS Microbiol. Ecol. 44:3-15. [DOI] [PubMed] [Google Scholar]
- 6.Cottrell, M. T., and D. L. Kirchman. 2000. Community composition of marine bacterioplankton determined by 16S rDNA clone libraries and fluorescence in situ hybridization. Appl. Environ. Microbiol. 66:5116-5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cottrell, M. T., and D. L. Kirchman. 2000. Natural assemblages of marine proteobacteria and members of the Cytophaga-Flavobacteria cluster consuming low- and high-molecular weight dissolved organic matter. Appl. Environ. Micrbiol. 66:1692-1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cottrell, M. T., and D. L. Kirchman. 2003. Contribution of major bacterial groups to bacterial biomass production (thymidine and leucine incorporation) in the Delaware estuary. Limnol. Oceanogr. 48:168-178. [Google Scholar]
- 9.Cottrell, M. T., and D. L. Kirchman. 2004. Single-cell analysis of bacterial growth, cell size and community structure in the Delaware estuary. Aquat. Microb. Ecol. 34:139-149. [Google Scholar]
- 10.DeLong, E. F., D. G. Franks, and A. L. Alldredge. 1993. Phylogenetic diversity of aggregate-attached vs. free-living marine bacterial assemblages. Limnol. Oceanogr. 38:924-934. [Google Scholar]
- 11.Eilers, H., J. Pernthaler, and R. Amann. 2000. Succession of pelagic marine bacteria during enrichment: a close look at cultivation-induced shifts. Appl. Environ. Microbiol. 66:4634-4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferguson, R. L., E. N. Buckley, and A. V. Palumbo. 1984. Response of marine bacterioplankton to differential filtration and confinement. Appl. Environ. Microbiol. 47:49-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuchs, B., M., G. Wallner, W. Beisker, I. Schwippl, W. Ludwig, and R. Amann. 1998. Flow cytometric analysis of the in situ accessibility of Escherichia coli 16S rRNA for fluorescently labeled oligonucleotide probes. Appl. Environ. Microbiol. 64:4973-4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuchs, B. M., M. V. Zubkov, K. Sahm, P. H. Burkill, and R. Amann. 2000. Changes in community composition during cultures of marine bacterioplankton as assessed by flow cytometric and molecular biological techniques. Environ. Microbiol. 2:191-201. [DOI] [PubMed] [Google Scholar]
- 15.Fuhrman, J. A., and M. Schwalbach. 2003. Viral influence on aquatic bacterial communities. Biol. Bull. 204:192-195. [DOI] [PubMed] [Google Scholar]
- 16.Fuhrman, J. A., and R. T. Noble. 1995. Viruses and protists cause similar bacterial mortality in coastal seawater. Limnol. Oceanogr. 40:1236-1242. [Google Scholar]
- 17.Gasol, J. M., P. A. del Giorgio, R. Massana, and C. M. Duarte. 1995. Active versus inactive bacteria: size dependence in a coastal marine plankton community. Mar. Ecol. Prog. Ser. 128:91-97. [Google Scholar]
- 18.Glöckner, F. O., B. M. Fuchs, and R. Amann. 1999. Bacterioplankton compositions of lake and oceans: a first comparison based on fluorescence in situ hybridization. Appl. Environ. Microbiol. 65:3721-3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.González, J. M., and M. A. Moran. 1997. Numerical dominance of a group of marine bacteria in the alpha subclass of the class Proteobacteria in coastal seawater. Appl. Environ. Microbiol. 63:4237-4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hahn, M. W., and M. G. Höfle. 2001. Grazing of protozoa and its effect on populations of aquatic bacteria. FEMS Microb. Ecol. 35:113-121. [DOI] [PubMed] [Google Scholar]
- 21.Jürgens, K., J. Pernthaler, S. Schalla, and R. Amann. 1999. Morphological and compositional changes in a planktonic bacterial community in response to enhanced protozoan grazing. Appl. Environ. Microbiol. 65:1241-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karner, M., and J. A. Fuhrman. 1997. Determination of active marine bacterioplankton: a comparison of universal 16S rRNA probes, autoradiography, and nucleoid staining. Appl. Environ. Microbiol. 63:1208-1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kirchman, D. L. 2002. The ecology of Cytophaga-Flavobacteria in aquatic environments. FEMS Microbiol. Ecol. 39:91-100. [DOI] [PubMed] [Google Scholar]
- 24.Kirchman, D. L., and J. Rich. 1997. Regulation of bacterial growth rates by dissolved organic carbon and temperature in the equatorial Pacific Ocean. Microb. Ecol. 33:11-20. [DOI] [PubMed] [Google Scholar]
- 25.Landry, M. R., and R. P. Hassett. 1982. Estimating the grazing impact of marine microzooplankton. Mar. Biol. 67:283-288. [Google Scholar]
- 26.Malmstrom, R. R., R. P. Kiene, and D. L. Kirchman. 2004. Identification and enumeration of bacterial assimilation of dimethylsulfoniopropionate (DMSP) in the North Atlantic and Gulf of Mexico. Limnol. Oceanogr. 49:597-606. [Google Scholar]
- 27.Manz, W., R. Amann, W. Ludwig, M. Vancanneyt, and K. H. Schleifer. 1996. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum Cytophaga-Flavobacter-Bacteroides in the natural environment. Microbiology 142:1097-1106. [DOI] [PubMed] [Google Scholar]
- 28.Manz, W., R. Amann, W. Ludwig, M. Wagner, and K. H. Schleifer. 1992. Phylogenetic oligoxynucleotide probes for the major subclasses of proteobacteria: problems and solutions. Syst. Appl. Microbiol. 15:593-600. [Google Scholar]
- 29.Massana, R., and K. Jürgens. 2003. Composition and population dynamics of planktonic bacteria and bacterivorous flagellates in seawater chemostat cultures. Aquat. Microb. Ecol. 32:11-22. [Google Scholar]
- 30.Morris, R. M., M. S. Rappé, S. A. Connon, K. L. Vergin, W. A. Siebold, C. A. Carlson, and S. J. Giovannoni. 2002. SAR11 clade dominates ocean surface bacterioplankton communities. Nature 420:806-810. [DOI] [PubMed] [Google Scholar]
- 31.Nagata, T., R. Fukuda, H. Fukuda, and I. Koike. 2001. Basin-scale geographic patterns of bacterioplankton biomass and production in the subarctic Pacific, July-September 1997. J. Oceanogr. 57:301-313. [Google Scholar]
- 32.Pernthaler, J., T. Posch, K. Šimek, J. Vrba, R. Amann, and R. Psenner. 1997. Contrasting bacterial strategies to coexist with a flagellate predator in an experimental microbial assemblage. Appl. Environ. Microbiol. 63:596-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pinhassi, J., and T. Berman. 2003. Differential growth response of colony-forming α- and γ-proteobacteria in dilution culture and nutrient addition experiments from Lake Kinneret (Israel), the Eastern Mediterranean Sea, and the Gulf of Eilat. Appl. Environ. Microbiol. 69:199-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Riemann, L., G. F., Steward, and F. Azam. 2000. Dynamics of bacterial community composition and activity during a mesocosm diatom bloom. Appl. Environ. Microbiol. 66:578-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Selje, N., M. Simon, and T. Brinkhoff. 2004. A newly discovered Roseobacter cluster in temperature and polar oceans. Nature 427:445-448. [DOI] [PubMed] [Google Scholar]
- 36.Šimek, K., P. Kojecká, J. Nedoma, P. Hartman, J. Vrba, and J. R. Dolan. 1999. Shifts in bacterial community composition associated with different microzooplankton size fractions in a eutrophic reservoir. Limnol. Oceanogr. 44:1634-1644. [Google Scholar]
- 37.Suzuki, M. T. 1999. Effect of protistan bacterivory on coastal bacterioplankton diversity. Aquat. Microb. Ecol. 20:261-272. [Google Scholar]
- 38.Suzuki, M. T., C. M. Preston, G. P. Chavez, and E. D. DeLong. 2001. Quantitative mapping of bacterioplankton populations in seawater: field tests across an upwelling plume in Monterey Bay. Aquat. Microb. Ecol. 24:117-127. [Google Scholar]
- 39.Tremaine, S. C., and A. L. Mills. 1987. Tests of the critical assumptions of the dilution method for estimating bacterivory by microeucaryotes. Appl. Environ. Microbiol. 53:2914-2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wallner, G., R. Erhart, and R. Amann. 1995. Flow cytometric analysis of activated sludge with rRNA-targeted probes. Appl. Environ. Microbiol. 61:1859-1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yokokawa, T., T. Nagata, M. T. Cottrell, and D. L. Kirchman. 2004. Growth rate of the major phylogenetic bacterial group in the Delaware estuary. Limnol. Oceanogr. 49:1620-1629. [Google Scholar]
- 42.Zarda, B., D. Hahn, A. Chatzinotas, W. Schonhuber, A. Neef, R. I. Amann, and J. Zeyer. 1997. Analysis of bacterial community structure in bulk soil by in situ hybridization. Arch. Microbiol. 168:185-192. [Google Scholar]
- 43.Zubkov, M. V., B. M. Fuchs, P. H. Burkill, and R Amann. 2001. Comparison of cellular and biomass specific activities of dominant bacterioplankton groups in stratified waters of the Celtic Sea. Appl. Environ. Microbiol. 67:5210-5218. [DOI] [PMC free article] [PubMed] [Google Scholar]