Abstract
Unicellular algae are the predominant microbial mat-forming phototrophs in the extreme environments of acidic geothermal springs. The ecology of these algae is not well known because concepts of species composition are inferred from cultivated isolates and microscopic observations, methods known to provide incomplete and inaccurate assessments of species in situ. We used sequence analysis of 18S rRNA genes PCR amplified from mat samples from different seasons and different temperatures along a thermal gradient to identify algae in an often-studied acidic (pH 2.7) geothermal creek in Yellowstone National Park. Fiber-optic microprobes were used to show that light for algal photosynthesis is attenuated to <1% over the 1-mm surface interval of the mat. Three algal sequences were detected, and each was present year-round. A Cyanidioschyzon merolae sequence was predominant at temperatures of ≥49°C. A Chlorella protothecoides var. acidicola sequence and a Paradoxia multisita-like sequence were predominant at temperatures of ≤39°C.
Nondescript spherical unicellular red algae (Rhodophyta) predominate in microbial mats in high-temperature (45 to 55°C) acidic (pH of <3) geothermal habitats in Yellowstone National Park and similar habitats worldwide (10, 49). Unicellular Chlorella-like algae (Chlorophyta) whose cells closely resemble those of thermophilic red algae are also present in acidic geothermal mats; however, Chlorella is not known to thrive at high temperatures (>45°C) (17). Knowledge of the ecology of these algae is based largely on traditional methods, i.e., cultivated species and microscopic descriptions of algal cells in environmental samples (27). However, microscopic identification of simple unicellular algal species is difficult (53). Indeed early studies refer to thermophilic red algae as Chlorella (3, 4). Moreover, cultivation-independent (molecular) analyses of microbial communities have repeatedly demonstrated that cultivated isolates are inadequate to describe the ecology of microbes in natural habitats (47, 48, 51, 52).
Recognizing the limitations of traditional algal classification schemes, researchers have used sequence analysis of 18S rRNA and ribulose bisphosphate carboxylase genes to establish phylogenetic relationships among cultivated strains of unicellular thermoacidophilic and Chlorella-like algae (1, 13, 24, 25, 30-33, 53). In many instances traditional generic designations based on cytomorphological and physiological characteristics are incongruent with genetic phylogeny, and generic nomenclature of some strains requires revision. In the case of the Chlorella-like algae, sequence analyses indicate that the ability to thrive in low-pH environments has evolved independently in diverse lineages of green algae (30) and that the genus “Chlorella” has been applied to phylogenetically divergent but morphologically similar unicellular algae (30-33, 53). In the case of thermoacidophilic red algae, three genera and at least four species have been described based on traditional characteristics, Galdieria maxima, Galdieria sulphuraria, Cyanidium caldarium, and Cyanidioschyzon merolae. In general, G. sulphuraria cells are spherical and indistinguishable from those of C. caldarium; however, G. sulphuraria strains are distinguished by their ability to grow readily under dark heterotrophic conditions. G. maxima strains grow slowly under dark heterotrophic conditions and are distinguished by large cell size. C. merolae is a strict autotroph distinguished by crescent- or elliptically shaped cells. Genetic studies indicate that all G. sulphuraria strains form a well-supported clade (nomenclature of some strains within this clade likely requires revision [25]) while species of the three remaining morphologically and physiologically diverse genera, C. caldarium, C. merolae, and G. maxima, are closely related to each other but genetically distinct from G. sulphuraria (13, 25). Genetic sequence analysis has become a generally acceptable method of identifying and classifying microorganisms. Much additional effort will be required to resolve ambiguities in generic concepts and nomenclature of algae based on traditional criteria and sequence data. However, the taxonomic ambiguities themselves and the discovery of new uncultivated thermoacidophilic algae in situ using molecular sequencing (13) indicate that sequence identification of “Chlorella” and thermoacidophilic algae inhabiting acidic geothermal environments is warrantable.
The Norris Geyser basin in Yellowstone National Park contains some of the most extensively studied acidic algal mat communities (10, 15). One acidic stream in particular, Nymph Creek, has been the focus of numerous investigations (7, 8, 10, 14, 16, 17, 19, 22, 38-40, 42, 43); however, microbial species in the Nymph Creek algal mat have been characterized using only traditional microbiological methods. We are using molecular analyses and microsensors to examine microbial populations and processes in the Nymph Creek mat (19, 42-44). Here, we use PCR amplification and 18S rRNA gene sequencing to identify thermoacidophilic red algae and Chlorella-like algae that predominate in the mat. We examine algal species composition over an annual cycle and across temperature gradients and use fiber-optic microprobes to describe the microenvironment for algal photosynthesis through the vertical aspect of the mat.
MATERIALS AND METHODS
Site description.
Nymph Creek is an acidic (pH, ∼2.7) stream located near the Norris Geyser basin of Yellowstone National Park, Wyoming. The creek originates from several permanent and ephemeral 56°C to 60°C hot springs. The shallow (1- to 2-cm-deep), warm spring effluents combine to form a creek that is about 1 to 2 m wide and flows for approximately 130 m before it empties into Nymph Lake. The upstream region of the creek is quite dynamic and changes considerably over time. Shallow springs emerge and recede due to seasonal changes in groundwater. In the shallow, warm effluent springs, temperature gradients from ∼52°C to 37°C can occur over short distances (∼10 cm) as measured from the center of the effluent channel to the stream edge. Sections of the algal mat that develop in the shallow, warm effluents are impacted by wide temperature changes caused by stochastic factors such as silt deposition, fallen arboreal debris, hail, or trampling by bison, all of which can alter warm water flow over sections of the mat. A temperature gradient also forms along the length of the mat as water flows downstream away from the thermal sources. Nondescript spherical green algal cells ∼2 μm to 4 μm in diameter are the predominant cells in mat samples. Near the high-temperature limit of algal growth (∼52°C) the “mat” is a thin green biofilm (<1 mm thick). At lower temperatures (∼49°C to 35°C) the mat is typically 3 to 5 mm thick.
Temperature data loggers.
Temperature data for an upstream and a downstream site in the mat were collected using HOBO XT data loggers (Onset Computer Corporation, Bourne, MA) recording at 2-hour intervals from small cylindrical probes (∼3-mm diameter) placed horizontally in the upper layer of the mat. The temperature at the upstream site when the probe was installed was ∼50°C. Another probe was installed ∼100 m downstream in a low-temperature region of the mat (∼30°C). The depth of the water over the probes at both sites was ∼1 cm. The probes were periodically repositioned due to shifts in the streambed or other disturbances.
Sample collection for molecular analyses and cultivation.
Samples for PCR analysis were collected in 2-ml plastic microcentrifuge tubes by scraping portions of the mat from the streambed using clean weighing spatulas. Samples were frozen on dry ice and stored at −20°C. To evaluate seasonal variations in algal species, samples were collected in December of 1999 and in April, June, and August of 2000 from upstream and downstream regions of the mat near areas containing the temperature probes. To examine the influence of temperature on algal species composition, a set of samples was collected on the same day in August 2001, from sites along the mat at increasing distances downstream from the warm thermal source springs where temperatures were 51°C, 49°C, 38°C, 35°C, and 30°C. Nucleic acids were obtained from mat samples and from cultivated algal isolates, using a bead beating cell lysis, nucleic acid extraction protocol as previously described (43).
Samples were collected from a ∼50°C region of the mat for enrichments of thermoacidophilic algae. These enrichments were performed in medium 17, pH 2.9, described on the website of the Culture Collection of Algae (SAG), Göttingen, Germany (http://www.epsag.uni-goettingen.de/html/culturemedia.html). Cultures were incubated at 50°C under an irradiance of 100 μmol photons m−2 s−1 provided by cool, white fluorescent lamps in 5% CO2 in air. Single algal colonies were repeatedly restreaked on SAG agar plates to obtain pure cultures. Pure culture isolates and algal mat samples from an ∼50°C site were incubated at 50°C in SAG medium supplemented with 0.2% d-glucose and 0.2% yeast extract without light for 2 weeks to test for the ability of thermoacidophilic algae to grow heterotrophically.
Chlorella-like isolates were obtained by spreading 10-fold serially diluted mat samples collected at ∼35°C onto CA agar (37) and successively picking portions of colonies and replating them to obtain pure cultures. The plates were incubated at room temperature under fluorescent lamps. A Nymph Creek Chlorella-like isolate, ROF/NC15, and a thermoacidophilic red algal isolate, PC15, were deposited in the University of Oregon Culture Collection of Microorganisms from Extreme Environments (Ecology and Evolution Program, Department of Biology, University of Oregon, Eugene).
18S rRNA gene PCR primers.
Unless otherwise indicated, sequences were amplified using the universal eukaryotic primers 5′-GTCAGAGGTGAAATTCTTGGATTTA-3′ and 5′-AGGGCAGGGACGTAATCAACG-3′ (25), which amplify an ∼700-bp region of the 18S rRNA gene from nucleotides 912 to 1593 of C. caldarium, GenBank reference AB090833.
PCR, cloning, sequencing, and taxonomic analysis.
Cloning of PCR-amplified rRNA gene sequences was performed using a TOPO-TA PCR cloning kit (Invitrogen, Carlsbad, Calif.), plasmid isolation was performed using a QIAprep Spin Miniprep kit (Qiagen, Valencia, Calif.), and sequencing was performed using a BigDye Ready Reaction Termination Mix (PE ABI, Foster City, Calif.) as previously described (18, 42). Raw sequence data were analyzed using Sequencher software (Gene Codes Corporation, Ann Arbor, Mich.). Preliminary identification of sequences was done using the BLASTn algorithm at the NCBI web site (5). Sequences were aligned using CLUSTAL X (11, 29). Dendrograms were generated using PAUP* 4.0b8 software (46) as described previously (28) using neighbor-joining analyses with uncorrected distance. Bootstrap values were from 1,000 replicates.
Microscale light measurements.
The penetration of photosynthetically active radiation (PAR) for oxygenic photosynthesis (λ = 400 to 700 nm) in the algal mat was measured with a fiber-optic scalar irradiance meter (34). Small, undisturbed core samples of the mat were transferred within 1 h to a nearby laboratory, where the light measurements were conducted. The sample was immersed in Nymph Creek water and illuminated with collimated light from a fiber-optic halogen lamp (Schott KL1500). Using a manually operated micromanipulator (Märtzhäuser MM33), a scalar irradiance microprobe (35) was inserted into the mat under a zenith angle of 135 and the intensity of PAR was measured in steps of 100-μm vertical distance (34).
Nucleotide sequence accession numbers.
GenBank reference numbers of 18S rRNA gene sequences detected in this study are AY422216 through AY422223.
RESULTS
Algal species detected.
Three algal 18S rRNA gene phylotypes designated types I, II, and III were repeatedly detected among clones from PCR-generated libraries of mat samples (Table 1). Sequence variation within each phylotype was <1%. Some of this variation may be attributed to unique microbial populations (20, 21); to artifacts associated with the PCR amplification, cloning, and sequencing approach to microbial community analysis (45); or to intragenomic sequence variation (9, 12). Since we have no evidence that suggests that the variation within each group represents distinct populations, we tentatively infer the presence of three algal species in Nymph Creek.
TABLE 1.
Cloned 18S rRNA gene sequences of algae detected from a microbial mat in an acidic geothermal creek in samples collected from a warm upstream (∼45 to 50°C) and a cool downstream (∼25 to 35°C) site at different times of the year and in samples collected at different temperatures along the length of the mat at increasing distances from the thermal source on a single day in August
| Type of algal 18S rRNA gene | No. of sequences detecteda
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Warm upstream site
|
Cool downstream site
|
Temp (°C)
|
|||||||||||
| Apr | Jun | Aug | Dec | Apr | Jun | Aug | Dec | 51 | 47 | 38 | 35 | 30 | |
| I | 9 | 7 | 8 | 7 | 0 | 0 | 0 | 0 | 12 | 7 | 0 | 0 | 0 |
| II | 0 | 0 | 3 | 3 | 2 | 6 | 26 | 5 | 0 | 1 | 5 | 13 | 3 |
| III | 0 | 0 | 1 | 2 | 15 | 6 | 5 | 6 | 0 | 0 | 1 | 2 | 7 |
Number of cloned algal sequences detected from seasonal samples collected over an annual cycle from upstream and downstream regions of the mat in April (Apr), June (Jun), August (Aug), and December (Dec). Transect samples were collected on the same day in August at different temperatures along the length of the mat.
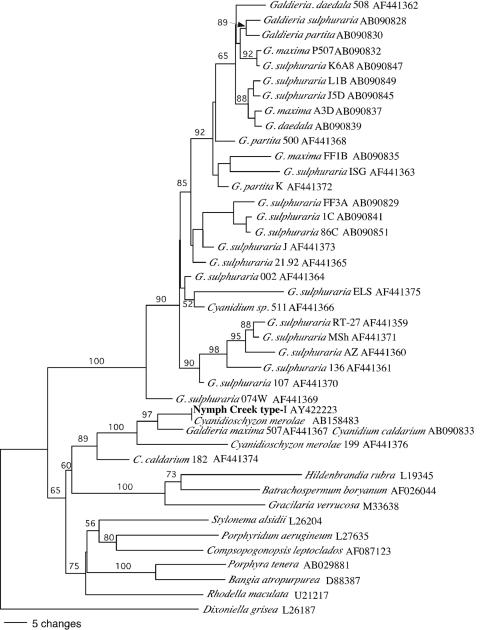
The type I sequence is most similar (>99% to 100% sequence similarity) to three (nonidentical) 18S rRNA gene sequences within the genome of Cyanidioschyzon merolae 10D (36) (GenBank accession no. AB158483, AB158484, and AB158485). The sequences of all algal isolates (six were sequenced) obtained from a 50°C mat sample from the upstream region of the mat were all type I. The spherical, 2- to 4-μm-diameter cells of each isolate were indistinguishable, by light microscopy, from cells in the mat. The dendrogram (Fig. 1) illustrates the type I sequence groups in a clade containing the sequences of C. merolae, C. caldarium, and Galdieria maxima (Fig. 1).
FIG. 1.
Dendrogram based on ∼680 nucleotides of 18S rRNA genes relating the predominant Nymph Creek thermoacidophilic alga (type I) to other red algal sequences. Bootstrap values (>50%) are indicated at nodes. GenBank reference numbers are listed next to species.
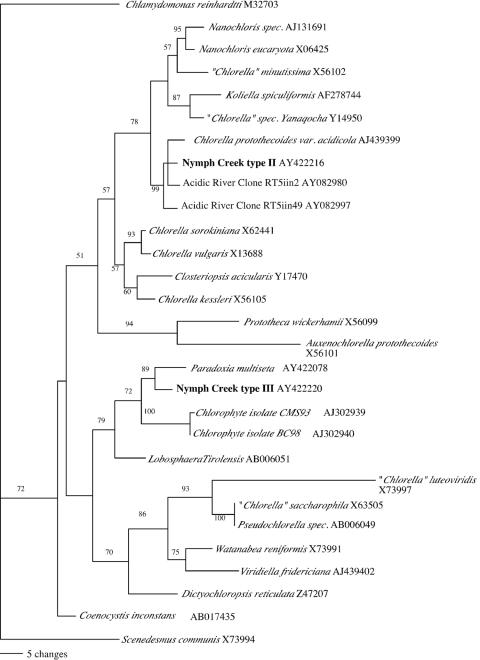
Type II sequences are most similar (>99% to 100% sequence similarity) to the 18S rRNA gene sequence of the acid-tolerant species “Chlorella” protothecoides var. acidicola (2, 30) (Fig. 2). Type II sequences are also nearly identical (>99% sequence similarity) to algal 18S rRNA gene sequences detected by PCR in an acidic river in Spain (6) (Fig. 2). Type III sequences are most similar (98% sequence similarity) to the 18S rRNA gene of Paradoxia multisita and to the 18S rRNA gene sequences of unidentified Chlorellaceae strains CMS93 and BC98 (Fig. 2). Cultivated algal isolates whose 18S rRNA gene sequences matched type II clones were obtained from a 35°C mat sample. Type II cells are spherical and indistinguishable by light microscopy from cells in the mat. Cultivated isolates representing type III 18S rRNA gene sequences were not obtained.
FIG. 2.
Dendrogram tree based on ∼680 nucleotides of 18S rRNA genes relating predominant Nymph Creek Chlorella-like algae (types II and III) to other Chlorella-like algal sequences. Bootstrap values (>50%) are indicated. GenBank reference numbers are listed next to species.
Absence of G. sulphuraria.
G. sulphuraria strains form a highly supported clade of thermoacidophilic algae that is distantly related to other thermoacidophilic algal species (13, 25) (Fig. 1). Traditional studies suggest that facultative heterotrophic G. sulphuraria-like thermoacidophilic algae are abundant in Nymph Creek and similar habitats in the Norris Geyser basin (10, 16, 17). However, none of the 18S rRNA gene sequences detected in our analyses grouped within the G. sulphuraria clade and type I algal isolates did not grow heterotrophically under conditions where a control G. sulphuraria strain (University of Texas at Austin Culture Collection of Algae strain UTEX 2393) grew readily.
It seemed unlikely that PCR primer bias explained the lack of G. sulphuraria-like algal sequences in the mat, since complementary priming sites exist on G. sulphuraria 18S rRNA gene sequences in public databases and the primer set amplified G. sulphuraria 18S rRNA gene sequences from control G. sulphuraria genomic DNA. Nonetheless, a second set of universal eukaryotic primers, NS1 and NS2 (50, 53), targeting an alternate region of the 18S rRNA gene was used to amplify DNA from a 48°C sample collected from the upstream region of the mat and DNA from a type I isolate. The sequences of all algal clones from the NS1/NS2 library (10 were sequenced) were >99% to 100% similar to the sequence of the type I isolate. No G. sulphuraria or other algal sequences were detected.
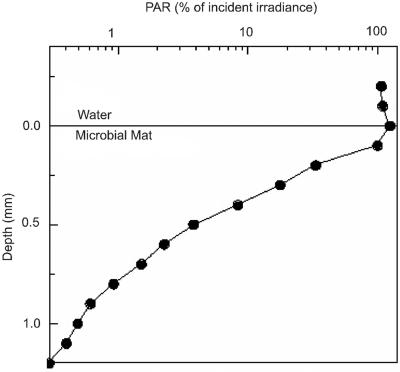
Template competition for universal eukaryotic primers likely favors detection of abundant algal species. Since G. sulphuraria sequences were not detected in our clone libraries, we hypothesized that G. sulphuraria cells are rare in the mat. G. sulphuraria is known to survive in dark endolithic environments via cryptic growth (26); we speculated that they might occupy a dark subsurface niche in the Nymph Creek mat. However, the extent of light (PAR) penetration in the Nymph Creek mat was unknown. We used a light microsensor to examine light penetration. The results show that light available for algal photosynthesis was strongly attenuated over the first 1-mm vertical interval of the mat (Fig. 3), indicating that an extremely low-light subsurface niche exists below this interval. This result is consistent with previous oxygen microsensor analyses of the Nymph Creek mat showing that oxygenic photosynthesis is restricted to the first 1-mm surface interval (38). We attempted to selectively enrich G. sulphuraria-like algae by inoculating heterotrophic enrichment broths with upstream (∼48°C) mat samples and incubating them under dark conditions. No algal growth was observed after 2 weeks. Control broths inoculated with G. sulphuraria grew well under the same conditions.
FIG. 3.
Light penetration depth profile of photon scalar irradiance (PAR, λ = 400 to 700 nm) available for oxygenic photosynthesis in the Nymph Creek mat. Data were normalized to the down-welling photon irradiance at the mat surface and are shown on a logarithmic intensity scale.
Temperature and algal species composition.
To examine the influence of temperature on algal species composition, we analyzed sequences in samples collected at different temperatures from sites at increasing distances downstream from the thermal source springs (Table 1). All 51 sequences obtained from the temperature distribution study were either type I, II, or III. Nineteen of 20 sequences from ≥47°C samples were type I. All 31 sequences from ≤38°C samples were type II or type III. This suggests that the distribution of the thermoacidophilic red algae is largely confined to the first ∼20 m of mat downstream from the thermal source springs, where temperatures are ≥39°C.
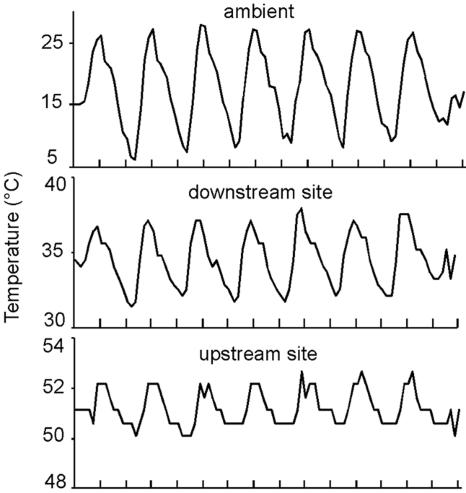
Geothermal streams can experience brief surges in temperature due to, for example, geyser eruptions or decreases in temperature caused by runoff from snowmelt (10). Sampling during or following such events can skew interpretations of temperature influence on algal population distributions (10). Data logger readings suggest that Nymph Creek temperature patterns were stable prior to collection of samples for algal temperature distribution analyses. Graphs of temperatures illustrate typical daily temperature dynamics for a week in August prior to sampling (Fig. 4). Diurnal peaks and troughs in stream temperatures correlate with those of ambient temperatures as previously reported (39). The temperature time series confirms that air temperature impacts mat temperatures even in the upstream region despite the continuous input of warm water into the creek channel. Changes in upstream and downstream temperature that occurred over extended time periods were also noted. These were mainly decreasing temperatures due to shifts in the flow of warm water over the probes caused by sediment deposition or due to cold winter conditions (10).
FIG. 4.
Example of typical August temperatures recorded every 2 hours by data loggers connected to sensors placed in the surface of the algal mat in an upstream high-temperature region of Nymph Creek, a downstream lower-temperature region, and a shaded site along the creek. Tick marks represent noon and midnight; peaks correspond to midday readings.
Seasonal algal species composition.
To explore the possibility that algal species composition varied over an annual cycle, we collected samples from both the warm upstream and the cooler downstream regions of the mat four times during an annual cycle (Table 1). Type I sequences were the most abundant clones in each of the four upstream seasonal samples and represented 31 of 40 total algal clones (Table 1). Nine Chlorella-like (six type II and three type III) sequences were also detected in upstream seasonal samples. Since Chlorella-like algae are not known to thrive at high temperatures, we suspect that the high proximity of cooler niches and shifts in warm water flows account for their sequences in upstream samples. Only Chlorella-like, type II and III sequences were detected in samples from the downstream site, 39 of type II and 32 of type III (Table 1). No other algal sequences were detected, and algal species in the Nymph Creek mat did not vary over an annual cycle despite variations in photoperiod that range from ∼8 daylight hours in December to ∼16 daylight hours in June.
DISCUSSION
Our analyses of 18S rRNA gene sequences from mat samples and cultivated isolates indicate that a single species of strictly photoautotrophic red algae with a spherical morphology, 2- to 4-μm cell diameter, and 18S rRNA gene sequence matching that of a Cyanidioschyzon merolae strain (sequence type I) is predominant in the upstream, high-temperature (47°C to 51°C) region of the Nymph Creek mat (Table 1). Unlike Cyanidioschyzon merolae, algal cells in the Nymph Creek mat, and isolates with type I sequences cultivated from the mat, are not oval, crescent, or club shaped (1, 41). This incongruity may suggest that not all C. merolae strains are aspherical; however, ambiguities between molecular taxonomic analyses and thermoacidophilic algal nomenclature based on determinative characteristics have been noted (1, 13, 25). For example, Galdieria maxima is a facultative heterotroph, and its cells are spherical and notably large (10- to 16-μm diameter) compared to those of other thermoacidophilic algal species (1); however, 18S rRNA (25) and rbcL (13) gene sequence analyses indicate that Galdieria maxima is much more closely related to Cyanidioschyzon merolae than to other Galdieria species (13). The nomenclature of thermoacidophilic algae is further confounded by instances of identical rbcL and 18S rRNA gene sequences that are associated with different genera/species in public databases, for example, Galdieria maxima and Cyanidium caldarium rbcL genes AY391370 and D63676 and 18S rRNA genes AB090833 and AF441367. Such ambiguities underscore the need for continued clarification of nomenclature and taxonomy of these algae. We tentatively refer to the Nymph Creek (type I) alga as Cyanidioschyzon merolae based on 18S rRNA gene sequence.
Nomenclature aside, a predominance of strictly autotrophic spherical thermoacidophilic algae in Nymph Creek is consistent with a traditional microbiological study describing predominantly strictly autotrophic, spherical thermoacidophilic algae in acidic springs in the Norris Geyser basin (15) and with a recent molecular analysis of a low-pH, endolithic algal community in the Norris Geyser basin describing abundant spherical algae with 18S rRNA gene sequences >99% similar to those of the closest type I relative, the G. maxima/C. caldarium sequence (Fig. 1) (46a). A predominance of strict autotrophic thermoacidophilic algae contrasts with other traditional studies suggesting that a single species of facultative heterotrophic (G. sulphuraria-like) thermoacidophilic algae predominates in Nymph Creek and springs throughout the Norris Geyser basin (10, 16, 17). We suspect that a bias toward the isolation of G. sulphuraria on heterotrophic plating medium (10) and the microscopic resemblance between G. sulphuraria and C. caldarium cells likely account for inferences of G. sulphuraria being widespread in the Norris Geyser basin.
Nymph Creek is routinely described in terms of a C. caldarium mat. It is notable that Chlorella-like algal sequences were the predominant type detected over much of the length of the Nymph Creek mat, ∼100 of 130 m downstream from the 38°C site (Table 1). The reason for the scarcity of type I sequences below the 38°C site is unclear. We speculate that a low washout rate of type I algal cells from upstream and competition from Chlorella-like algae at lower temperatures are significant factors. A shift from red algae to Chlorella-like algae at ∼39°C is consistent with a traditional study of a similar Yellowstone stream showing a sharp transition from thermoacidophilic red algae to “Chlorella” species at ∼38°C due to seasonal shifts in temperature (17). Abrupt transitions from red algae (green-pigmented mat) to diatoms (brown-pigmented mat) at 39°C have also been noted in geothermal springs in Japan (10).
Although Chlorella-like algae have been noted in Nymph Creek and other acidic areas of the Norris Geyser basin, species have not been identified (10, 39). The 18S rRNA gene sequence of the type II strain suggests that it is Chlorella protothecoides var. acidicola, an acid-tolerant alga that was originally isolated from low-pH soil (2, 30). Sequences nearly identical to type II have also been detected in an acidic river in Spain (6). This suggests that C. protothecoides-like algae may be widespread in low-pH aquatic habitats (23). A second Chlorella-like alga (type III) also appears to be abundant in the Nymph Creek mat. The 18S rRNA gene sequence of this alga suggests that it is a relative of Paradoxia multisita AY422078 (98% sequence similarity). However, the alga harboring the type III sequence has not been isolated. Sequence information suggests that neither Nymph Creek Chlorella-like alga is a “Chlorella” species sensu stricto (30, 32, 33).
Molecular analyses are beginning to provide the first few cultivation-independent assessments of algal species that thrive in acidic geothermal habitats (13, 25). Accurate information about the identity of algae and other microbial species in situ, and an understanding of their distribution, provides a base on which realistic concepts of the ecology of these extreme ecosystems can be constructed. Knowledge of species composition will likely enhance interpretations of past and future studies of bulk properties in the often-studied Nymph Creek mat.
Acknowledgments
We thank the National Science Foundation (Microbial Observatory Grant 9977922), the Research Institute for Children at New Orleans, the Danish Natural Science Research Council, and the Thermal Biology Institute at Montana State University for financial support.
This project was conducted under the direction of the Yellowstone Center for Resources following the guidelines for research in Yellowstone National Park.
We thank John Varley and Christie Hendrix for assistance.
REFERENCES
- 1.Albertano, P., C. Ciniglia, G. Pinto, and A. Pollio. 2000. The taxonomic position of Cyanidium, Cyanidioschyzon, and Galdieria: an update. Hydrobiologia 433:137-143. [Google Scholar]
- 2.Albertano, P., and R. Taddei. 1984. Chlorella protothecoides Krüger var. acidocola, a new variety from very low pH environments. Algol. Stud. 37:401-408. [Google Scholar]
- 3.Allen, M. B. 1954. Studies of a blue-green Chlorella. Proc. 8th Int. Bot. Congr. 17:41-42. [Google Scholar]
- 4.Allen, M. B. 1959. Studies with Cyanidium caldarium, an anomalously pigmented chlorophyte. Arch. Mikrobiol. 32:270-277. [DOI] [PubMed] [Google Scholar]
- 5.Altschul, S., T. Madden, A. Schaeffer, J. Zhang, Z. Zhang, W. Miller, and D. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amaral Zettler, L. A., F. Gomez, E. Zettler, B. G. Keenan, R. Amils, and M. L. Sogin. 2002. Eukaryotic diversity in Spain's River of Fire. Nature 417:137. [DOI] [PubMed] [Google Scholar]
- 7.Ball, J. W., R. B. McCleskey, D. K. Nordstrom, J. M. Holloway, and P. L. Verplanck. 2002. Water-chemistry data for selected springs, geysers, and streams in Yellowstone National Park, Wyoming, 1999-2000. USGS open-file report 02-382, p. 104. U.S. Geological Survey, Reston, Va.
- 8.Belly, R. T., M. R. Tansey, and T. D. Brock. 1973. Algal excretion of 14C-labeled compounds and microbial interactions in Cyanidium caldarium mats. J. Phycol. 9:123-127. [Google Scholar]
- 9.Boucher, Y., C. J. Douady, A. K. Sharma, M. Kamekura, and W. F. Doolittle. 2004. Intragenomic heterogeneity and intergenomic recombination among haloarchaeal rRNA genes. J. Bacteriol. 186:3980-3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brock, T. D. 1978. The genus Cyanidium, p. 255-302. In T. D. Brock (ed.), Thermophilic microorganisms and life at high temperatures. Springer, New York, N.Y.
- 11.Chenna, R., H. Sugawara, T. Koike, R. Lopez, T. J. Gibson, D. G. Higgins, and J. D. Thompson. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31:3497-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cilia, V., B. Lafay, and R. Christen. 1996. Sequence heterogeneities among 16S ribosomal RNA sequences, and their effect on phylogenetic analyses at the species level. Mol. Biol. Evol. 13:451-461. [DOI] [PubMed] [Google Scholar]
- 13.Ciniglia, C., H. S. Yoon, A. Pollio, G. Pinto, and D. Bhattacharya. 2004. Hidden biodiversity of the extremophilic Cyanidiales red algae. Mol. Ecol. 13:1827-1838. [DOI] [PubMed] [Google Scholar]
- 14.Cockell, C. S., and L. J. Rothschild. 1999. The effects of UV radiation A and B on diurnal variation in photosynthesis in three taxonomically and ecologically diverse microbial mats. Photochem. Photobiol. 69:203-210. [DOI] [PubMed] [Google Scholar]
- 15.De Luca, P., R. Gambardella, and A. Merola. 1979. Thermoacidophilic algae of North and Central America. Bot. Gaz. 140:418-427. [Google Scholar]
- 16.Doemel, W. N., and T. D. Brock. 1971. The physiological ecology of Cyanidium caldarium. J. Gen. Microbiol. 67:17-32. [Google Scholar]
- 17.Doemel, W. N., and T. D. Brock. 1970. The upper temperature limit of Cyanidium caldarium. Arch. Mikrobiol 72:326-332. [DOI] [PubMed] [Google Scholar]
- 18.Ferris, M. J., M. Kuhl, A. Wieland, and D. M. Ward. 2003. Cyanobacterial ecotypes in different optical microenvironments of a 68°C hot spring mat community revealed by 16S-23S rRNA internal transcribed spacer region variation. Appl. Environ. Microbiol. 69:2893-2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferris, M. J., T. S. Magnuson, J. A. Fagg, R. Thar, M. Kuhl, K. B. Sheehan, and J. M. Henson. 2003. Microbially mediated sulphide production in a thermal, acidic algal mat community in Yellowstone National Park. Environ. Microbiol. 5:954-960. [DOI] [PubMed] [Google Scholar]
- 20.Ferris, M. J., G. Muyzer, and D. M. Ward. 1996. Denaturing gradient gel electrophoresis profiles of 16S rRNA-defined populations inhabiting a hot spring microbial mat community. Appl. Environ. Microbiol. 62:340-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferris, M. J., and D. M. Ward. 1997. Seasonal distributions of dominant 16S rRNA-defined populations in a hot spring microbial mat examined by denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 63:1375-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fishbain, S., J. G. Dillon, H. L. Gough, and D. A. Stahl. 2003. Linkage of high rates of sulfate reduction in Yellowstone hot springs to unique sequence types in the dissimilatory sulfate respiration pathway. Appl. Environ. Microbiol. 69:3663-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Franken, M., and W. Franken. 1977. Limnologische Untersuchungen am grossen Bullensee, einem sauren Heidesee Norddeutschlands I. Chemie, Hydrologie, Phytoplankton. Arch. Hydrobiol. Suppl. 53:364-403. [Google Scholar]
- 24.Gross, W. 1999. Revision of comparative traits for the acido- and thermophilic red algae Cyanidium and Galdieria, p. 439-446. In J. Seckbach (ed.), Enigmatic microorganisms and life in extreme environments. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 25.Gross, W., I. Heilmann, D. Lenze, and C. Schnarrenberger. 2001. Biogeography of the Cyanidiaceae (Rhodophytes) based on 18S ribosomal RNA sequence data. Eur. J. Phycol. 36:275-280. [Google Scholar]
- 26.Gross, W., J. Kuver, G. Tischendorf, N. Bouchaala, and W. Busch. 1998. Cryptoendolithic growth of the red alga Galdieria sulphuraria in volcanic areas. Eur. J. Phycol. 33:25-31. [Google Scholar]
- 27.Gross, W., and C. Oesterhelt. 1999. Ecophysiological studies on the red alga Galdieria sulphuraria isolated from southwest Iceland. Plant Biol. 1:694-700. [Google Scholar]
- 28.Hall, B. 2001. Phylogenetic trees made easy: a how-to manual for molecular biologists. Sinauer Associates, Inc., Sunderland, Mass.
- 29.Higgins, D. G., and P. M. Sharp. 1988. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene 73:237-244. [DOI] [PubMed] [Google Scholar]
- 30.Huss, V. A., C. Ciniglia, P. Cennamo, S. Cozzolino, G. Pinto, and A. Pollio. 2002. Phylogenetic relationships and taxonomic position of Chlorella-like isolates from low pH environments (pH < 3.0). BMC Evol. Biol. 2:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huss, V. A., and M. L. Sogin. 1990. Phylogenetic position of some Chlorella species within the chlorococcales based upon complete small-subunit ribosomal RNA sequences. J. Mol. Evol. 31:432-442. [DOI] [PubMed] [Google Scholar]
- 32.Huss, V. A. R., C. Frank, E. C. Hartman, M. Hirmer, A. Kloboucek, B. M. Seidel, P. Wenzeler, and E. Kessler. 1999. Biochemical taxonomy and molecular phylogeny of the genus Chlorella sensu lato (Chlorophyta). J. Phycol. 35:587-598. [Google Scholar]
- 33.Karsten, U., T. Friedl, R. Schumann, K. Hoyer, and S. Lembcke. 2005. Mycosporine-like amino acids and phylogenies in green algae: parasiola and its relatives from the trebouxiophyceae (Chlorophyta). J. Phycol. 41:557-566. [Google Scholar]
- 34.Kühl, M., C. Lassen, and N. P. Revsbech. 1997. A simple light meter for measurements of PAR (400-700 nm) with fiber-optic microprobes: application for P vs. I measurements in microbenthic communities. Aquat. Microb. Ecol. 13:197-207. [Google Scholar]
- 35.Lassen, C., H. Plough, and B. B. Jørgensen. 1992. A fibre-optic scalar irradiance microsensor: application for spectral light measurements in sediments. FEMS Microbiol. Ecol. 86:247-254. [Google Scholar]
- 36.Matsuzaki, M., O. Misumi, I. T. Shin, S. Maruyama, M. Takahara, S. Y. Miyagishima, T. Mori, K. Nishida, F. Yagisawa, Y. Yoshida, Y. Nishimura, S. Nakao, T. Kobayashi, Y. Momoyama, T. Higashiyama, A. Minoda, M. Sano, H. Nomoto, K. Oishi, H. Hayashi, F. Ohta, S. Nishizaka, S. Haga, S. Miura, T. Morishita, Y. Kabeya, K. Terasawa, Y. Suzuki, Y. Ishii, S.Asakawa, H. Takano, N. Ohta, H. Kuroiwa, K. Tanaka, N. Shimizu, S. Sugano, N. Sato, H. Nozaki, N. Ogasawara, Y. Kohara, and T. Kuroiwa. 2004. Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature 428:653-657. [DOI] [PubMed] [Google Scholar]
- 37.Nishihare, N., S. Horiike, T. Takahashi, T. Kosaka, Y. Shigenaka, and H. Hosoya. 1998. Cloning and characterization of endosymbiotic algae isolated from Paramecium bursaria. Protoplasma 203:91-99. [Google Scholar]
- 38.Revsbech, N. P., and D. M. Ward. 1983. Oxygen microelectrode that is insensitive to medium chemical composition: use in an acid microbial mat dominated by Cyanidium caldarium. Appl. Environ. Microbiol. 45:755-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rothschild, L. J. 2001. Algal physiology at high temperature, low pH and variable pCO2: implications for evolution and ecology, p. 51-63. In A.-L. Reysenbach, M. Voytek, and R. Mancinelli (ed.), Thermophiles: biodiversity, ecology and evolution. Kluwer Academic/Plenum Publishers, New York, N.Y.
- 40.Rothschild, L. J. 1994. Elevated CO2: impact on diurnal patterns of photosynthesis in natural microbial ecosystems. Adv. Space Res. 14:285-289. [DOI] [PubMed] [Google Scholar]
- 41.Seckbach, J. 1994. The natural history of Cyanidium (Geitler 1933): past and present perspectives, p. 99-112. In J. Seckbach (ed.), Evolutionary pathways and enigmatic algae: Cyanidium caldarium (Rhodophyta) and related cells. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 42.Sheehan, K. B., J. A. Fagg, M. J. Ferris, and J. M. Henson. 2003. PCR detection and analysis of the free-living amoeba Naegleria in hot springs in Yellowstone and Grand Teton National Parks. Appl. Environ Microbiol. 69:5914-5918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sheehan, K. B., M. J. Ferris, and J. M. Henson. 2003. Detection of Naegleria in a thermal acidic spring. J. Eukaryot. Microbiol. 50:263-265. [DOI] [PubMed] [Google Scholar]
- 44.Sheehan, K. B., J. M. Henson, and M. J. Ferris. 2005. Legionella species diversity in an acidic biofilm community in Yellowstone National Park. Appl. Environ. Microbiol. 71:507-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Speksnijder, A. G. C. L., G. A. Kowalchuk, S. D. Jong, E. Kline, J. R. Stephen, and H. J. Laanbroek. 2001. Microvariation artifacts introduced by PCR and cloning of closely related 16S rRNA gene sequences. Appl. Environ. Microbiol. 67:469-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Swofford, D. L. 1998. PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 4. Sinauer Associates, Sunderland, Mass.
- 46a.Walker, J. J., J. R. Spear, and N. R. Pace. 2005. Geobiology of a microbial endothilic community in the Yellowstone geothermal environment. Nature 434:1011-1014. [DOI] [PubMed] [Google Scholar]
- 47.Ward, D. M. 1998. A natural species concept for prokaryotes. Curr. Opin. Microbiol. 1:271-277. [DOI] [PubMed] [Google Scholar]
- 48.Ward, D. M., M. M. Bateson, R. Weller, and A. L. Ruff-Roberts. 1992. Ribosomal RNA analysis of microorganisms as they occur in nature. Adv. Microb. Ecol. 12:219-286. [Google Scholar]
- 49.Ward, D. M., R. Weller, J. Shiea, R. W. Castenholz, and Y. Cohen. 1989. Hot spring microbial mats: anoxygenic and oxygenic mats of possible evolutionary significance, p. 3-15. In Y. Cohen and E. Rosenberg (ed.), Microbial mats—physiological ecology of benthic microbial communities. American Society for Microbiology, Washington, D.C.
- 50.White, T. J., T. Bruns, S. Lee, and J. Taylor. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p. 315-322. In M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (ed.), PCR protocols: a guide to methods and applications. Academic Press, San Diego, Calif.
- 51.Woese, C. R. 1992. Prokaryote systematics: the evolution of a science, p. 3-17. In A. Balows, H. G. Truper, M. Dworkin, W. Harder, and K.-H. Schleifer (ed.), The prokaryotes, vol. 2. Springer-Verlag, New York, N.Y. [Google Scholar]
- 52.Woese, C. R., O. Kandler, and M. L. Wheelis. 1990. Towards a natural system of organisms: proposal for the domains archaea, bacteria, and eukarya. Proc. Natl. Acad. Sci. USA 87:4576-4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu, H., R. Hseu, and L. Lin. 2001. Identification of Chlorella spp. isolates using ribosomal DNA sequences. Bot. Bull. Acad. Sin. 42:115-121. [Google Scholar]