Abstract
Molecular analysis of grassland rhizosphere soil has demonstrated complex and diverse bacterial communities, with resultant difficulties in detecting links between plant and bacterial communities. These studies have, however, analyzed “bulk” rhizosphere soil, rather than rhizoplane communities, which interact most closely with plants through utilization of root exudates. The aim of this study was to test the hypothesis that plant species was a major driver for bacterial rhizoplane community composition on individual plant roots. DNA extracted from individual roots was used to determine plant identity, by analysis of the plastid tRNA leucine (trnL) UAA gene intron, and plant-related bacterial communities. Bacterial communities were characterized by analysis of PCR-amplified 16S rRNA genes using two fingerprinting methods: terminal restriction fragment length polymorphisms (T-RFLP) and denaturing gradient gel electrophoresis (DGGE). Links between plant and bacterial rhizoplane communities could not be detected by visual examination of T-RFLP patterns or DGGE banding profiles. Statistical analysis of fingerprint patterns did not reveal a relationship between bacterial community composition and plant species but did demonstrate an influence of plant community composition. The data also indicated that topography and other, uncharacterized, environmental factors are important in driving bacterial community composition in grassland soils. T-RFLP had greater potential resolving power than DGGE, but findings from the two methods were not significantly different.
Molecular techniques have demonstrated the complexity and heterogeneity of soil bacterial communities, with considerable diversity in 16S rRNA gene sequences amplified from soil DNA and RNA (12, 14, 31) and significant small-scale spatial heterogeneity (9, 37, 38). Although bacterial communities have been characterized, little is known of the factors driving diversity, in part due to the complexity of communities but also because of functional redundancy, the abundance of inactive or dormant organisms, and the heterogeneity of the soil physicochemical environment. Plants are likely to be a major factor influencing bacterial communities through, for example, root exudation, with potential feedback effects on plant growth. A number of studies have attempted to draw links between plants and below-ground bacterial diversity, but most have focused on plants growing in monocultures or on agricultural soils. These studies have provided evidence for the influence of a number of factors on bacterial community composition, including plant species (25, 33, 44, 47) and soil characteristics (5, 15, 21), while others indicate that the importance of the plant community depends on soil type (1, 28). Bagayoko et al. (2) have shown that plant species exert different effects on soil pH and nutrient availability, but these authors did not characterize microbial communities, whereas Hedlund (19) showed that sowing plant seed mixtures in soils affects microbial growth compared to unseeded soil.
The majority of studies of nonagricultural plant communities have focused on rhizosphere soil communities, defined as those influenced by plant roots (3, 4, 11, 16, 17, 22, 31, 32, 46); few have investigated rhizoplane communities, those colonizing the root surface, as a distinct ecological niche (22, 26, 27). However, any influence of plants is likely to be mediated through root exudates and is therefore likely to be greatest on the rhizoplane, where specific nutritional selection will occur, prior to diffusion of less plant-specific breakdown products into rhizosphere soil. In addition, the high root density in grassland systems prevents isolation of the influence of specific plant rhizospheres in a complex plant community.
The aim of the present study was to assess the interaction between plant species and the community composition of bacteria colonizing the rhizoplane in a grazed, upland Scottish grassland soil. The plant community at this site has previously been shown (41) to be dominated by seven grass species. To relate bacterial communities to the rhizoplane of specific plants, DNA was extracted from individual root fragments. Plant species were determined by analysis of the plastid tRNA leucine (trnL) UAA gene intron (41), and associated bacterial communities were determined by two fingerprinting methods on the same samples: denaturing gradient gel electrophoresis (DGGE) and terminal restriction fragment length polymorphism (T-RFLP) of 16S rRNA gene fragments amplified from the root-associated DNA. DGGE has been used frequently to characterize natural bacterial communities (35) and separates sequences on the basis of differences in denaturing characteristics, and hence migration distance, in a chemical gradient. T-RFLP separates sequences on the basis of restriction site polymorphism and thus, after digestion, base-pair sizes of labeled terminal fragments are detected on an automated sequencer (29). Fingerprinting methods allow rapid comparison of samples and can be used to detect differences in bacterial communities. The study therefore aimed to test the hypothesis that bacterial rhizoplane communities would be specific to plant species composition and also enabled comparison of the respective abilities of two commonly used fingerprinting methods to discriminate bacterial communities associated with different grass species.
MATERIALS AND METHODS
Root samples and DNA extraction.
Plant material was collected at Fasset Hill (UK National Grid Reference NT 852207), an upland grazed grassland at the Sourhope Research Station in the Scottish Borders (41) consisting of brown forest soil, with a pH (CaCl2) of 5.5 to 6.9 and an organic matter content of 0.33 to 0.37 mg C g dry weight−1 (32). Plots were grazed by sheep during the summer months but received no N, P, or K fertilizer. Forty, spatially referenced soil cores (10-cm diameter, ∼8-cm depth) were taken from a 14-m-by-12-m plot belonging to the U4a vegetation category (national vegetation classification) (43). A block of approximately 1 cm3 was removed from the center of the organic layer of each soil core, which was identified by measurement after dissection of the core, using a palette knife to remove the thatch and mineral layers. Soil was removed physically, by shaking. A total of 24 root fragments were then selected randomly from each core, and an average of 14 (range, 9 to 21) fragments were used in the study. Roots that could not be identified or from which no PCR product could be obtained were not used. DNA from individual root fragments was extracted, purified with the QIAGEN DNeasy 96 Plant Kit (QIAGEN, Cologne, Germany) as described previously (41) and was stored at −80°C until used. Each root fragment was identified to genus or species level by PCR-RFLP analysis of the plastid tRNA leucine (trnL) UAA gene intron (41). The relative abundances of the main plant species identified at the site were as follows: Agrostis capillaris, 61.3%; Agrostis vinealis, 11.4%; Deschampsia cespitosa, 7.8%; Festuca rubra, 7.3%; and Poa pratensis, 5.4% (41). The vegetation composition of each core was calculated as species relative abundance and used to calculate the vegetation similarity among cores.
T-RFLP analysis of amplified DNA.
For T-RFLP analysis, first round products were PCR-amplified using the primer set 28F (5′-GAGTTTGATYMTGGC TC-3′) and 1494r (5′-TACGGYTACCTTGTTACGAC-3′). Products of first-round PCR were used as templates for nested PCR, using the primer set 63F-FAM (5′-AGG CCT AAC ACA TGC AAG TC-3′) and 1087R (5′-CTC GTT GCG GGA CTT ACC CC-3′), which generated a 1,064-base product of the 16S rRNA gene. 63F-FAM was labeled at the 5′ end with phosphoramidite fluorochrome 5-carboxyfluorescein (5′ 6-FAM) which was synthesized by Oligo QIAGEN. The standard reaction mixture contained, in a total volume of 100 μl, 1× NH4 reaction buffer (Bioline, London, United Kingdom), 1.5 mM MgCl2, 0.25 mM concentrations of each deoxynucleoside triphosphate, 5 pM concentrations of each forward and reverse primer, 20 μg of bovine serum albumin (Roche Diagnostics, Lewes, United Kingdom), 2.5 U of Biotaq DNA polymerase (Bioline), and 1 μl of DNA template. The PCR program used for the first-round PCR amplification was as follows: 5 min at 95°C (1 cycle); 45 s at 95°C, 45 s at 55°C, 160 s at 72°C (35 cycles); 10 min at 72°C (1 cycle). Aliquots (5 μl) of 16S rRNA amplicons were analyzed by gel electrophoresis on 1% agarose gel after staining with ethidium bromide. PCR products were purified with the GeneElute PCR clean-up kit (Sigma-Aldrich Company, Ltd., Dorset, United Kingdom).
Prior to digestion, PCR product concentrations were determined spectrophotometrically. Initially, four different restriction enzymes (HhaI, RsaI, MvnI, and TacI) were used for 25 randomly selected samples to determine the best enzymes for further experiments. HhaI produced more peaks and gave higher consistency, and all further samples were therefore digested with HhaI only. Each 20 μl of reaction mixture contained purified DNA (1 μg), 1× restriction buffer, 20 U of HhaI (Promega, Southampton, United Kingdom), and 0.2 μg of acylated BSA with the remainder made up of autoclaved Milli-Q-water. Digestion was carried out at 37°C for 3 h, and the reaction was stopped by incubating samples at 95°C for 15 min. Digested DNA was diluted with sterile Milli-Q-water to achieve a concentration of 2 ng μl−1 for fragment size analysis. Samples to be analyzed were mixed with formamide and Applied Biosystems GS500 size standard as described in the manufacturer's instructions, except that 0.05 μl of standard was used per sample and run under standard conditions on an ABI 3700 automated sequencer. Output was analyzed using Genescan and Genotyper to define band distribution, followed by conversion to presence/absence for statistical analysis.
DGGE analysis of PCR-amplified DNA.
Two strategies were used to PCR amplify a 160-base section of the V3 hypervariable region of the bacterial 16S rRNA gene using bacterial primers for subsequent DGGE analysis. The first strategy involved direct amplification with primers p3 and p2 (36), generating a fragment of 194 bases. The second utilized a nested approach with a first-round PCR amplification using primers 27f (23) and Pfr (8) as forward and reverse primers, respectively, amplifying a region of approximately 1,000 bases. These amplification products were then subjected to a second PCR amplification using primer set p3 and p2 (36). The nested PCR approach was used to provide higher PCR amplification yields and better band resolution on DGGE gels. All amplification reactions were carried out in a 50-μl PCR mixture (final volume) containing the following: 1× NH4 reaction buffer (Bioline, London, United Kingdom), 1.5 mM MgCl2, 0.25 mM concentrations of each deoxynucleoside triphosphate, 4 pM concentrations of each forward and reverse primer, 0.25 μg of T4 gene 32 protein (Roche Diagnostics, Lewes, United Kingdom), 1 U of Biotaq DNA polymerase (Bioline), 1.2 U Bio-X-ACT polymerase (Bioline), and 1 μl of DNA template. The template for the first PCR round was genomic DNA, and the template for the second PCR round was a 1:50 dilution of PCR product from the first-round amplification. T4 gene 32 protein was used to neutralize the effects of the PCR inhibitors that were present in the DNA extracted from the root fragments (45). PCR amplification and yields were checked under UV illumination on ethidium bromide-stained agarose gels (1.2% [wt/vol]). The PCR program used for the first-round PCR amplification was as follows: 5 min at 95°C (1 cycle); 30 s at 94°C, 30 s at 55°C, and 180 s at 72°C (10 cycles); 30 s at 92°C, 30 s at 55°C, and 180 s at 72°C (25 cycles); and 10 min at 72°C (1 cycle). The same PCR program was used for the PCR with p3 and p2 primers but with the extension time reduced to 45 s. The T4 gene 32 protein was omitted from the nested PCR mix.
Amplified 16S rRNA gene fragments were analyzed by DGGE (36) on 8% (wt/vol) bisacrylamide gels with a denaturing gradient of 38 to 68% (100% denaturing being defined as 40% [vol/vol] formamide and 7 M urea). Electrophoresis was performed with a DCode System (Bio-Rad Laboratories, Hercules, CA) for 12 h at 100 V and 60°C, after which gels were stained with silver nitrate and scanned at a 1200 dpi resolution with an Epson GT-9600 scanner. The staining procedure was as follows (24, 31). Gels were fixed by shaking for 2 h in a fixing solution (10% ethanol [vol/vol], 0.5% glacial acetic acid [vol/vol]) and were incubated with shaking in silver nitrate solution (0.2% [wt/vol], in fixing solution) for 20 min. After a rinse to remove silver nitrate solution, the gels were shaken in a developing solution (3% NaOH [wt/vol] and 1.3% formaldehyde, prepared in water) until DNA bands appeared (30 to 60 min). The gels were then preserved in ethanol-glycerol preservative solution (25% ethanol, 10% glycerol, 65% H2O) for at least 15 min and stored in sealed plastic bags at room temperature. DGGE banding patterns of the digitized gel images were analyzed with Phoretix 1D Advanced Software (version 4.01; NonLinear Dynamics, Ltd., Newcastle-upon-Tyne, United Kingdom) by manual identification of individual bands and quantification, by the software, of the relative intensity of each band within individual profiles of each sample. Bands in each gel were normalized for variations in DNA loading by detecting the lane with the lowest amount of DNA (i.e., lowest total band intensity) and then the faintest band within that lane. The percentage intensity of the faintest band in the lane with the lowest loading was taken as the limit of detection. Bands with a lower percentage of total band volume in all other lanes were excluded from the analysis.
Statistical analysis.
Statistical analysis of the samples was based on the complete sample profiles as expressed either by the pattern of DGGE gel bands or by the pattern of T-RFLP bands. The analysis consisted of three stages. First, the similarity between every pair of samples was derived. This was calculated as the proportion of common bands for the DGGE gels or shared terminal fragments for T-RFLP. Second, the similarities were converted to principal coordinates, thus allowing the original samples to be scored on a small number of axes, or dimensions, which account for most of the interesting sample similarity information. Finally, these new sample scores were used as the variables to answer the basic biological questions, such as finding relationships between community composition and pH, determining differences among plant root species and among core locations, and identifying spatial patterns. At this stage more traditional statistical methods were used, namely, regression, residual maximum likelihood (REML), and semivariance analysis. REML was used to identify significant differences among plant species, because the data were unbalanced, i.e., the relative abundances of the plant species differed and therefore the number of observations was not the same for each combination of factor levels. Differences in DGGE gel resolution, due to denaturant gradient formation and electrophoresis conditions, led to gel to gel variation, which was accounted for in analyses by introducing a “gel” term to the REML analysis. Relationships between community composition in samples and intersample separation distance were quantified by using semivariograms. The semivariogram measures the spatial variation of a phenomenon by describing how sample data are related as a function of distance and direction. In general, two closely neighboring points are more likely to have similar values than two points farther apart. Therefore, semivariogram values tend to increase with distance until a plateau (sill) is reached, after which values fluctuate randomly about the sill. The sill is the sample variance. The distance at which the sill is reached is termed the range and represents the average intersample distance beyond which samples are no longer spatially correlated. Semivariograms usually exhibit a discontinuity at the origin, the nugget effect, which is due to measurement error or to variation occurring at scales below the minimum sampling distance and therefore not accounted for. Root fragments from the same core were assigned the same coordinates because their location within the core could not be determined. Therefore, variability among communities associated with root fragments from the same core contributed to the nugget effect observed in the semivariograms. Both omnidirectional and directional semivariograms were determined. Omnidirectional semivariograms were determined by calculating intersample semivariance regardless of direction across the sampling site. Directional semivariograms (semivariance calculated between samples in a single direction) were calculated to identify anisotropy. A property is anisotropic if it exhibits different values or patterns when measured in different directions. For the present study, this involved investigation of the influence of the slope of the plot on bacterial community composition.
In addition to assessing the influence of individual root fragments on bacterial composition, the influence of vegetation composition was determined, i.e., the influence of plant species composition of roots in the immediate vicinity of a root fragment with which a bacterial community was associated. Semivariance analysis was also used to identify patterns among rhizoplane bacterial communities as a function of the vegetation profiles (relative abundance of plant species) of the cores from which root fragments were sampled. Rather than using the physical distance among cores to calculate the semivariograms, the distance among vegetation profiles was used. The distance between pairs of vegetation profiles was calculated by measuring the similarity of vegetation profiles of cores using the ecological coefficient of similarity and subtracting similarity values from 1. Thus, the more similar the vegetation profiles of pairs of cores, the smaller the distance between the cores. Semivariogram plots calculated in this way had the same characteristics as the traditional semivariograms described above. However, in this case interpretation of the semivariogram range is not the physical distance beyond which there is no longer spatial correlation among samples but rather the level of vegetation similarity below which samples are uncorrelated. Directional semivariograms based on vegetation profiles were not calculated because such profiles have no underlying concept of direction.
Due to differences in DGGE gel resolution, semivariograms were calculated for individual gels separately and then averaged across gels to determine effects of spatial location and of core vegetation composition on bacterial community composition. Semivariogram plots were produced for the first 10 principal coordinate analysis (PCO) dimensions of the DGGE and T-RFLP data, accounting for approximately 50 and 30% of the total distance information, respectively.
All statistical analyses were carried out by using Genstat 6 (2002; VSN International, Ltd., Hemel Hempstead, United Kingdom) or Isatis 4.0.1 (Geovariances, Avon, France).
RESULTS
T-RFLP analysis of community composition.
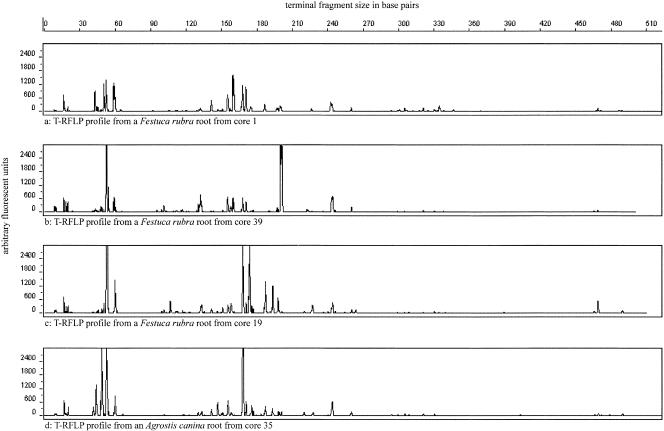
T-RFLP analysis of 16S rRNA genes was used to compare rhizoplane bacterial communities. Profiles were typical of those obtained for communities of soil bacteria, in terms of complexity, and examples are illustrated in Fig. 1. The distribution of terminal fragments detected showed a steep decline below 30, and samples with less than 30 fragments were excluded from analysis. Samples analyzed therefore had a minimum of 30 and a maximum of 89 fragments (mean = 57, standard error = 1). PCO was used to convert sample T-RFLP similarity matrices into a small number of linear dimensions, which are expected to account for much of the biological information, in order to identify factors influencing the similarity among bacterial communities. In the new linear dimensions each of the original samples is represented by a principal coordinate score. Sample scores in the first 10 dimensions were related to factors such as plant species, pH and the vegetation composition of cores. REML analysis showed that there were no significant differences (P < 0.05) in T-RFLP patterns among plant species. Regression analysis found significant relationships (P < 0.05) between pH and dimensions 1, 2, and 4. Scatter diagrams showed that the data were in fact highly dispersed, with the exception of the relationship between pH and dimension 1, which was highly significant (r2 = 0.3; P < 0.001). Although dimension 1 accounted for only a small part (5.5%) of the total distance information, the highly significant nature of the relationship suggests that this is a small but real effect.
FIG. 1.
Examples of T-RFLP profiles of rhizoplane communities. Cores 1 and 39 have a high similarity in plant species composition, and cores 19 and 35 have a low similarity in plant species composition. Roots were randomly chosen from selected cores. Note that the T-RFLP comparisons were made on a presence or absence basis.
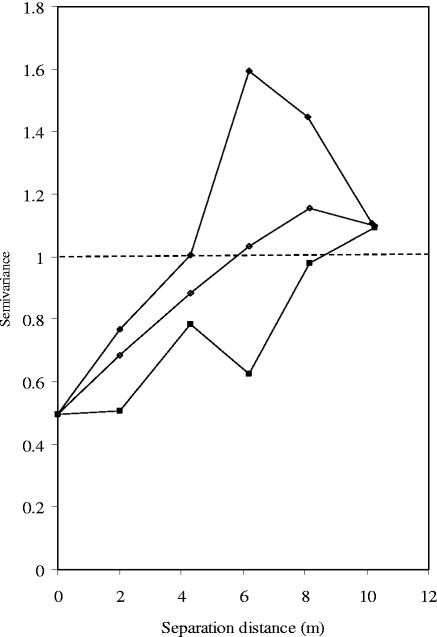
Evidence of spatial correlation was only found in dimension 1 (Fig. 2). Variance between samples in this dimension increased as a function of physical distance across plot. This means that bacterial communities from the samples that were in close proximity to one another tended to be more closely related than communities from samples further apart. The community compositions of samples separated by distances greater than the range (approximately 5 m; Fig. 2) were not correlated with one another. This spatial pattern was related to the direction of physical separation across the plot, as directional semivariograms indicated the presence of anisotropic structures. Thus, semivariance across the plot slope did not increase as rapidly as up-slope semivariance (Fig. 2). The up-slope range was approximately 4 m, while that across slope was approximately 8 m. This means that the factors contributing to variance in the first PCO dimension, such as pH or the position on the slope, varied less as a function of intersample distance across the slope than up-slope and that samples separated by a distance of <4 m up-slope but 8 m across slope were spatially correlated. Beyond these distances changes in the semivariance can be viewed as random fluctuations. Since there was just one pH measurement per core (thus 40 values in total), there were not sufficient samples to analyze pH spatial distribution. However, pH variance across the slope was lower than that up-slope. These data confirm the view that, although the PCO dimension accounted for only a small part of the total distance information, it represented a real interaction between bacterial communities and their environment.
FIG. 2.
Standardized semivariograms for first PCO dimension. Omni-directional (○), across slope (▪), and up-slope (•) semivariograms. Dashed line indicates total sample variance.
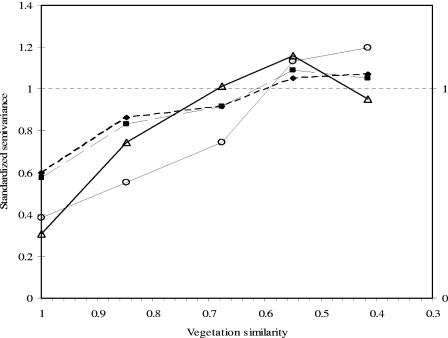
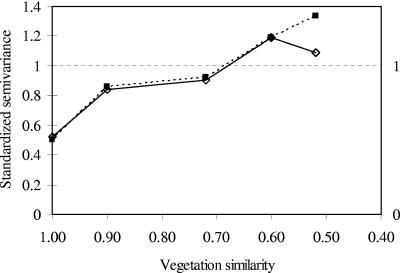
PCO semivariance as a function of vegetation profile distance exhibited structure in many dimensions (i.e., dimensions 2, 4, 6, 8, and 9; Fig. 3), all of which accounted for 14% of total distance information. Thus, rhizoplane bacterial community composition from cores with a similar vegetation composition was more similar than that from cores with a different vegetation composition, regardless of their respective locations within the plot. Variability between cores in dimensions 4 and 6 represented <40% of the variance in these dimensions but in dimensions 2, 8, and 9 intracore variability represented >57% of total variance (Fig. 3). The “range” was generally between similarity values of 0.6 and 0.7 (Fig. 3). Thus, the similarity between pairs of rhizoplane bacterial communities from cores that had similar plant communities (at least 60 to 70% similarity) was on average greater than between pairs from cores with less-similar plant communities. Although 14% also represents a relatively small percentage, these data demonstrate that vegetation composition was the most important factor influencing rhizoplane communities of those tested. However, it is clear that other factors, not measured, also influenced rhizoplane bacterial community composition.
FIG. 3.
Examples of standardized semivariograms in vegetation composition space. Semivariograms for dimensions 2 (⧫) and 9 (▪) showed large intracore variability, whereas in dimensions 4 (○) and 6 (▵) the intracore variability was low.
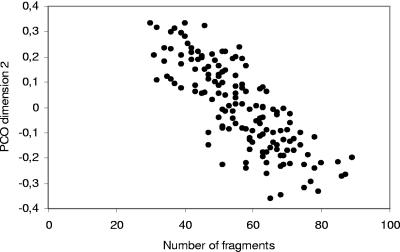
There was a highly significant negative relationship (r = −0.8; P < 0.001) between the number of terminal restriction fragments and PCO dimension 2 (Fig. 4). Since the second PCO dimension was related to the vegetation profile of the cores, this suggests that the number of T-RFLP peaks was influenced by the vegetation profile of the cores. Variability within individual cores ranged from 27 to 60% of total variance in the PCO dimensions studied, suggesting that an important portion of the variance among communities occurred at scales below the scale of analysis, i.e., root fragment scale.
FIG. 4.
Relationship between second PCO dimension and number of terminal restriction fragments in T-RFLP data.
DGGE analysis of community composition.
Rhizoplane bacterial community composition was also investigated by using DGGE analysis of 16S rRNA genes amplified from rhizoplane DNA, and typical profiles are provided in Fig. 5. Two PCR strategies, direct amplification and nested amplification, were used to generate 16S rRNA gene fragments for DGGE analysis. The mean number of bands detected per sample was 39 (standard error = 0.86) for DGGE gels loaded with nested PCR products. The minimum number of bands was 21, and the maximum was 69. In contrast, DNA amplified directly with primers p3 and p2 generated a mean of 31 detectable bands (standard error = 0.77), with minimum and maximum numbers of 13 and 57 bands. PCO analysis was also used to convert similarities among sample DGGE profiles to scores in linear dimensions. The scores were then analyzed as described above to identify factors influencing community composition. Bacterial communities represented by DGGE profiles obtained by using direct amplification showed no significant differences among plant species. For bacterial communities represented by DGGE profiles generated by using nested PCR amplification, PCO dimension 7 (3% total distance information) showed significant differences (P < 0.05) between Deschampsia cespitosa and all other grasses except for Agrostis canina. In addition, bacterial communities colonizing Festuca rubra were significantly different from Festuca juncifolia, Agrostis canina, and Poa pratensis. Thus, these data tend to confirm the T-RFLP analysis, indicating little or no effect of individual plant species on rhizoplane bacterial composition.
FIG. 5.
Typical DGGE profiles of 16S rRNA genes amplified from DNA extracted from rhizoplanes of each of the plants colonizing the study site.
Semivariance analysis of data from direct PCR amplification showed that dimension 2 was spatially structured (i.e., not completely randomly distributed), with a range of approximately 4 m. This means that community composition was influenced by environmental factors that were also spatially structured. PCO semivariance as a function of vegetation profile distance showed structure in the first dimension with a “range” of ca. 0.7 (Fig. 6). Thus, community composition in cores with vegetation compositions that were >70% similar was more similar than community composition in cores with more varied vegetation profiles. Dimension 1 accounted for 14% and dimension 2 accounted for 10% of the total distance information in this analysis. PCO semivariance of samples analyzed using data from nested PCR amplification did not show any clear evidence of spatial structure. There was, however, structure as a function of vegetation profile distance in the second and sixth dimensions with “ranges” of ca. 0.7 (Fig. 6). These dimensions accounted for 16% of the total distance information.
FIG. 6.
Standardized semivariograms for DGGE data in vegetation composition space. Semivariograms for dimension 1 (▪) after direct PCR amplification and for dimension 2 (⋄) after nested PCR amplification.
For profiles of direct PCR products, within-core variation represented 50 to ≥80% of total variance, as was found with T-RFLP. Within core variance for DGGE gels generated by using nested PCR amplification accounted for 45 to ≥75% of total variance. Generally, the DGGE data corroborate the T-RFLP data, which indicate that both the vegetation profile of the cores and the location of the cores within the site affect rhizoplane bacterial community composition but that the most important effects on community composition occur at scales below the root fragment scales.
DISCUSSION
This study aimed to determine whether plant species and bacterial communities were related in grassland soil containing mixed plant communities by combined molecular analysis of plant roots and associated rhizoplane bacterial communities, which are likely to influenced most by root exudates. Community profiles were analyzed by PCO, REML, and geostatistics. PCO was used to reduce the dimensionality of the data sets and to enable detection of biological patterns in the community profiles. REML was used to estimate the amount of variation within and between cores and to estimate the effect of plant species on community profiles. Geostatistical analysis was used to identify patterns of variation in community profiles throughout the plot and in relation to the vegetation composition of cores.
Comparison of DGGE and T-RFLP analyses.
Although DGGE and T-RFLP analyses of bacterial communities led to similar general conclusions, the technique used did influence analysis of data. The major factor influencing DGGE analysis was the inability to run all samples on a single gel and the potential for variation between gels. As a consequence, each dimension accounted for a greater percentage of total distance for DGGE profiles than for T-RFLP patterns, despite modification of statistical analysis to account for intergel variation. Therefore, in DGGE analysis, each dimension may represent the effect of a number of different factors or influences, reducing resolution or masking some information (in particular of weaker effects). Resolution was further reduced, but not greatly, by detection of slightly fewer bands on DGGE gels than peaks on T-RFLP traces, although resolution of DGGE gels can be increased by optimization of gradients. A major advantage of DGGE analysis is putative identity of the origin of bands of interest, through sequence analysis, but this was not used in the present study. Although comparison of restriction patterns with those predicted from database sequences provides an equivalent identification method for T-RFLP, this approach is less reliable. However, the major advantage of T-RFLP is its ability to analyze large numbers of samples rapidly, which was of benefit in the present study. In this respect, comparative analysis of the two fingerprinting techniques is in agreement with the findings of Moeseneder et al. (34).
Links between plant communities and rhizoplane bacterial communities.
Pot studies of monocultures and mixed cultures or studies in agriculture soils indicate a relationship between plant and rhizosphere microbial communities (16, 22, 44), but such relationships are much more difficult to detect in undisturbed natural communities (3, 4, 31, 32). In assessing links between plant species and rhizoplane bacterial community composition, analysis of data from DGGE and T-RFLP gave broadly similar findings. REML analysis indicated that plant species had little direct effect on bacterial community composition, i.e., the plant to which bacterial community was directly associated did not have an important influence on bacterial community composition. However, intracore variance was lower than total variance, i.e., bacterial communities on root fragments from the same core were more similar than those on fragments from different cores. This suggests that there was a “local environment” associated with the combined rhizoplanes of specific plants.
Rhizoplane communities would be influenced by factors other than plant species, for example, the composition of vegetation in the vicinity of the communities (i.e., from the same core), which will also supply bacterial communities with substrate and affect other soil parameters such as the nutrient status of the surrounding soil and factors such as pH. To test hypotheses regarding the influence of these factors, geostatistical analysis of intersample variance in physical space and vegetation composition space was carried out. Spatial patterns and the scale of spatial patterns identified by geostatistical analysis can shed light on the variables driving community composition (40). For example, evidence of spatial correlation in community profiles at scales above 1 m (i.e., samples separated by distances greater than 1 m are spatially correlated) is likely to be caused by interactions with variables that are themselves spatially correlated at scales above 1 m.
The spatial pattern found in first dimension (Fig. 2) shows that the location of a sample within a plot influenced bacterial community composition. The pattern was similar to those found previously for total P, total N, total C, and NH4+ at this site (42), suggesting that soil properties such as these affect community composition. Furthermore, intersample semivariance in the first dimension of the T-RFLP data across the slope was lower than that up-slope, suggesting that factors influencing community composition were themselves affected by the slope of the plot. Soil properties such as water content, pH, and particle size distribution are known to be affected by topography (30) and therefore may be variables driving bacterial community composition. The significant relationship between pH and the first PCO dimension of the T-RFLP data confirms this. Spatial structure was not found at the intercore scale in any of the other PCO dimensions of the T-RFLP or DGGE data analyzed, although many showed a local environment effect. This suggests that other driving variables were exerting influence at scales below 2 m and within cores.
Patterns as a function of vegetation profile distance show that the species composition of roots in the vicinity of bacterial communities is associated with their structure (Fig. 3 and 6). Bacterial community profiles from cores with similar vegetation compositions were generally more similar than bacterial community profiles from cores with less-similar vegetation compositions. Thus, the composition of mixed plant communities may be more important than the possible influences of individual plant species on bacterial communities. This could be because both are determined by common physicochemical factors, with no direct causal relationship, or because of similarities in available energy sources from mixed root exudates, reflecting the complexity of the plant communities in upland grasslands. Bacterial communities from cores with vegetation composition similarities of less than 60 to 70% showed no correlation (Fig. 3 and 6). This could have been because the composition of substrate available to the bacterial communities in these cores was different as a result of different root exudates (6, 20). The second dimension of PCO analysis of the T-RFLP data was also strongly correlated with the number of bands (Fig. 4), suggesting that the richness of communities was influenced by vegetation composition.
The major conclusions from both T-RFLP and DGGE data were that bacterial communities are very variable, since each PCO dimension accounts for a low percentage of the total distance, and are subject to a wide range of influences (plant species, local plant composition, and soil characteristics), none of which was found to be dominant. A significant amount of within core variance between bacterial communities associated with root fragments was present in each dimension. This was not due to a plant species effect, since significant differences among species were not found to account for any of the total variability. It is not surprising to find differences in community composition among samples within cores, since bacterial populations are known to be spatially structured at scales below 1 mm (7, 18, 38, 39). This means that bacterial populations are subject to factors driving their development at scales below 1 mm and that these factors are not homogeneous. It is possible that differences in structure among bacterial communities within the 1 cm3 from which the root fragments were extracted were due to differences in available substrate which, in turn, may be due to variations in root composition throughout the 1 cm3. In an experiment established to simulate diffusion of root exudates, Falchini et al. (10) found that the concentration of 14C-labeled glucose and glutamic acid diminished rapidly beyond 4 mm from the source and that the concentration of oxalic acid was virtually nonexistent beyond 2 mm. Furthermore, the influence of decomposing plant material on microbial activity is known to extend only 4 mm for wheat and to 5 mm for rye leaves (13). This hypothesis is not possible to verify since the root fragments were not spatially referenced within cores. Other possible sources of this variance are interactions with other microorganisms, root age, experimental error, and factors that were not measured such as, for example, water content and soil structure. In addition, the study targeted DNA, rather than RNA, increasing complexity resulting from analysis of both active and dormant organisms.
The study therefore provides further evidence for the complexity of bacterial communities in grassland soils and resultant difficulties in distinguishing plant-mediated effects. Nevertheless, statistical analysis of data generated by different fingerprinting techniques demonstrated an influence of plant community composition on bacterial communities but also indicated significant influences of other factors.
Acknowledgments
This study was carried out as part of the MICRONET project funded by the Scottish Executive Rural Affairs Department (SEERAD).
We thank Julie Squires (Scottish Crop Research Institute, Dundee) as well as Karyn Ridgway and Irene Watson (University of York) for assistance in collection of root samples, and we are grateful to Karyn Ridgway for DNA extraction and PCR-RFLP analysis of root fragments.
REFERENCES
- 1.Alvey, S., C. H. Yang, A. Buerkert, and D. E. Crowley. 2003. Cereal/legume rotation effects on rhizosphere bacterial community structure in West African soils. Biol. Fertil. Soils 37:73-82. [Google Scholar]
- 2.Bagayoko, M., S. Alvey, G. Neumann, and A. Buerkert. 2000. Root-induced increases in soil pH and nutrient availability to field-grown cereals and legumes on acid sandy soils of Sudano-Sahelian West Africa. Plant Soil 225:117-127. [Google Scholar]
- 3.Brodie, E., S. Edwards, and N. Clipson. 2002. Bacterial community dynamics across a floristic gradient in a temperate upland grassland ecosystem. Microb. Ecol. 44:260-270. [DOI] [PubMed] [Google Scholar]
- 4.Broughton, L. C., and K. L. Gross. 2000. Patterns of diversity in plant and soil microbial communities along a productivity gradient in a Michigan old-field. Oecologia 125:420-427. [DOI] [PubMed] [Google Scholar]
- 5.Buckley, D. H., and T. M. Schmidt. 2003. Diversity and dynamics of microbial communities in soils from agro-ecosystems. Environ. Microbiol. 5: 441-452. [DOI] [PubMed] [Google Scholar]
- 6.Curl, E. A., and B. Truelove. 1986. The rhizosphere. Springer-Verlag, Berlin, Germany.
- 7.Dechesne, A., C. Pallud, D. Debouzie, J. P. Flandrois, T. M. Vogel, J. P. Gaudet, and G. L. Grundmann. 2003. A novel method for characterizing the microscale 3D spatial distribution of bacteria in soil. Soil Biol. Biochem. 35:1537-1546. [Google Scholar]
- 8.Edwards, U., T. Rogall, H. Blocker, M. Emde, and E. C. Bottger. 1989. Isolation and direct complete nucleotide determination of entire genes: characterization of a gene coding for 16S-rRNA. Nucleic Acids Res. 17:7843-7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellingsoe, P., and K. Johnsen. 2002. Influence of soil sample sizes on the assessment of bacterial community structure. Soil Biol. Biochem. 34: 1701-1707. [Google Scholar]
- 10.Falchini, L., N. Naumova, P. J. Kuikman, J. Bloem, and P. Nannipieri. 2003. CO2 evolution and denaturing gradient gel electrophoresis profiles of bacterial communities in soil following addition of low molecular weight substrates to simulate root exudation. Soil Biol. Biochem. 35:775-782. [Google Scholar]
- 11.Fang, C. W., M. Radosevich, and J. J. Fuhrmann. 2001. Characterization of rhizosphere microbial community structure in five similar grass species using FAME and BIOLOG analyses. Soil Biol. Biochem. 33:679-682. [Google Scholar]
- 12.Felske, A., and A. D. L. Akkermans. 1998. Spatial homogeneity of abundant bacterial 16S rRNA molecules in grassland soils. Microb. Ecol. 36:31-36. [DOI] [PubMed] [Google Scholar]
- 13.Gaillard, V., C. Chenu, S. Recous, and G. Richard. 1999. Carbon, nitrogen and microbial gradients induced by plant residues decomposing in soil. Eur. J. Soil Sci. 50:567-578. [Google Scholar]
- 14.Gelsomino, A., A. C. Keijzer-Wolters, G. Cacco, and J. D. van Elsas. 1999. Assessment of bacterial community structure in soil by polymerase chain reaction and denaturing gradient gel electrophoresis. J. Microbiol. Methods 38:1-15. [DOI] [PubMed] [Google Scholar]
- 15.Girvan, M. S., J. Bullimore, J. N. Pretty, A. M. Osborn, and A. S. Ball. 2003. Soil type is the primary determinant of the composition of the total and active bacterial communities in arable soils. Appl. Environ. Microbiol. 69:1800-1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grayston, S. J., S. Q. Wang, C. D. Campbell, and A. C. Edwards. 1998. Selective influence of plant species on microbial diversity in the rhizosphere. Soil Biol. Biochem. 30:369-378. [Google Scholar]
- 17.Grayston, S. J., G. S. Griffith, J. L. Mawdsley, C. D. Campbell, and R. D. Bardgett. 2001. Accounting for variability in soil microbial communities of temperate upland grassland ecosystems. Soil Biol. Biochem. 33:533-551. [Google Scholar]
- 18.Grundmann, G. L. 2004. Spatial scales of soil bacterial diversity: the size of a clone. FEMS Microbiol. Ecol. 48:119-127. [DOI] [PubMed] [Google Scholar]
- 19.Hedlund, K. 2002. Soil microbial community structure in relation to vegetation management on former agricultural land. Soil Biol. Biochem. 34:1299-1307. [Google Scholar]
- 20.Hodge, A., E. Paterson, S. J. Grayston, C. D. Campbell, B. G. Ord, and K. Killham. 1998. Characterisation and microbial utilization of exudate material from the rhizosphere of Lolium perenne grown under CO2 enrichment. Soil Biol. Biochem. 30:1033-1043. [Google Scholar]
- 21.Johnson, M. J., K. Y. Lee, and K. M. Scow. 2003. DNA fingerprinting reveals links among agricultural crops, soil properties, and the composition of soil microbial communities. Geoderma 114:279-303. [Google Scholar]
- 22.Kowalchuk, G. A., D. S. Buma, W. de Boer, P. G. L. Klinkhamer, and J. A. van Veen. 2002. Effects of above-ground plant species composition and diversity on the diversity of soil-borne microorganisms. Antonie Leeuwenhoek 81:509-520. [DOI] [PubMed] [Google Scholar]
- 23.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. Academic Press, Ltd., Chichester, England.
- 24.Mahmood, S., T. E. Freitag, and J. I. Prosser. Comparison of PCR primer-based strategies for characterisation of ammonia oxidizer communities in environmental samples. FEMS Microbiol. Ecol., in press. [DOI] [PubMed]
- 25.Maloney, P. E., A. H. C. van Bruggen, and S. Hu. 1997. Bacterial community structure in relation to the carbon environments in lettuce and tomato rhizospheres and in bulk soil. Microb. Ecol. 34:109-117. [DOI] [PubMed] [Google Scholar]
- 26.Marilley, L., and M. Aragno. 1999. Phylogenetic diversity of bacterial communities differing in degree of proximity of Lolium perenne and Trifolium repens roots. Appl. Soil Ecol. 13:127-136. [Google Scholar]
- 27.Marilley, L., G. Vogt, M. Blanc, and M. Aragno. 1998. Bacterial diversity in the bulk soil and rhizosphere fractions of Lolium perenne and Trifolium repens as revealed by PCR restriction analysis of 16S rDNA. Plant Soil. 198:219-224. [Google Scholar]
- 28.Marschner, P., C. H. Yang, R. Lieberei, and D. E. Crowley. 2001. Soil and plant specific effects on bacterial community composition in the rhizosphere. Soil Biol. Biochem. 33:1437-1445. [Google Scholar]
- 29.Marsh, T. L. 1999. Terminal restriction fragment length polymorphism (T-RFLP): an emerging method for characterizing diversity among homologous populations of amplification products. Curr. Opin. Microbiol. 2: 323-327. [DOI] [PubMed] [Google Scholar]
- 30.Marshall, T. J., J. W. Holmes, and C. W. Rose. 1996. Soil physics. Cambridge University Press, Cambridge, United Kingdom.
- 31.McCaig, A. E., L. A. Glover, and J. I. Prosser. 2001. Numerical analysis of grassland bacterial community structure under different land management regimens by using 16S ribosomal DNA sequence data and denaturing gradient gel electrophoresis banding patterns. Appl. Environ. Microbiol. 67:4554-4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCaig, A. E., L. A. Glover, and J. I. Prosser. 1999. Molecular analysis of bacterial community structure and diversity in unimproved and improved upland grass pastures. Appl. Environ. Microbiol. 65:1721-1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miethling, R., K. Ahrends, and C. C. Tebbe. 2003. Structural differences in the rhizosphere communities of legumes are not equally reflected in community-level physiological profiles. Soil Biol. Biochem. 35:1405-1410. [Google Scholar]
- 34.Moeseneder, M. M., J. M. Arrieta, G. Muyzer, C. Winter, and G. J. Herndl. 1999. Optimization of terminal-restriction fragment length polymorphism analysis for complex marine bacterioplankton communities and comparison with denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 65:3518-3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muyzer, G., and K. Smalla. 1998. Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) in microbial ecology. Antonie Leeuwenhoek 73:127-141. [DOI] [PubMed] [Google Scholar]
- 36.Muyzer, G., E. C. Dewaal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nicol, G. W., L. A. Glover, and J. I. Prosser. 2003. Spatial analysis of archaeal community structure in grassland soil. Appl. Environ. Microbiol. 69: 7420-7429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nunan, N., K. J. Wu, I. M. Young, J. W. Crawford, and K. Ritz. 2003. Spatial distribution of bacterial communities and their relationships with the micro-architecture of soil. FEMS Microbiol. Ecol. 44:203-215. [DOI] [PubMed] [Google Scholar]
- 39.Nunan, N., K. Wu, I. M. Young, J. W. Crawford, and K. Ritz. 2002. In situ spatial patterns of soil bacterial populations, mapped at multiple scales, in an arable soil. Microb. Ecol. 44:296-305. [DOI] [PubMed] [Google Scholar]
- 40.Parkin, T. B. 1993. Spatial variability of microbial processes in soil: a review. J. Environ. Qual. 22:409-417. [Google Scholar]
- 41.Ridgway, K. P., J. M. Duck, and P. W. Young. 2003. Identification of roots from grass swards using PCR-RFLP and FFLP of the plastid trnL (UAA) intron. BMC Ecol. 3:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ritz, K., W. McNicol, N. Nunan, S. Grayston, P. Millard, D. Atkinson, A. Gollotte, D. Habeshaw, B. Boag, C. D. Clegg, B. S. Griffiths, R. E. Wheatley, L. A. Glover, A. E. McCaig, and J. I. Prosser. 2004. Spatial structure in soil chemical and microbiological properties in an upland grassland. FEMS Microbiol. Ecol. 49:191-205. [DOI] [PubMed] [Google Scholar]
- 43.Rodwell, J. S. 1992. British plant communities. Cambridge University Press., Cambridge, United Kingdom.
- 44.Smalla, K., G. Wieland, A. Buchner, A. Zock, J. Parzy, S. Kaiser, N. Roskot, H. Heuer, and G. Berg. 2001. Bulk and rhizosphere soil bacterial communities studied by denaturing gradient gel electrophoresis: plant-dependent enrichment and seasonal shifts revealed. Appl. Environ. Microbiol. 67: 4742-4751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tebbe, C. C., and W. Vahjen. 1993. Interference of humic acids and DNA extracted directly from soil in detection and transformation of recombinant-DNA from bacteria and a yeast. Appl. Environ. Microbiol. 59:2657-2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Westover, K. M., A. C. Kennedy, and S. E. Kelley. 1997. Patterns of rhizosphere microbial community structure associated with co-occurring plant species. J. Ecol. 85:863-873. [Google Scholar]
- 47.Wieland, G., R. Neumann, and H. Backhaus. 2001. Variation of microbial communities in soil, rhizosphere, and rhizoplane in response to crop species, soil type, and crop development. Appl. Environ. Microbiol. 67:5849-5854. [DOI] [PMC free article] [PubMed] [Google Scholar]