Abstract
Spores of the arbuscular mycorrhizal fungi (AMF) Glomus geosporum and Glomus constrictum were harvested from single-spore-derived pot cultures with either Plantago lanceolata or Hieracium pilosella as host plants. PCR-denaturing gradient gel electrophoresis analysis revealed that the bacterial communities associated with the spores depended more on AMF than host plant identity. The composition of the bacterial populations linked to the spores could be predominantly influenced by a specific spore wall composition or AMF exudate rather than by specific root exudates. The majority of the bacterial sequences that were common to both G. geosporum and G. constrictum spores were affiliated with taxonomic groups known to degrade biopolymers (Cellvibrio, Chondromyces, Flexibacter, Lysobacter, and Pseudomonas). Scanning electron microscopy of G. geosporum spores revealed that these bacteria are possibly feeding on the outer hyaline spore layer. The process of maturation and eventual germination of AMF spores might then benefit from the activity of the surface microorganisms degrading the outer hyaline wall layer.
Arbuscular mycorrhizal fungi (AMF) play a key role in facilitating nutrient uptake by crops in low-input farming systems, a prerequisite to maintain sufficient productivity under these conditions (3). AMF spores provide a long-term reservoir of inoculum and are the only AMF propagules that can be identified to the species level (33). The spore wall of Glomus geosporum is composed of three layers: an outer hyaline layer that decays until it sloughs off, leaving a granular surface; a laminated yellow-brown to orange-brown middle layer; and a more rigid inner layer that is often adherent to the middle layer (14). The thin hyaline layer is composed mainly of chitin (32) and has been found to be often colonized by microorganisms in several Glomus species (5). The Glomus constrictum spore wall is composed of only two layers, a decomposing outer hyaline layer that is absent in older spores and a rigid, laminated orange-brown to reddish-black dark layer (14).
An optimal colonization of plant roots, particularly in disturbed habitats such as agricultural fields, depends not only on the presence of extraradical hyphae or mycorrhizal root debris but, mainly, on the survival and well-timed germination of AMF spores in the soil. This process can be altered by various abiotic and biotic factors, in particular by the association with soil microorganisms (38). Indeed, some bacterial populations, called mycorrhiza helper bacteria, have beneficial effects on AMF growth not only by improving mycorrhizal root colonization and stimulating extraradical hyphal growth but also by facilitating AMF spore germination (11, 12). The latter effect has been shown for Actinomycetes (2, 7, 22), Pseudomonas and Corynebacterium (21), and Bacillus (38) spp.
Bacteria associated with AMF spores colonize mainly the outer wall layer and rarely penetrate into the inner layers (5, 9, 19, 36). Nevertheless, some bacteria have been found in the cytoplasm of AMF spores (4, 18). The role of AMF spore-associated bacteria is not clear. They could stimulate spore germination by eroding spore walls (9, 19), by producing stimulatory compounds such as CO2 and other volatiles (7), or by influencing AMF phosphorus acquisition (30).
Root exudation could enhance spore germination by stimulating the growth of bacteria beneficial for AMF (21). However, since the quantity and composition of exudates differ from one plant to another (17), different bacterial populations could be stimulated, depending on their preference for distinct plant exudates.
In most of the previous studies on spore-associated microorganisms, the bacteria were isolated upon culturing. However, bacteria not cultivable on ordinary media could represent a significant part of the bacterial community associated with AMF spores. Indeed, only a small fraction (1 to 10%) of the total bacterial community is cultivable (1). Direct molecular approaches that avoid a cultivation step give a broader picture of bacterial communities. PCR-denaturing gradient gel electrophoresis (PCR-DGGE) analysis of the 16S rRNA gene permits fingerprinting of the dominant bacteria of a given sample (10, 23). The detection of populations representing as little as 0.1 to 1% of the total target organisms is feasible. In the present study, the bacterial community associated with spores of Glomus geosporum BEG 18 and G. constrictum BEG 19 was assessed with PCR-DGGE analysis. In order to find out whether specific root exudates or, rather, the fungal species determine the bacterial community structure, spores of the two Glomus species were harvested from both pot cultures with Plantago lanceolata (Plantaginaceae) and pot cultures with Hieracium pilosella (Asteraceae) as host plants.
MATERIALS AND METHODS
Mycorrhizal inoculum.
The AMF used in this study were Glomus geosporum (BEG 18) and Glomus constrictum (BEG 19), originating from the same calcareous grassland at Nenzlingen, Switzerland (35). The single-spore-derived cultures were maintained by subculturing in pots under the same conditions, using Plantago lanceolata and Hieracium pilosella as host plants. The seeds were purchased from FENACO (Winterthur, Switzerland). The growth substrate was TerraGreen-sand-loess (2:2:1) (TerraGreen was American aluminum oxide, oil dry US special, type III R, <0.125 mm, from Lobbe Umwelttechnik, Iserlohn, Germany; sand was Quartz d'Alsace, K30, from Kaltenhouse, France; and loess was from a local site near Basel, Switzerland). The pots were cultivated in a greenhouse with ambient natural light and temperature conditions and irrigated with deionized water by using an automated watering system (Tropf-Blumat; Weninger GmbH, Telfs, Austria).
Experimental setup.
Twelve 1-liter plastic pots were filled with sterile substrate composed of TerraGreen, quartz sand, and loess (5:4:1) and moistened with water. In each pot, a small hole was drilled, in which a teaspoon of mycorrhizal inoculum was placed. On top of the inoculum, a few seeds of Hieracium pilosella or Plantago lanceolata were sown and covered with sterile quartz sand. The Tropf-Blumat watering system was installed, and the cultures were grown in a greenhouse in spring and summer under ambient conditions. The following four combinations of symbionts were cultivated in triplicates: H. pilosella with either G. geosporum or G. constrictum and P. lanceolata with either G. geosporum or G. constrictum.
Sampling.
Six 15-ml soil cores were sampled from each pot after 170 days of growth in the case of H. pilosella and 247 days of growth in the case of P. lanceolata in order to obtain a sufficient amount of spores. The soil cores were wet sieved through 250- and 63-μm meshes. The suspension of residue gained from the 63-μm mesh sieve was centrifuged at 900 × g for 2 min in a sterile density gradient with a 70% (wt/vol) sucrose layer at the bottom. The spores were collected from the gradient interphase, placed in glass plates, and rinsed three times with sterile milli Q water. For the DNA extraction, 200 spores per pot were individually recovered under a stereomicroscope with a micropipette. For scanning electron microscopy (SEM) analysis, the sampling procedure from the G. geosporum/P. lanceolata replicates was repeated to recover more than 100 spores.
DGGE analysis.
DNA extraction for the DGGE analysis was performed with 200 spores per pot, using the FastDNA spin kit for soil (Bio101, Vista, Calif.) according to the manufacturer's protocol and a bead beater (Fast-Prep model FP 120; Bio101). A double-step PCR was used to amplify the V3 region, a fragment of about 200 bp of the bacterial 16S rRNA gene, as described by Weisskopf et al. (37). A composite mix of bacterial 16S rRNA gene fragments from Pseudomonas fluorescens ATCC 27663, Acidovorax facilis DSM 550, Bacillus subtilis ATCC 14893, Sinorhizobium meliloti DSM 1981, and Aquaspirillum dispar ATCC 27650 was added on each side of the DGGE gel as a reference DGGE pattern. DGGE was performed using an 8% (wt/vol) acrylamide-bisacrylamide gel (37.5:1; Qbiogene, Illkirch, France) with a 30 to 60% linear urea-formamide (Fluka, Buchs, Switzerland; Qbiogene) denaturing gradient (100% denaturant corresponds to 40% formamide plus 7 M urea). Five hundred nanograms of the PCR product was electrophoresed in 1× Tris-acetate-EDTA buffer (Qbiogene) at 60°C with a constant voltage of 150 V for 5.5 h using the Bio-Rad D-Code electrophoresis system (Bio-Rad Inc., Richmond, Calif.). The gels were stained in the dark for 20 min in 0.01% Sybr Green I (Molecular Probes, Leiden, The Netherlands) in 1× Tris-acetate-EDTA solution. The gels were photographed with the Multi-Analyst package (Bio-Rad Inc.). The DGGE fingerprints were normalized according to the reference patterns and were compared using the GelCompar software (Applied Maths, Kortrijk, Belgium). DGGE banding patterns were then converted into a numerical matrix used in the statistical analysis. Each band was considered a single bacterial population, and the band intensity was representative of the relative abundance of the population (10). The bands whose average relative contributions were below 1% were discarded.
Band sequencing.
The DGGE profiles were more similar among H. pilosella pot replicates. Therefore, 10 DGGE bands in the profile of one replicate of H. pilosella/G. geosporum and 10 more bands in the profile of one replicate of H. pilosella/G. constrictum were cut out for sequencing. DNA was recovered and purified as follows. The selected bands were cut out and placed in a 1.5-ml Eppendorf tube containing 100 μl 10 mM Tris-HCl, pH 7.5, and incubated at 4°C for 3 days. The supernatant was then recovered in a new Eppendorf tube. One volume of iced isopropanol (−20°C) and 1/10 volume of 3 M sodium acetate were added, and this mixture was incubated at −20°C for 1 day. After centrifugation at 13,000 rpm at 4°C for 30 min, the supernatant was discarded. The pellet containing the DNA was washed with 1 volume of 100% ethanol and then centrifuged at 13,000 rpm for 30 min. The supernatant was completely removed, and the pellet was air dried for 15 min. The DNA was resuspended in 50 μl 10 mM Tris-HCl, pH 7.5. The V3 region of the DNA was then reamplified according to the PCR protocol described above. Again, the amplified products were loaded on a DGGE gel to improve DNA yield and check band purity. If the band on this second gel matched the previously selected one, it was cut out, purified, and reamplified the same way. The amplified products were then purified with the NUCLEOTRAP-CR kit (Macherey-Nagel, Düren Germany) according to the manufacturer's protocol. The DNA fragments were ligated using the pGEM-T vector system (Promega), following the protocol of the manufacturer. Transformation was performed by electroporation using the Bio-Rad Gene Pulser XCell and PC module into E. coli XLI-Blue. The transformed bacterial cells were then plated onto Luria-Bertani (LB) agar containing ampicillin (150 μg/ml), X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) (0.1 mM) and IPTG (isopropyl-β-d-thiogalactopyranoside) (0.2 mM). Plasmids were recovered from white colonies by using the NucleoSpin plasmid kit (Macherey-Nagel) according to the manufacturer's protocol. The resulting DNA fragments were sequenced by Macrogen Corp., South Korea. Three clones per band were sequenced, and only the bands having similar sequences in two out of these three clones are presented in Results. The 16S rRNA gene sequences were aligned using the ClustalX software (34), and the phylogenetic trees were constructed using the neighbor-joining method (31) with the Njplot software (ftp://pbil.univ-lyon1.fr/pub/mol_phylogeny/njplot) (25). The topology of the distance tree was tested by resampling data with 100 bootstraps (8) to provide confidence estimates for tree topologies.
SEM analysis.
The spores were fixed using 1% OsO4 and air dried. After being coated with gold, the samples were examined with a Phillips XL 30 scanning electron microscope with an acceleration voltage of 10 kV.
Statistical analysis.
To analyze the relationships between the DGGE patterns of the different samples, correspondence analysis (CA) was used. This ordination method is adapted to analyze presence/absence or abundance data tables and is well suited for populations with unimodal distributions along environmental gradients (10). To perform the CA, a data matrix was composed of rows of objects representing the culture condition replicate and columns of species representing a DGGE band position along the vertical gel gradient. The relative abundance of a species in a sample corresponded to the DGGE band's relative intensity with regard to the sum of all band intensities in a pattern. The CA was then applied on the basis of numerical data matrices converted using the program Progiciel R (16). From the association matrix obtained, the characteristic values associated with the characteristic vectors were calculated using a multidimensional dispersion cloud of the data with the Canoco 4.0 software (Microcomputer Power, Ithaca, N.Y.). Variation partitioning analysis (6) enables display of the variability of patterns constrained by the factors of interest. Therefore, this analysis was used to display the contributions of an AMF species or plant species to the bacterial community profiles. The significance of the results was tested with the Monte Carlo permutation test. Variation partitioning analysis was performed with the software R (26).
Nucleotide sequence accession numbers.
The DGGE band sequences were submitted to the EMBL nucleotide sequence database and assigned accession no. AJ864379 to AJ864393.
RESULTS
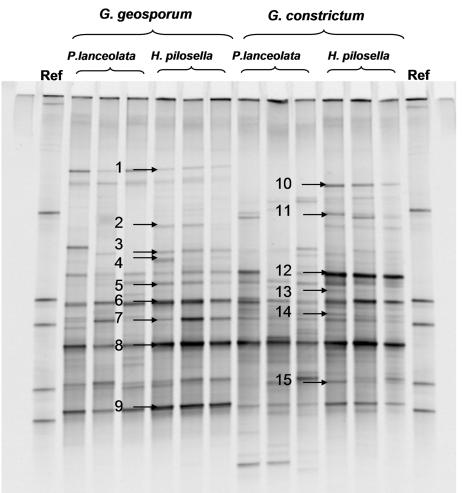
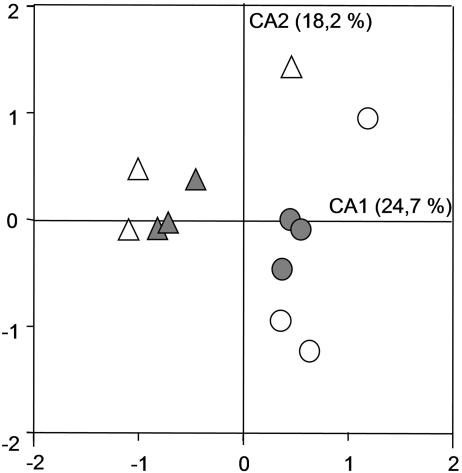
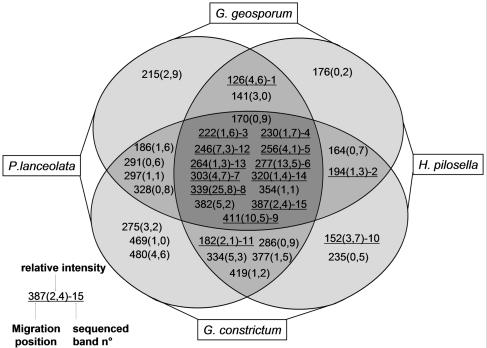
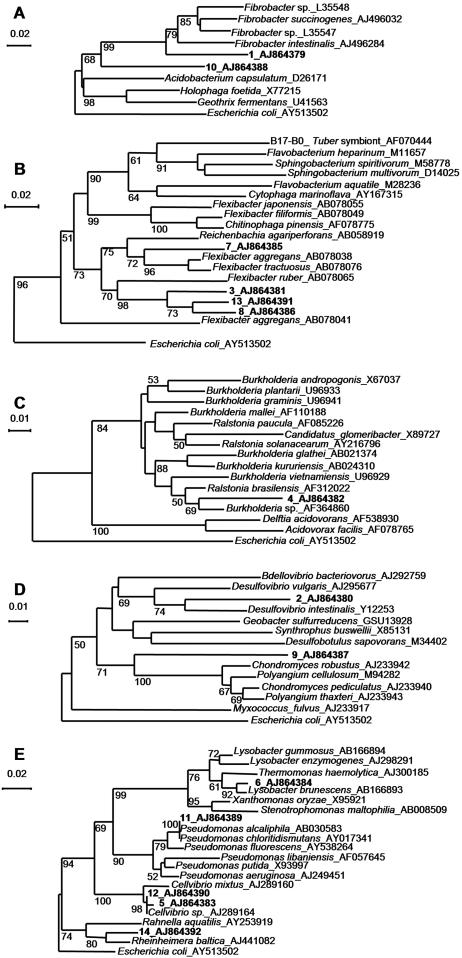
The homogeneity among replicates of DGGE patterns of bacterial communities associated with AMF spores was higher in cultures on H. pilosella than in those on P. lanceolata. This was obvious by optical observation (Fig. 1) and was confirmed by correspondence analysis (Fig. 2). The distances among the samples of two different host plants within one AMF species were shorter than the distances among the samples of the two AMF strains with one plant species, indicating that the bacterial community was structured mainly by the AMF species (Fig. 2). This observation is supported by variation partitioning analysis, which revealed that the host plant explained 12.1% (P = 0.001) of the variation of the bacterial DGGE patterns, that the AMF species explained 21.6% (P = 0.001) of the variation, and that there was no cross-variation (P = 0.12). A strong proportion of bands were common to all the culture conditions (Fig. 3), showing that many bacterial populations were always associated with AMF spores whatever the fungal species and the host plant were. In addition, many of these common bands had a high relative intensity, indicating that they were probably the most dominant populations on the AMF spores. To determine their affiliations, 11 bands in common (bands 3, 4, 5, 6, 7, 8, 9, 12, 13, 14, and 15) and four bands belonging to particular culture conditions (bands 1, 2, 10, and 11) were excised, cloned, and sequenced. Because of the higher homogeneity of the replicates, only DGGE bands obtained from H. pilosella cultures were selected. Band 1_AJ864379 was present only with G. geosporum and was affiliated with the phylum Fibrobacteres (Fig. 4A), band 2_AJ864380 was present only with H. pilosella and was related to the genus Desulfovibrio (Fig. 4D), band 10_AJ864388 was present only in G. constrictum and H. pilosella cultures and was affiliated with the phylum Fibrobacteres (Fig. 4A). Band 11_AJ864389 was present only with G. constrictum and was affiliated with the genus Pseudomonas (Fig. 4E). Three bands were present under all of the culture conditions but with a relative abundance much higher in the case of G. geosporum: band 6_AJ864384 was related to the genus Lysobacter (Fig. 4E), band 7_AJ864385 was related to the genus Flexibacter (Fig. 4B), and band 9_AJ864387 was related to the genus Chondromyces (Fig. 4D). Finally, eight bands were found under all culture conditions and with similar relative abundances: bands 3_AJ864381, 8_AJ864386, and 13_AJ864391 were related to the genus Flexibacter (Fig. 4B); bands 5_AJ864383 and 12_AJ864390 were related to the genus Cellvibrio (Fig. 4E); band 4_AJ864382 was related to the genus Burkholderia (Fig. 4C); band 14_AJ864392 was related to the genus Rheinheimera (Fig. 4E); and band 15_AJ864393 was related to Cyanobacteria (data not shown). Interestingly, most of the genera identified are bacteria that can hydrolyze biopolymers such as proteins, cellulose, and chitin (15, 24, 27, 28, 29). When the mean values of the relative intensities of all the band sequences related to these biopolymer-degrading genera are added up, they represent 60% of the overall intensity in G. geosporum/P. lanceolata, 84% in G. geosporum/H. pilosella, 53% in G. constrictum/P. lanceolata, and 73% in G. constrictum/H. pilosella cultures. Therefore, these polymer-degrading bacteria probably represent the main populations contributing to the bacterial community associated with AMF spores.
FIG. 1.
DGGE gels of the 16S rRNA gene V3 region for bacterial communities associated with the spores of G. geosporum and G. constrictum. Ref, reference pattern composed of five known bacterial sequences. The amplified product of each of the three replicates per culture condition was loaded on the gel. Bands cut and sequenced are indicated with arrows and labeled 1 to 15.
FIG. 2.
Ordination plot generated by correspondence analysis, representing the relationships between AMF spore-associated bacterial communities defined by the DGGE patterns. Three replicates per culture condition were integrated. Circles, G. geosporum; triangles, G. constrictum; open symbols, P. lanceolata; gray symbols, H. pilosella. Values on the axes indicate the percentage of the total variation explained by the axes. CA1, correspondence analysis axis 1; CA2, correspondence analysis axis 2.
FIG. 3.
Distribution of the bands composing the DGGE profiles among the different culture conditions. The bands are named after their vertical position along the DGGE gel. The relative intensity as a mean value of the intensities of the bands obtained for the corresponding culture conditions is indicated in parentheses. Bands excised, cloned, and sequenced are underlined, and their assigned number is given.
FIG. 4.
Affiliation of the sequences retrieved from the DGGE bands with existing 16S rRNA gene sequences, using neighbor-joining trees. Bootstrap values of below 50 are not indicated. The sequences from the database are indicated in italic with their accession numbers. The sequenced bands are shown in boldface. A, Fibrobacteres-Acidobacteria group; B, Cytophaga-Flexibacter-Bacteroides group; C, Betaproteobacteria; D, Deltaproteobacteria; E, Gammaproteobacteria.
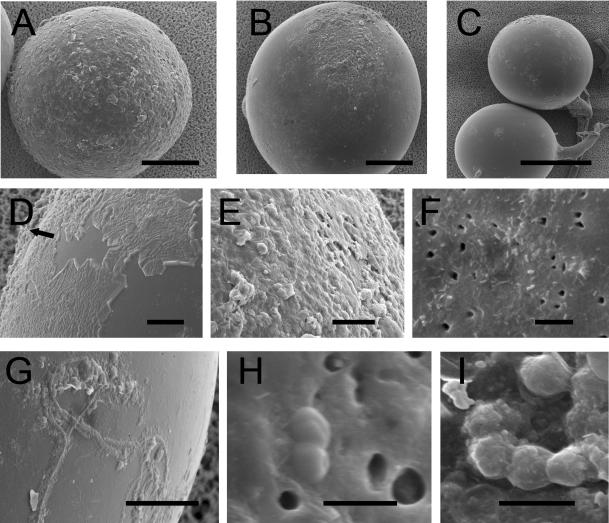
Prior to being prepared for microscopy, G. geosporum spores were divided into the following three consecutive maturity stages discernible under the dissecting microscope: the youngest spores, which were light yellow-brown without or containing a few dark patches; medium-brown spores with many patches; and dark orange-brown spores with many patches or one giant patch. SEM observations revealed that the thin outer hyaline wall layer of G. geosporum spores was gradually degraded and replaced by mucilaginous products (Fig. 5D and E). The rough surface composed of the degraded and mucilated outer hyaline layer was present to different extents depending on the spore maturity stage. Out of 38 light-colored spores observed, only 6 had a smooth surface, 18 were covered with roughness of up to half of the visible surface by SEM, and the surfaces of 14 were entirely rough (Fig. 5A). Out of 24 medium-brown spores, 16 were virtually smooth, 8 were covered with a rough material on 50% of the surface, and 8 were entirely rough (Fig. 5B). Finally, out of 17 dark-colored spores examined, 15 were entirely smooth and only 2 were slightly rough (Fig. 5C). Bacterial cells of different sizes were present either in the sloughing hyaline layer or on the surface of the second, laminated wall layer (Fig. 5F). Decaying material complicated the observation of bacterial cells because they appeared to be covered with their own mucilage. On the smooth laminated surfaces, bacterial filaments were observed covered with mucilage products (Fig. 5G). The spore surface also contained many holes, possibly corresponding to lysis zones (Fig. 5F and H). Finally, many coccus-shaped cells were also present (Fig. 5H and I).
FIG. 5.
SEM images of the surface of G. geosporum spores. (A) Young, light yellow-brown spore with its sloughed and eroded outer hyaline layer covering the whole surface; (B) older, medium-brown spore with a residual outer hyaline layer; (C) old, dark orange-brown spore that has lost almost all its outer hyaline layer; (D) outer hyaline layer starting to “peel off” and being replaced by mucilaginous products (arrow); (E) mucilaginous outer hyaline layer; (F) bacterial cells of various shapes adhering to the surface of the laminated layer, with holes possibly corresponding to lysis zones in the spore wall; (G) filamentous bacterial cells adhering to the laminated layer covered with mucilaginous products; (H) coccus-shaped bacteria in division state on the spore surface; (I) chain of coccus-shaped bacteria covered with mucilaginous products. Bars (in micrometers): A and B, 50; C, 100; D, E, F, and G, 10; H and I, 1.
DISCUSSION
The bacterial community associated with the Glomus spores was more influenced by the AMF identity (G. geosporum or G. constrictum) than by that of the host plant (H. pilosella or P. lanceolata). Despite the impact of the root on its surrounding environment and consequently on the microbial community, the plant did not predominantly affect the spore-associated bacterial community structure. Moreover, there was good homogeneity within replicates. The AMF spores thus seem to provide a microhabitat with particular conditions for the development of specific bacterial populations. The difference in composition of the spore walls or of exudates of these two Glomus species may have played a major role in the selection of bacterial populations living on the spore. In addition, the two Glomus species were isolated from the same site (35) and subcultured under the same conditions. The subculturing process may have enriched spore-associated bacterial populations adapted either to G. geosporum or to G. constrictum, which could have increased the discrepancies between the spore-associated bacterial community structures of the two fungi at the time of analysis.
Roughly one-third of the DGGE bands, among which were some exhibiting the highest relative intensities, were found in the profiles of cultures from all conditions. As a whole, these bands represented more than 50% of the relative intensity of the entire profiles. They comprised sequences affiliated mainly with genera with hydrolytic representatives (Cellvibrio, Chondromyces, Flexibacter, Lysobacter, and Pseudomonas). These biopolymer-degrading bacteria are possibly feeding on the outer hyaline spore layer that is present in both species and consists mainly of chitin, a straight-chain polymer of N-acetylglucosamine (32). Filippi et al. (9) actually demonstrated that many bacteria were attached to the hyaline wall layer of Glomus mosseae spores and that up to 107 CFU/g chitinolytic microorganisms were present on the sporocarp surface. Moreover, Maia and Kimbrough (19) observed bacteria colonizing the walls of a Glomus sp., which they apparently degraded until some remnants were left over. Finally, Walley and Germida (36) observed that the presence and activity of bacteria were generally limited to the outer spore wall surface. In our study, evidence of a bacterial saprophytic activity was suggested by SEM observations of G. geosporum spores, showing that the spore's outer hyaline layer was strongly eroded and that the spore surface was covered with mucilaginous products. Moreover, the outer hyaline layer of the light-colored, juvenile spores showed an early stage of degradation, whereas in the more mature, darker spores this layer was in most cases completely degraded. We also observed that many bacterial cells were either adhering to the laminated layer or embedded in the sloughing hyaline layer. Using transmission electron microscopy, Filippi et al. (9) found bacterial forms in holes within the outer layer of the Glomus mosseae spores and suggested that the holes were formed by the bacterial lytic activity. We also observed many holes on the spore, but as only the surface is seen by SEM, we were not able to confirm the presence of bacteria in these holes.
The most dominant bacterial population we identified was constituted by the genus Flexibacter. This genus is well known for its ability to degrade biomacromolecules in various habitats (27). Some Flexibacter species can form long threads of up to 50 μm long (27). In our study, SEM observations revealed that many long filaments were present on the spore surface. However, these filaments could also be actinobacteria. Actinobacteria were often found to be associated with AMF spores. For example, Mugnier and Mosse (22) reported that G. mosseae spores germinated in vitro only in the presence of microorganisms, including Streptomyces orientalis. Ames and coworkers (2) found that out of 190 spores examined, 100 were colonized by one or more chitin-decomposing microorganisms; 82% were colonized by actinomycetes, 17% by bacteria, and 1% by fungi. Carpenter-Boggs et al. (7) demonstrated a positive correlation between higher germination rate and the amount of production of geosmin, CO2, and 2-methylisoborneol by the actinomycetes. In our study, none of the DGGE bands sequenced were affiliated with actinobacteria. As not all the discrete bands of the DGGE profiles were sequenced, we may have missed this group of microorganisms. However, the studies mentioned above were performed with isolation-cultivation techniques. It is probable that the more abundant spore-associated bacteria might have been neglected, as only 1 to 10% of the total bacterial community is cultivable (1). Moreover, Walley and Germida (36) observed many dwarf cells (<0.3 μm in diameter) embedded within the spore walls, which could represent bacteria in a viable but not cultivable state (20).
Several bands sequenced were affiliated with genera capable of cellulolytic activity. The presence of cellulolytic bacteria on the spore surface indicates that microorganisms attached to the spores may also degrade plant material around them (e.g., cellulose from sloughed off cortical root cells). Gryndler et al. (13) have reported that an amendment of cellulose, if incubated in the soil for a long time, increased the number of bacteria and saprophytic fungi in the soil and also stimulated AMF growth. They suggested that this AMF stimulation could result from an AMF uptake of nutrients released from the decomposing saprophytic microflora. Root exudates also provide the microorganisms with readily assimilable organic substrates (17) and thus stimulate the growth of the biopolymer-degrading populations that would in turn accelerate the decay of the outer spore walls. Indeed, the outer hyaline layers, which are generally the first component of the spore wall synthesized in juvenile spores, are rarely present on mature spores in the soil (14). The presence of active biopolymer-degrading bacterial populations on the spore surface could support also spore germination by releasing nutrients or degrading toxic compounds that inhibit germination. Thus, the process of maturation and eventual germination of AMF spores might benefit from the activity of the surface microorganisms degrading the outer hyaline layer.
Acknowledgments
We thank Nicole Jeanneret, Prasun Ray, and Deepak Pant for technical assistance; Jérôme Hamelin and Florian Kohler for statistical analysis expertise; and Anne Smiejan-Roesti for critical reading of the paper.
This study was supported by the Swiss Agency for Development and Cooperation (SDC) in the framework of the Indo-Swiss Collaboration in Biotechnology (ISCB) program and the National Centre of Competence in Research (NCCR) in Plant Survival, University of Neuchâtel.
REFERENCES
- 1.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ames, R. N., K. L. Mihara, and H. G. Bayne. 1989. Chitin-decomposing Actinomycetes associated with a vesicular arbuscular mycorrhizal fungus from a calcareous soil. New Phytol. 111:67-71. [Google Scholar]
- 3.Atkinson, D. J. A., Baddeley, N. Goicoechea, J. Green, M. Sanchez-Diaz, and C. A. Watson. 2002. Arbuscular mycorrhizal fungi in low input agriculture, p. 211-222. In S. Gianinazzi, H. Schüepp, J. M. Barea, and K. Haselwandter (ed.), Mycorrhizal technology in agriculture: from genes to bioproducts. Birkhäuser Verlag, Basel, Switzerland.
- 4.Bianciotto, V., E. Lumini, L. Lanfranco, D. Minerdi, P. Bonfante, and S. Perotto. 2000. Detection and identification of bacterial endosymbionts in arbuscular mycorrhizal fungi belonging to the family Gigasporaceae. Appl. Environ. Microbiol. 66:4503-4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonfante-Fasolo, P., and A. Schubert. 1987. Spore wall architecture of Glomus spp. Can. J. Bot. 65:539-546. [Google Scholar]
- 6.Borcard, D., P. Legendre, and P. Drapeau. 1992. Partialling out the spatial component of ecological variation. Ecology 73:1045-1055. [Google Scholar]
- 7.Carpenter-Boggs, L., T. E. Loynachan, and P. D. Stahl. 1995. Spore germination of Gigaspora margarita stimulated by volatiles of soil-isolated Actinomycetes. Soil Biol. Biochem. 27:1445-1451. [Google Scholar]
- 8.Felsenstein, J. 1985. Confidence-limits on phylogenies—an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 9.Filippi, C., G. Bagnoli, A. S. Citernesi, and M. Giovannetti. 1998. Ultrastructural spatial distribution of bacteria associated with sporocarps of Glomus mosseae. Symbiosis 24:1-12. [Google Scholar]
- 10.Fromin, N., J. Hamelin, S. Tarnawski, D. Roesti, K. Jourdain-Miserez, N. Forestier, S. Teyssier-Cuvelle, F. Gillet, M. Aragno, and P. Rossi. 2002. Statistical analysis of denaturing gel electrophoresis (DGE) fingerprinting patterns. Environ. Microbiol. 4:634-643. [DOI] [PubMed] [Google Scholar]
- 11.Garbaye, J. 1994. Helper bacteria—a new dimension to the mycorrhizal symbiosis. New Phytol. 128:197-210. [DOI] [PubMed] [Google Scholar]
- 12.Gryndler, M., H. Hrselova, and D. Striteska. 2000. Effect of soil bacteria on hyphal growth of the arbuscular mycorrhizal fungus Glomus claroideum. Folia Microbiol. 45:545-551. [DOI] [PubMed] [Google Scholar]
- 13.Gryndler, M., M. Vosatka, H. Hrselova, I. Chvatalova, and J. Jansa. 2002. Interaction between arbuscular mycorrhizal fungi and cellulose in growth substrate. Appl. Soil Ecol. 19:279-288. [Google Scholar]
- 14.International Culture Collecion of (Vesicular) Arbuscular Mycorrhizal Fungi. [Online.] http://invam.caf.wvu.edu.
- 15.Kersters, K., P. de Vos, M. Gillis, J. Swings, P. Vandamme, and E. Stackerbrand. 2003. Introduction to the Proteobacteria. In M. Dworkin, (ed.), The prokaryotes: an evolving electronic resource for the microbiological community, 3rd ed., release 3.0. [Online.] Springer-Verlag, New York, N.Y. http://link.springer-ny.com/link/service/books/10125.
- 16.Legendre, P., and A. Vaudor. 1991. Le progiciel R. Analyse multidimensionelle, analyse spatiale. Université de Montréal, Québec, Canada.
- 17.Lynch, J. M., and J. M. Whipps. 1991. Substrate flow in the rhizosphere, p. 15-24 In D. L. Keister and P. B. Cregan (ed.), The rhizosphere and plant growth. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 18.MacDonald, R. M., and M. R. Chandler. 1981. Bacterium-like organelles in the vesicular-arbuscular mycorrhizal fungus Glomus caledonius. New Phytol. 89:241-246. [Google Scholar]
- 19.Maia, L. C., and J. W. Kimbrough. 1998. Ultrastructural studies of spores and hypha of a Glomus species. Int. J. Plant Sci. 159:581-589. [Google Scholar]
- 20.Mascher, F., C. Hase, Y. Moënne-Loccoz, and G. Défago. 2000. The viable-but-nonculturable state induced by abiotic stress in the biocontrol agent Pseudomonas fluorescens CHA0 does not promote strain persistence in soil. Appl. Environ. Microbiol. 66:1662-1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mayo, K., R. E. Davis, and J. Motta. 1986. Stimulation of germination of spores of Glomus versiforme by spore-associated bacteria. Mycologia 78:426-431. [Google Scholar]
- 22.Mugnier, J., and B. Mosse. 1987. Spore germination and viability of a vesicular arbuscular mycorrhizal fungus, Glomus mosseae. Trans. Br. Mycol. Soc. 88:411-413. [Google Scholar]
- 23.Muyzer, G., E. C. Dewaal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S ribosomal RNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nagarajkumar, M., R. Bhaskaran, and R. Velazhahan. 2004. Involvement of secondary metabolites and extracellular lytic enzymes produced by Pseudomonas fluorescens in inhibition of Rhizoctonia solani, the rice sheath blight pathogen. Microbiol. Res. 159:73-81. [DOI] [PubMed] [Google Scholar]
- 25.Perriere, G., and M. Gouy. 1996. WWW-Query: an on-line retrieval system for biological sequence banks. Biochimie 78:364-369. [DOI] [PubMed] [Google Scholar]
- 26.R Development Core Team. 2004. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Online.] http://www.R-project.org.
- 27.Reichenbach, H. 1999. The order Cytophagales. In M. Dworkin, (ed.), The prokaryotes: an evolving electronic resource for the microbiological community, 3rd ed., release 3.0. [Online.] Springer-Verlag, New York, N.Y. http://link.springer-ny.com/link/service/books/10125.
- 28.Reichenbach, H. 1999. The genus Lysobacter. In M. Dworkin, (ed.), The prokaryotes: an evolving electronic resource for the microbiological community, 3rd ed., release 3.0. [Online.] Springer-Verlag, New York, N.Y. http://link.springer-ny.com/link/service/books/10125.
- 29.Reichenbach, H. 1999. The ecology of the myxobacteria. Environ. Microbiol. 1:15-21. [DOI] [PubMed] [Google Scholar]
- 30.Ruiz-Lozano, J. M., and P. Bonfante. 2000. Intracellular Burkholderia of the arbuscular mycorrhizal fungus Gigaspora margarita possesses the vacB gene, which is involved in host cell colonization by bacteria. Microb. Ecol. 39:137-144. [DOI] [PubMed] [Google Scholar]
- 31.Saitou, N., and M. Nei. 1987. The neighbor-joining method—a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 32.Sbrana, C., L. Avio, and M. Giovannetti. 1995. The occurrence of Calcofluor and lectin-binding polysaccharides in the outer wall of arbuscular mycorrhizal fungal spores. Mycol. Res. 99:1249-1252. [Google Scholar]
- 33.Smith, S. E., and D. J. J. Read. 1997. Mycorrhizal Symbiosis. Academic Press, London, United Kingdom.
- 34.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van der Heijden, M. G. A., J. N. Klironomos, M. Ursic, P. Moutoglis, R. Streitwolf-Engel, T. Boller, A. Wiemken, and I. R. Sanders. 1998. Mycorrhizal fungal diversity determines plant biodiversity, ecosystem variability and productivity. Nature 396:69-72. [Google Scholar]
- 36.Walley, F. L., and J. J. Germida. 1996. Failure to decontaminate Glomus clarum NT4 spores is due to spore wall-associated bacteria. Mycorrhiza 6:43-49. [Google Scholar]
- 37.Weisskopf, L., N. Fromin, N. Tomasi, M. Aragno, and E. Martinoia. 2005. Secretion activity of white lupin's cluster roots influences bacterial abundance, function and community structure. Plant Soil 268:181-194.
- 38.Xavier, L. J. C., and J. J. Germida. 2003. Bacteria associated with Glomus clarum spores influence mycorrhizal activity. Soil Biol. Biochem. 35:471-478. [Google Scholar]