Abstract
Frequent high-amplitude redox fluctuation may be a strong selective force on the phylogenetic and physiological composition of soil bacterial communities and may promote metabolic plasticity or redox tolerance mechanisms. To determine effects of fluctuating oxygen regimens, we incubated tropical soils under four treatments: aerobic, anaerobic, 12-h oxic/anoxic fluctuation, and 4-day oxic/anoxic fluctuation. Changes in soil bacterial community structure and diversity were monitored with terminal restriction fragment length polymorphism (T-RFLP) fingerprints. These profiles were correlated with gross N cycling rates, and a Web-based phylogenetic assignment tool was used to infer putative community composition from multiple fragment patterns. T-RFLP ordinations indicated that bacterial communities from 4-day oxic/anoxic incubations were most similar to field communities, whereas those incubated under consistently aerobic or anaerobic regimens developed distinctly different molecular profiles. Terminal fragments found in field soils persisted either in 4-day fluctuation/aerobic conditions or in anaerobic/12-h treatments but rarely in both. Only 3 of 179 total fragments were ubiquitous in all soils. Soil bacterial communities inferred from in silico phylogenetic assignment appeared to be dominated by Actinobacteria (especially Micrococcus and Streptomycetes), “Bacilli,” “Clostridia,” and Burkholderia and lost significant diversity under consistently or frequently anoxic incubations. Community patterns correlated well with redox-sensitive processes such as nitrification, dissimilatory nitrate reduction to ammonium (DNRA), and denitrification but did not predict patterns of more general functions such as N mineralization and consumption. The results suggest that this soil's indigenous bacteria are highly adapted to fluctuating redox regimens and generally possess physiological tolerance mechanisms which allow them to withstand unfavorable redox periods.
Oxygen is the primary terminal electron acceptor for the respiratory processes of most upland soil microorganisms (41); it is a critical determinant of both soil redox status and the physiological pathways available to bacteria and fungi mediating C and N cycles. As soil redox (pE) decreases, dominant element transformations are generally assumed to shift in a well-defined succession from high-energy-yield processes to those that release less energy for microbial growth. This idea, that biological metabolism should follow the electrochemical constraints of a system, is a relatively old concept; it was first described in 1960 (1) and later supported by studies of waterlogged sediments, aquifers (11, 32), and rice paddies (56). In all of these cases, changes in redox potential occur gradually over time and/or space.
In upland humid tropical forests, soils have been found to experience rapid redox fluctuations (51). In these ecosystems, the combination of high C availability, warm temperatures, abundant rainfall, clay soils with high water-holding capacity, and high metabolic activity can lead to conditions where O2 is rapidly and completely consumed from both liquid- and gas-filled pore spaces (19). Low O2 concentrations are associated with increased labile C (38) and reductions of redox potential that are frequently sufficient for methanogenesis (28, 51). Wet upland tropical soils are less likely to exhibit the slow uniform progression from more oxidized to more reduced processes that is commonly observed in continually waterlogged sediments and subsurface aquifers (32). Instead, these tropical soils are likely to be highly dynamic, with dominant electron couples changing rapidly during frequent cycles of wetting and drying or periods of high biological activity. In these soils, high net primary productivity (62) and daily rainfall (totaling 3.5 to 4.5 m annually) lead to high availability of reductant and redox conditions that change markedly over relatively short time scales.
The physiological implications of fluctuating oxygen availability for bacterial communities have rarely been studied in upland soils. This is due to both the methodological difficulties of soil redox measurements (54) and an assumption that aerobic processes dominate upland soils. However, the frequency of redox shifts may have a strong effect on community development and function, analogous to the effects that drying and rewetting events have on soil microbial biomass and activity (18) or that temperature fluctuations have on heterotrophic respiration and methane production (59). Several recent studies have shown that shifting redox patterns can affect ecosystem processes such as methane efflux (47), glucose respiration (43), and nitrification and denitrification (16). These redox shifts can occur on very short (hourly or daily) (47) to longer (monthly or seasonally) time scales (57); however, the sensitivity of microbial communities to repeated redox cycling has not been investigated.
In high-Eh, well-oxygenated habitats, obligate aerobes depend on oxidative respiration and phosphorylation to maintain their energy and nutrient requirements. While these organisms generally do not find anoxic conditions toxic, many are inactivated or starved as O2 becomes limiting (17). Conversely, many obligate anaerobes such as methanogens, iron and sulfate reducers, find O2 toxic because they lack superoxide dismutase and catalase enzymes necessary to degrade O2− and peroxides (17, 25, 58). Free O2 toxicity may also occur due to the disruption of enzymes essential to anaerobic energy metabolism, such as hydrogenases and pyruvate:ferredoxin oxidoreductase (17). Because of the importance of soil oxygen availability to microbial metabolism, spatial shifts in microbial physiology are traditionally thought to occur along gradients of soil redox potential. As soil redox potential decreases, the dominance of functional groups shifts (1, 43, 58). Ludemann et al. (33) demonstrated such a pattern in paddy soils by using terminal restriction fragment length polymorphism (T-RFLP); others have also measured similar patterns in seasonal bacterial communities and functional changes in salt marshes (16, 27). Studies of such changes in uplands soils are rare, although Picek et al. (43) observed that microbial biomass, C mineralization, and glucose consumption were sensitive to 48-h redox shifts in a Czech field soil.
There is substantial current interest in microbial community composition and diversity in below-ground ecosystems; this is particularly true for tropical communities (5). A growing body of evidence indicates that microbial community composition significantly affects ecosystem processes (10, 60), just as above-ground plant community composition has been shown to affect processes such as N cycling (64) and decomposition (24). An important measure of the functional significance of microbial community composition is ecosystem nitrogen processing. The transformations in the nitrogen cycle are particularly susceptible to changes in soil (pE), since redox potential acts as a master switch between microbe-catalyzed processes that sequester N and those that emit N trace gases.
The objective of this study was to examine microbial community dynamics in a soil environment characterized by fluctuating redox potential. We hypothesized that the intensity and duration of soil O2 fluctuations affect the phenotypic plasticity of bacterial communities, since fluctuation should logically select for physiologies that are tolerant of extremes and resume activity rapidly. Frequent soil redox fluctuation may also make dormancy a strategy for bacteria which need to endure redox periods incompatible with their primary mechanisms of energy generation. Alternatively, a fluctuating redox system might be dominated by facultative metabolisms, such as those that can switch between O2 and NO3 or O2 and fermentation.
To determine the survival strategies important in humid tropical forest soils, we incubated soil under static aerobic and anaerobic conditions as well as fluctuating conditions and then used multiple-enzyme T-RFLP analyses to quantify bacterial community responses and diversity. We used a Web-based (in silico) tool (29) to analyze multiple T-RFLP profiles from standardized enzyme digests. By comparing results of multiple restriction enzyme digests to those of known bacterial sequences, this tool increases the specificity of putative phylogenetic assignments and was used to infer how our imposed redox treatments affected community composition. Lastly, to garner more information on the relationship between community structure and function, we compared T-RFLP profiles with gross N cycling rates.
MATERIALS AND METHODS
Soil collection.
Soil samples were collected from the Colorado forest type of the Luquillo Experimental Forest, which is part of the Long-Term Ecological Research program in Puerto Rico (18°18′N, 65°50′ W). This lower montane wet tropical forest occurs between 600 and 900 m above sea level (38) and has an aseasonal climate, a mean annual temperature of 18.5°C, a relative humidity of 98%, and annual precipitation of 4,500 mm (63). The clay-loam ultisols are rich in kaolinite, biotite, and goethite derived from volcanoclastic sandstones with quartz diorite intrusions (39, 65). In the 0- to 10-cm soil depth, total N averages 0.2%, organic matter averages 8%, the pH is 4.81, and the bulk density is 0.55 g cm−3. These soils are frequently saturated, and gravimetric moisture content averages 44% (38). Bulk O2 concentrations measured in the 0- to 10-cm depth average 13% yet fluctuate between 3 and 17%. Long-term monitoring has shown that O2 fluctuation in these soils occurs on a time scale of days to weeks; only on rare occasions do soil O2 levels remain below 10% for longer than 1 month (51). While soil organic C is spatially variable in these soils, a positive correlation between soil moisture and high soil organic C at small spatial scales has been established (12, 61).
Approximately 75 soil cores were collected from the 0- to 10-cm depth by using 6- by 12-cm polyvinyl chloride tubes; samples were collected from a 10- by 10-m area. Cores were kept intact, immediately placed in sealed ziplock bags, and transported to the University of California, Berkeley, at ∼20°C. Within 24 h of sampling, cores were extruded and quickly homogenized in a 15-gallon cooler to create a single large composite sample. This sample was split into portions for initial analysis and for incubation treatments. For the initial analysis, 3-g subsamples were transferred to sterile microcentrifuge tubes and stored at −80°C for molecular community analysis. Soil moisture and microbial biomass (via chloroform fumigation incubation) were measured with the remaining soil. A 15N pool dilution was also conducted by adding 15NO3 and 15NH4 to separate soil portions and measuring 15NO3, 15NH4, 15N2O, 15N2, and 15N-microbial biomass after 0, 3, 6, and 14 h. These measurements allowed determination of inorganic N pools and rates of gross N processing, including nitrification, mineralization, denitrification, dissimilatory nitrate reduction to ammonium (DNRA), NO3/NH4 consumption, and residence time (see references 13, 22, 42, and 52 for detailed methods).
Redox incubations.
Using the composite soil, 170 g oven dry equivalent soil was added to 1,000-ml jars and divided into four treatments with five replicates each. Jars were capped with gas-tight lids fitted with a Hungate septa and a 5-in. piece of tygon tubing on the inside of the jar. Hydrated gases were delivered to the bottom of each jar (vented with a syringe needle) at a rate of 66 ml/min, giving a headspace turnover time of approximately 8 min. Soil moisture was maintained at 59% (±0.01%). Treatments included (i) “static aerobic” jars, which were constantly flushed with medical-grade air; (ii) “static anoxic” jars, which received N2 gas; (iii) “short-term fluctuation” jars, where flushing alternated between air and N2 every 12 h; and (iv) “long-term fluctuation” jars, where flushing alternated every 4 days. Jars were incubated under these treatments at 18°C and harvested after 3 weeks. At this point, each jar was subdivided; a 3-g portion was used for molecular community analysis and the remainder for 15N-based process analyses (52) as described above. The timing of flushing was designed so that jars from both fluctuating treatments were anaerobic just prior to harvesting. Periodic measurement of headspace O2, N2O, and CH4 gas concentrations indicated that N2 flushing had the desired effect in lowering soil redox, as evidenced by dramatically increased CH4 and N2O fluxes and zero O2.
T-RFLP analysis.
We extracted whole-community DNA from 0.5 g soil with the Fast DNA spin kit for soil (Bio 101 Systems, Carlsbad, CA) according to the manufacturer's instructions and pooled three extracts per sample to limit effects of random extraction bias (30). We confirmed the presence of high-molecular-weight DNA (6 to 10 kb) on 0.8% agarose gels; samples from all treatments had comparable DNA concentrations (80 ng/μl). We performed PCR amplification using 1 μl of a 1:10 dilution of soil extracts as template. PCR mixtures (50-μl final volume) contained 1× reaction buffer, 1.5 mM MgCl2, 250 μM each deoxynucleoside triphosphate, 20 μg bovine serum albumin (bovine serum albumin), 2.5 units of Taq DNA polymerase, and 400 nM primers (27F [AGAGTTTGATCCTGGCTCAG] and 1492R [TACGGYTACCTTGTTACGACTT]). The forward primer 27F was labeled with the fluorescent moiety 6-carboxyfluorescein at the 5′ end for detection by capillary electrophoresis. Primers were synthesized by QIAGEN Operon (Alameda, CA), deoxynucleoside triphosphates and bovine serum albumin were obtained from Roche Molecular Systems (Alameda, CA), and all other PCR reagents were purchased from Promega (Madison, WI). PCR was performed in a Perkin-Elmer 9600 thermocycler with an initial denaturation step of 94°C for 3 min, followed by 27 cycles of 94°C for 30 s, 53°C for 30 s, and 72°C for 1 min and a final extension at 72°C for 7 min. Three replicate PCR products per sample were pooled and purified with an Ultra Clean PCR clean-up kit (MoBio, Solana Beach, CA) according to the manufacturer's instructions.
Restriction digests were standardized by using 400 ng of purified PCR product for each sample. They were performed with four separate reactions per sample, using the restriction endonucleases MspI (CĈGG), AluI (AGĈT), RsaI (GTÂC), and HhaI (GCGĈ) (New England Biolabs Inc.) and associated buffers under conditions recommended by the manufacturer. Digests were incubated for 24 h at 37°C, desalted, and prepared for sequencing according to established procedures (6). Terminal restriction fragment (T-RF) lengths were measured on an ABI 3100 capillary electrophoresis system, and electropherograms were analyzed with Gene Scan software for fragment lengths of between 50 and 500 bp. The precision of a simultaneously run GeneScan ROX500 (ABI) standard was consistently ±0.5 bp. Sample profiles with less than 10,000 total fluorescence units were discarded and rerun (as suggested in reference 4). Peak patterns from different samples were manually aligned by size sorting and manually grouping (binning) peaks within 0.5 base pairs of fragment length. This technique, though time-consuming, avoids errors associated with automated rounding algorithms (30) and ensures that “double peaks” are identified and accounted for only once. Peak heights were relativized based on the proportion of total sample abundance (as advocated in references 4 and 30). Only fragments occurring in at least two sample profiles were included in community analysis. Approximately 20% of identified peaks were present in only one out of five replicates and were removed from analysis. Reproducibility of profiles was between 92 and 97% for a given sample replicate (extracted, amplified, digested, and sequenced on separate occasions). The term T-RF is used to represent individual fragments, although we recognize that one T-RF may incorporate genetic material from one or several distinct bacterial ribotypes (29).
Indicator species analysis (ISA) was used to identify individual T-RFs unique to a given redox treatment(s); we analyzed both T-RF fidelity (frequency) and T-RF exclusivity (abundance). This technique, based on Dufrêne and Legendre's method (15), combines information on the concentration of “species” (in this case, a T-RF) abundance in a particular group and the faithfulness of occurrence of a species in a particular group. It produces indicator values for each species in each group; a perfect indicator species occurs within a particular group or treatment without fail and is assigned an “indicator value” of 100. Indicator value assignments are tested for statistical significance using a Monte Carlo technique. Unlike the commonly used TWINSPAN analysis, ISA is more sensitive and does not assumes the existence of a strong a priori gradient dominating the data (15). Further, ISA helps identify T-RF assemblages that best characterize treatments or groups of treatments (15).
Using T-RFLP fingerprints, phylogenetic assignments were inferred using an automated Web-based tool (PAT) that compares T-RF sizes to the University of Idaho's Microbial Community Analysis (MiCA) database of fragments produced by known 16S rRNA gene sequences from over 2,000 bacterial ribotypes (29; http://trflp.limnology.wisc.edu/index.jsp). This and other in silico approaches (35) can generate putative identifications for uncultured members of a microbial community and has been successfully compared to clone libraries in characterizing samples from lakes (29), soils (45, 46), and the human colon (36). The benefits of this technique are its capacity to rapidly generate phylogenetic assignments from submitted T-RFLP profiles; however, because experimental errors may occur in fragment size determination, these assignments may be less precise than those of a clone library (29, 30). Although many phylogenetically similar species may contribute to the same T-RF (66), the comparison of multiple enzyme digests for each sample helps to improve the specificity of PAT assignments (29). Therefore, we chose to analyze output at the level of phylum-class as opposed to the species level (where errors in assignment are more likely). As an additional conservative measure, we set the fragment bin tolerance window for the PAT algorithm at ±1 bp for fragments of 0 to 250 bp in length and ±2 bp for 250- to 500-bp fragments.
Multiple T-RFLP digests (MspI, AluI, RsaI, and HhaI) were used to increase the phylogenetic resolution of ribotype assignments (29). We used the PAT method instead of creation of a clone library due to our large sample size, the highly diverse bacterial community, and problems associated with comparing community composition among different samples' clone libraries (48, 53). The results of PAT phylogenetic assignments were not used for statistical analyses, but instead are summarized in Fig. 3 and 4 to give an overview of treatment effects on the composition and phylogenetic distribution of community members. In cases where the PAT tool identified different strains of the same bacterial species multiple times, these were consolidated into one listing for analysis. In the rare cases where a set of T-RFs matched more than one species, only the genus was included as a single listing. Species matches that occurred in only one replicate out of five for a given treatment were also excluded. Phylogenetic assignments were classified according to Bergey's Manual of Systematic Bacteriology (20).
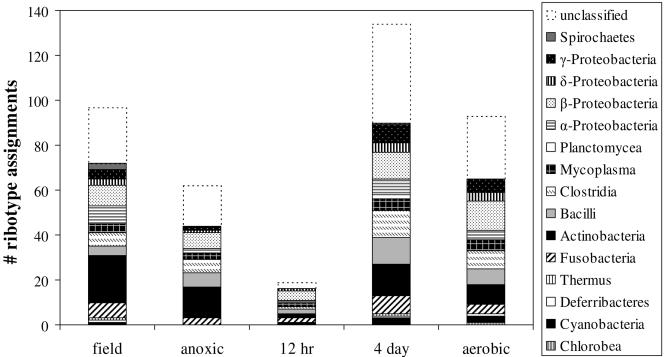
FIG. 3.
Bacterial phyla and classes detected by T-RFLP and an in silico phylogenetic assignment tool in soils incubated under four redox regimens. The plots indicate the number of ribotypes per treatment that were assigned to each phylogenetic class. The unclassified group contains unnamed clones, strains, and symbionts whose 16S rRNA sequences are known.
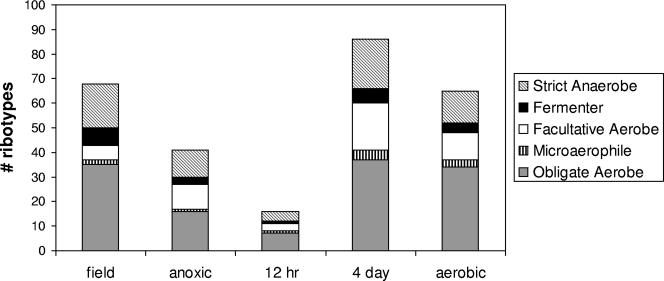
FIG. 4.
Number of bacterial ribotypes identified by an in silico PAT technique in soils incubated under four redox regimens and classified by redox functional group.
Statistical analysis.
T-RFLP data from the MspI digest were analyzed with PCORD v4 (MJM Software Design, Gleneden Beach, OR). Fragment length and relative peak height were analyzed using both parametric (principal-component analysis [PCA]), and nonparametric (nonmetric multidimensional scaling) ordination techniques. The primary purpose of PCA ordination is to reduce the number of variables down to the few that best account for sample variability (37). Principal-component axes summarize how samples most differ from one another along an arbitrary gradient; a secondary analysis (such as analysis of variance or multiple regression) needs to be used to test whether the axes correlate with treatment differences. While some authors advocate the analysis of presence/absence data in ordinations (4), we found no significantly different results from relativized peak height data and chose to present the latter. Only principal components with eigenvectors of >0.25 (9) were examined, and/or where principal components cumulatively explained 75% of the data variance (21). Sample outliers (>2 standard deviations from the mean) were removed from the analysis.
We chose to use an indirect gradient ordination (PCA as opposed to canonical correspondence analysis) because this analysis maximizes community variance among samples, regardless of treatment (37). While some community data do not meet PCA's assumptions, this data set does, as it has a roughly monotonic increasing linear relationship between T-RF abundance and the treatment gradient. Therefore we focus on the PCA results, as opposed to nonmetric multidimensional scaling, where the loss of statistical power yielded similar, though less interpretable, results (37, 42).
Cluster analysis and the multiresponse permutation procedure (37) were used to test for significant differences between treatment groups. The multiresponse permutation procedure “A” statistic is a measure of within-group homogeneity. When all items within a group are identical, A = 1.0. Typical values for “A” in community ecology are below 0.1; while an A value of >0.3 is considered a relatively large value (37). Diversity measures calculated for T-RFs included richness (based on the number of unique fragments that appeared in each sample) (31), Shannon's diversity index (50), and evenness (Shannon's index divided by the natural log of richness) (23). While the T-RFLP method does not give a complete estimate of sample bacterial diversity (only a fraction of bacterial species are represented in each identified terminal fragment), there should be a strong correlation between the T-RFLP terminal fragment diversity (the number of unique peaks and relative abundance of each peak) and the number of bacterial species in a given sample (31).
Analysis of variance and Tukey pairwise comparison tests were used to test for significant differences between treatments for diversity indices and PCA ordination of MspI-based T-RFLP data. To examine correlations between environmental variables/N-cycling rates and microbial community structure (represented by principal components), we used multiple regression with JMP software (SAS Institute, Inc., Cary, North Carolina) and evaluated the ability of the environmental variables to predict microbial T-RFLP profiles summarized as principal components (for further discussions of this technique, see references 2, 3, and 55). A separate multiple regression analysis was performed for each principal component. Significance was determined as a P value of <0.05 unless otherwise noted.
RESULTS
The soil bacterial community was most diverse in freshly sampled, nonincubated soil; 139 restriction fragments were detected after an MspI digestion of field soil, while 179 unique T-RFs were found from the field and treatment soils combined. AluI digestion yielded a similarly large number of fragments (184), while RsaI produced slightly fewer (148). While mean T-RF diversity declined with soil incubation, soils exposed to 4-day redox fluctuation retained significantly higher T-RF diversity than those exposed to static anoxic or 12-h fluctuation conditions (Table 1) (P < 0.0001). Soils kept consistently aerobic or anaerobic had intermediate T-RF richness diversity. Evenness, which indicates whether all T-RFs from a treatment occur at similar abundances, was significantly higher in the anoxic and 12-h fluctuation treatments (P < 0.0001).
TABLE 1.
Diversity indices of T-RFs generated by 16S rRNA amplification and MspI digestion of Luquillo Experimental Forest, P.R., soils
| Index | Valuea under the indicated conditions
|
||||
|---|---|---|---|---|---|
| Field | Anoxic | 12 h | 4 day | Aerobic | |
| Richnessb | 139 a | 74 c | 25 d | 92 b | 76 bc |
| Evennessc | 0.82 a | 0.97 b | 0.98 b | 0.87 a | 0.83 a |
| Shannon diversityd | 4.0 a | 2.7 b | 2.8 bc | 3.2 c | 2.7 bc |
| Indicator valuee | 42.6 a | 5.3 b | 19.7 c | 36.5 d | 5.9 b |
Significant treatment differences are indicated by different lowercase letters.
Mean number of unique terminal fragments per treatment, a measure of diversity.
Equitability, equal to Shannon index/ln richness.
Diversity of terminal fragments in a sample unit.
An index of both T-RF frequency and abundance, higher values indicate better indicator T-RFs.
The MRRP test of within-group homogeneity shows clearly that T-RFs from individual treatments cluster together (A = 0.68; P < 0.0001), an indication that grouping the data according to the imposed treatments is more accurate than any other random expectation. Hierarchical cluster analysis also suggests that based on community similarity, the incubation treatments are the best T-RF groupings; it indicates that at a gross scale (50% data variance explained), field, 4-day, and aerobic communities cluster together, while anoxic and 12-h communities fall in a separate cluster. Only at finer resolution (25% data variance explained) do field communities diverge from the 4-day and aerobic communities. This pattern was frequently observed throughout the analysis.
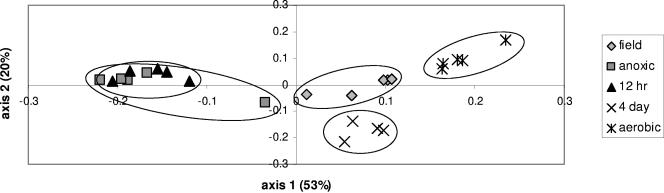
Ordination of MspI-derived T-RFLP patterns shows that communities exposed to static redox treatments changed substantially compared to the original soil community (Fig. 1). Along the first principal component (PC1) (53% variance explained; eigenvalue = 0.52), soils treated with a 4-day redox fluctuation cycle retained a bacterial community composition that was statistically indistinguishable from that of the field soil, whereas soils with the anoxic/12-h and aerobic treatments moved sharply apart in ordination space (Fig. 1). Analysis of variance indicated significant treatment differences (P < 0.001), with the following pairwise differences: (anoxic/12-h) ≠ (field/4-day) ≠ (aerobic). Along PC2 (20% variance explained), there is an indication that the field and 4-day communities are distinct; however, the strength of this pattern, while statistically significant, is muted due to a lower eigenvalue (0.2). Examination of individual T-RFs shows that most terminal fragments present in the original community persisted in one of two treatment groupings: either the aerobic/4-day fluctuation soils or the anoxic/12-h fluctuation soils. Of the 179 total T-RFs detected, only 3 from the field soil persisted in all four incubation treatments. While 34 fragments were found only in the field soil and were not detectable following incubation, 16 T-RFs not found in the original community were promoted by the incubation treatment. The majority of these incubation-induced T-RFs were promoted by the anoxic and 12-h treatments.
FIG. 1.
Principal-component analysis of bacterial T-RFLP fragment patterns of microbial communities from Luquillo Experimental Forest soils, Puerto Rico. Soil communities were analyzed fresh from the field (field) or after 3 weeks of incubation under four redox regimens: no O2 (static anaerobic), 12 h (aerobic/anaerobic fluctuation), 4 day (aerobic/anaerobic fluctuation), and O2 (static aerobic). Replicate samples circled, and axes are scaled to percent variance explained.
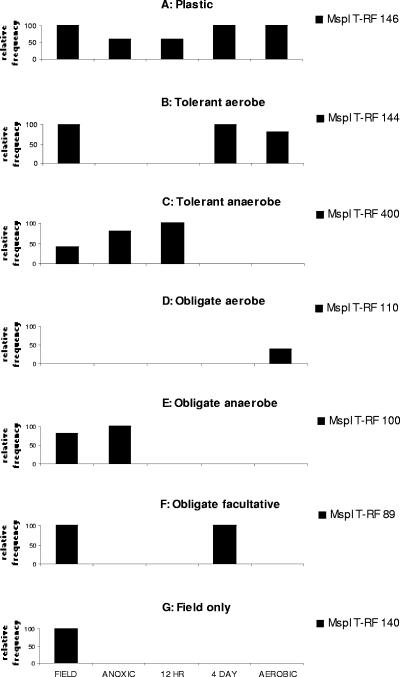
Based on this analysis, each MspI T-RF pattern was classified according to seven basic survival strategies/profiles (Table 2); examples from this data set are shown in Fig. 2. We defined the following strategies: “plastic,” organisms that tolerate all redox treatments; “tolerant aerobe,” organisms competitive under aerobic conditions but tolerant of brief anoxic periods; “tolerant anaerobe,” organisms competitive under anoxic conditions but tolerant of brief oxic periods; “obligate anaerobe” and “obligate aerobe,” organisms intolerant of all but static redox conditions; “facultative advantage,” organisms most competitive under a fluctuating redox regimen; and “field only,” organisms eliminated by lab conditions. Based on their higher T-RF representation, the “tolerant” and “facultative advantage” strategies appear to dominate in these soils (Table 2).
TABLE 2.
Number of terminal restriction fragmentsa classified according to seven basic strategies (see text)
| Profile | Strategy | Treatment(s) for which strategy is expected | No. of T-RFs |
|---|---|---|---|
| A | Plastic | Anoxic, 12 h, 4 day, aerobic | 3 |
| B | Tolerant aerobe | Aerobic, 4 day | 32 |
| C | Tolerant anaerobe | Anoxic, 12 h | 15 |
| D | Obligate aerobe | Aerobic | 1 |
| E | Obligate anaerobe | Anoxic | 2 |
| F | Facultative advantage | 12 h, 4 day | 90 |
| G | Field only (lab intolerant) | None | 34 |
Fragments generated from T-RFLP analysis of Luquillo Experimental Forest soils incubated under two static and two fluctuating redox treatments.
FIG. 2.
Seven T-RF patterns representative of alternative redox fluctuation survival strategies defined in the text. Relative frequency is based on occurrence out of five replicates.
Indicator species analysis integrates both frequency and abundance T-RF data and allowed us to evaluate the statistical significance of indicator values via a random Monte Carlo simulation. A “perfect indicator” T-RF occurs in a particular treatment without error, with an indicator value of 100. Mean indicator values (Table 1) were highest in field soils and were significantly higher in fluctuating than in static treatments. At a P value of <0.0001, we observed the following number of perfect indicator T-RFs for each treatment: field, 5; anoxic, 2; 12-h, 0; 4-day, 1; and aerobic, 0. This suggests that no one redox fluctuation treatment exclusively met the habitat needs of the majority of this soil bacterial community.
The in silico PAT analysis (Fig. 3) gives an indication of the diversity of community composition in these soils; however, due to the incomplete nature of the master ribosomal database, the results do not necessarily indicate relative abundance. After consolidation, the four restriction digests and PAT analysis generated a list of 145 species matches for T-RFs isolated from these soils. The PAT tool was unable to assign a species to many T-RFs; the percentages of unmatched fragments were 52% (MspI), 57% (HhaI), 64% (RsaI), and 71% (AluI). Addition of a fourth restriction enzyme to the analysis did not dramatically affect the composition of the output; however, the first, second, and third enzymes each significantly refined the species list. Assignments according to treatment are shown in Fig. 3, classified by phylum or class (for particularly diverse phyla). Also included are an additional number of clones, strains, and symbionts identified by the PAT program but unnamed. The community composition of these soils varied with incubation condition; the 12-h treatment had significantly fewer ribotypes identified by the PAT program (P < 0.0001), while aerobic soils and those with 4-day redox variation retained a richness and distribution of ribotypes similar to that for the field soil. While a diverse assemblage of organisms was identified, high-GC gram-positive bacteria and Proteobacteria dominated the list; the most highly represented groups included Actinobacteria (particularly the Micrococcus and Streptomycetes subgroups), Firmicutes-“Bacilli,” Firmicutes-“Clostridia,” and Betaproteobacteria-Burkholderia, with greater than six ribotypes each. Ordination analysis showed that the lower diversity identified in the 12-h fluctuating treatment was due to a lower proportion or complete lack of “Fusobacteria,” Actinobacteria, “Clostridia,” Mycoplasma, Gammaproteobacteria-enterobacteria, and Alphaproteobacteria-rhizobia. In contrast, the 4-day aerobic/anaerobic soils were matched to prominent assemblages of “Bacilli,” “Clostridia,” Burkholderia, cyanobacteria, Planctomycea, and a large number of unclassified organisms (Fig. 3). It is interesting to note the prominence of bacteria from the Actinobacteria in these wet, low-pH, frequently anoxic soils. Bacteria from this class have commonly been associated with dry, high-pH soil environments in the field of soil microbiology (34, 41).
To determine the relationships between community structure and soil functioning, environmental process variables were regressed against the first principal component (PC1) from the MspI-based T-RFLP PCA ordination (Table 3), followed by the second principal component (PC2). Environmental and process variables included in multiple regression analyses were soil moisture, initial field NO3 and NH4 pools, microbial biomass N, mean NO3/NH4 residence time, N gas flux, and rates of mineralization, nitrification, denitrification, DNRA, and NO3/NH4 consumption (42). All rates were measured over 0 to 24 h following 15N label addition. We removed soil moisture and residence time from the analysis, as they were nonsignificant and highly autocorrelated with other variables. Both bacterial community composition indices (PC1 and PC2) were highly correlated with initial soil NO3 concentration. There was significant correlation between PC1 and nitrification, DNRA, and denitrification (listed in order of decreasing r2). The second principal component axis (PC2) was significantly, though more weakly, correlated with nitrification, DNRA, and NH4 consumption. There was no evidence of a link between T-RFLP-based community indices and microbial biomass or 15N biomass uptake, nor was there a correlation between bacterial communities and NH4 pools or mineralization.
TABLE 3.
Regressions between T-RFLP principal-component ordination and soil chemistry/N-processing ratesa
| Parameter | PC1
|
PC2
|
||
|---|---|---|---|---|
| r2 | P | r2 | P | |
| Initial NO3 pool (μg/g) | 0.749 | 0.000 | 0.353 | 0.006 |
| Nitrification (μg/g/day) | 0.419 | 0.013 | 0.208 | 0.044 |
| DNRA (μg/g/day) | 0.363 | 0.025 | 0.191 | 0.053 |
| 15N2 flux | 0.228 | 0.049 | 0.026 | 0.493 |
| Total N gas flux (denitrification) | 0.225 | 0.050 | 0.025 | 0.508 |
| 15N2O flux | 0.185 | 0.085 | 0.033 | 0.440 |
| Initial NH4 pool (μg/g) | 0.142 | 0.124 | 0.013 | 0.631 |
| Microbial biomass N | 0.073 | 0.310 | 0.025 | 0.504 |
| 15N microbial biomass uptake | 0.032 | 0.444 | 0.001 | 0.970 |
| Mineralization (μg/g/day) | 0.029 | 0.417 | 0.104 | 0.166 |
| NH4 consumption (μg/g/day) | 0.001 | 0.605 | 0.371 | 0.004 |
| PC2 | 0.000 | 0.830 | NAb | NA |
Data are from soil samples collected in the Luquillo Experimental Forest and incubated under four redox regimens. Significant correlations (P < 0.05) are indicated in boldface; those with r2 values of >0.2 are underlined. Separate multiple regression was performed for each principal component. All rates were measured over 0 to 24 h following 15N labeling.
DISCUSSION
In lower montane wet forest soils of Puerto Rico, there is clear evidence of rapid and extreme redox fluctuation based on measures of below-ground oxygen concentrations and trace gas fluxes (51). Mimicking these field conditions in laboratory incubations, we found that fluctuating redox had a strong effect on microbial community composition and diversity. The similarity of bacterial communities in incubated soils that experienced 4-day redox fluctuation suggests that fluctuation on a multiday time scale may best approximate field conditions in the upper reaches of the Luquillo Mountains. The majority of previous research on O2 fluctuation has been carried out within the conceptual paradigm of the “oxic-anoxic” interface (7). The soil systems we discuss here are variable upland systems where there may be no persistent or consistent boundary between oxic and anoxic zones. Instead, a fluid or possibly stochastic redox environment exists that is heterogeneous and dependent on highly variable diffusive influx and consumption of oxygen and other terminal electron acceptors.
During redox incubations, a large number of native ribotypes became undetectable via T-RFLP. This could be due to the disturbance of soil microsites or to gross-scale changes in soil pE caused by homogenization. We note, however, that the PAT output identified a greater number of species in the 4-day treatment soils than in the field soils. Four-day fluctuation soil communities retained high bacterial diversity, and after 3 weeks of lab incubation, most resembled the original field communities in terms of species diversity and identity. Ordination of these bacterial communities shows a distinct divergence between organisms exposed to static redox extremes. We suspect that this is caused by selective deletions from the original community, as organisms that are competitive in a fluctuating environment are no longer detectable when their habitat suddenly becomes static. Additionally, obligate organisms that were sheltered in semipermanent microsites may cease to function competitively as their environment segues to an incompatible redox state. Obligate anaerobes in particular may quickly drop out of a soil community as the environment is forced to become wholly aerobic, due to the effects of O2 toxicity (17). Indeed, in this experiment, a significant loss of microbial biomass occurred in the static anoxic and aerobic soils after 6 weeks of incubation (42). This may in part explain the divergence in ordination space between static redox treatments and the field soil and also the great number of T-RFs lost from the static soils relative to the original soils. In addition, individual T-RF analysis shows that different terminal fragments were lost depending on whether soil was under anoxic/12-h conditions versus aerobic conditions.
In these profiles, the anaerobic soil community consistently tended to cluster with the 12-h fluctuation soils. Previous research has shown that high C availability and high soil moisture (51, 61) cause extremely high biological O2 demand in these soils, such that any O2 that does diffuse into microsites is quickly utilized and exhausted. We speculate that 12 h is not long enough for O2 to completely resaturate microsites; however, 4 days may be long enough to allow periodic aerobic respiration. This suggests that the 4-day soil redox fluctuated in a fairly dramatic sense (beyond the microaerophilic boundary), whereas the 12-h soil likely remained poised at a reduced pE for much of the time.
In a number of cases, T-RFs that were dominant in the original soil became lower-ranking fragments after the treatment, whereas some T-RFs that were rare in the original soil came to dominate following redox treatment. While T-RFLP, being PCR based, is a largely qualitative assay, the patterns reported here were replicated over both separate samples and separate PCR amplifications. It appears that by physically disturbing and then radically changing redox regimens in these soils, the competitive balance between organisms has been altered, promoting different portions of the original community in the process. These results highlight the possibility that shifts in soil redox patterns (a result of predicted changes in tropical climate and precipitation patterns) could directly affect microbial community structure.
To our knowledge, ISA has not been previously used for T-RFLP analysis. However, it is a useful tool in data exploration, giving a sense of “representative diversity” that is perhaps a better measure than simple species richness in indicating the typical species assemblage diversity or “effective diversity” of a given treatment (15). Absence of an indicator value or a low index value is also ecologically relevant in that it may indicate a situation where a species in never present. Bacteria in field and fluctuating soils, with relatively high indicator values, occurred more consistently in treatment replicates than organisms from static redox soils. While perfect indicator T-RFs were rare, those that occurred were primarily associated with field soils and likely are organisms inhibited by lab incubation conditions. The relatively high fidelity of field and fluctuating soils, yet low exclusivity, indicates that a significant portion of the native community has evolved physiological mechanisms to contend with a variable redox environment. For example, in a variable-pE soil, microbes may have developed defense mechanisms for withstanding the redox stress due to both the presence (toxicity) and absence (starvation) of O2 (44), including antioxidant enzymes such as superoxide reductase (26, 40). They may also employ substrate storage mechanisms similar to those studied in aerobic-anaerobic activated sludge, where bacteria store polyphosphates and volatile fatty acids for delayed use as energy sources (8, 49).
We evaluated four noncompeting hypotheses regarding the physiological survival strategies of bacteria in this fluctuating redox soil. These strategies and the T-RF pattern evidence for each are presented below (Table 2).
(i) Bacteria in fluctuating redox soils are physiologically plastic and able to compete under all redox conditions. This strategy fits profile “A” (Table 2; Fig. 2) and was not supported by the data. Only 3 fragments fit this profile out of the total of 179 fragments, suggesting that few organisms in this soil are able to remain ecologically competitive under both aerobic and anaerobic conditions.
(ii) Bacteria in this soil are specifically adapted to fluctuating redox through tolerance to brief periods of unfavorable redox conditions. This is consistent with profiles “B” and “C,” in which organisms persist in either a primarily aerobic or primarily anaerobic regimen, but not both. Our data show that 27% of the T-RFs found in this study followed this strategy.
(iii) Bacteria in this fluctuating redox soil are strict aerobes and anaerobes that are intolerant of incompatible redox periods in their immediate environment and likely exist in spatially isolated or discrete microsites. This incorporates profiles “D” and “E.” We saw no strong evidence for this strategy in our data. Only three T-RFs were exclusive to the static treatments. However, we did find many T-RFs in the field soil that were no longer detectable in any of the treated soils (profile “G”). It is possible that some of these organisms were such strict anaerobes that they were killed by the brief physical mixing period that preceded incubation.
(iv) In this soil, bacteria either require fluctuation or are so highly adapted to redox fluctuation that they are eliminated under static conditions. This is supported by profile “F.” A large proportion (50%) of the T-RFs generated in this study followed this profile.
We conclude that hypotheses ii and iv are the most likely explanations of bacterial community patterns and redox fluctuation tolerance in this soil. As there are so few T-RFs found solely in the static treatments, there is little evidence to support the presence of either highly competitive obligate anaerobes or aerobes.
The results gleaned from the PAT analysis are a preliminary step in the phylogenetic characterization of bacteria in these soils. While it is intriguing to note the diversity of organisms identified, the technique is unlikely to positively imply phylogenetic groups for this soil at any finer resolution than the order/genus level. Erroneous assignments may occur as a result of errors in fragment size calling. However, in the interest of constructing some functional implications from the species matches generated by the PAT analysis, we compiled a list of the redox preferences of each identified species according to the National Center for Biotechnology Information taxonomy website (http://www.ncbi.nlm.nih.gov/Taxonomy) and published literature (34). By this ad hoc test, approximately half the identifiable groups in the field soil were aerobes, a third were anaerobes, and the remaining groups were split between fermenters and facultative organisms (Fig. 4). By contrast, the facultative community had tripled in size in the 4-day fluctuating soils. While such results are inherently simplified and could be made more robust with the development of a clone library or microarray analysis (14), we advocate for the PAT-type analysis as an important exploratory technique that may yield further avenues of investigation. This may be particularly true in tropical soils where efficient extraction of community DNA and PCR amplification are nontrivial due to high levels of humic materials and other contaminants.
As evidenced by the large number of terminal restriction fragments that remained unmatched, PAT-type analysis is hindered by the incomplete nature of the 16S rRNA databases (e.g., the Ribosomal Database Project [35] and the Microbial Community Analysis website [29]) which form the basis for exploratory searches. Unmatched fragments may also be a result of T-RFLP artifacts. We saw little evidence that the large proportion of unmatched fragment patterns was due to insufficient sequence data to make a match (e.g., too few restriction digests) (29). Instead, we strongly suspect that these unmatched fragments represent previously uncharacterized bacteria; hence, we are pursuing development of clone libraries for these tropical soils, which as a whole remain microbiologically uncharacterized.
In an attempt to integrate community composition and function data (through multiple regression), we saw a close correlation between the bacterial community structure of these soils and N-cycling functions, including nitrification, DNRA, and denitrification. All three of these energy-conserving processes are particularly sensitive to redox; the distinction between anaerobic/12-h soils, field/4-day soils, and aerobic soils seen in ordination space may be in part driven by the selective deletion and inclusion of organisms with highly specific N-based metabolisms. Some possible examples identified in the PAT output include Nitrosospira multiformis (nitrification), Shewanella spp. and Clostridium spp. (DNRA), and Pseudomonas spp. (denitrification). As might be expected, gross N mineralization and biomass N uptake were insensitive to the imposed redox regimens, confirming that these processes proceed with relative disregard for soil redox status.
A combination of complementary community analysis techniques (ordination, indicator species analysis, and a Web-based phylogenetic assignment tool) show that soil communities experiencing anoxic/oxic fluctuation on the scale of 4 days remain most similar to native field communities. This fluctuation-adapted community is likely dominated by organisms that retain physiological tolerance mechanisms which allow them to withstand energetically unfavorable redox periods. Two analyses linking community structure and function (PAT analysis and ordination-multiple regression) suggest that climate change-induced shifts in the variability of O2 regimens may affect specific N-cycling functions such as nitrification and denitrification in tropical soils through preferential promotion of specific bacterial functional groups. Ongoing modeling and analysis of future tropical soil climate change should be alert to its effects on a bacterial community that appears to be highly adapted to the current environment.
Acknowledgments
This research was supported by the Andrew W. Mellon Foundation, NSF grants DEB-0089783 and LTER BSR-8811902, CA AES project grants 7069-MS (to W. Silver) and 6117-H (to M.K.F.), and a graduate fellowship from the DOE Global Change Education Program.
This work would not have been possible with out the generous support of W. Silver, who provided site access and much assistance with 15N analysis. We also thank A. Thompson, D. Herman, E. Marin-Spiotta, M. Waldrop, K. DeAngelis, V. Eviner, M. Rusli, C. Hawkes, Eric Dubinsky, M. McGroddy, S. Baek, R. Jackson, R. Ostertag, and P. Templer for their help with laboratory analyses and technical assistance. E. Brodie provided particularly useful advice on molecular technique and manuscript preparation.
REFERENCES
- 1.Baas-Becking, L. G. M., I. R. Kaplan, and D. Moore. 1960. Limits of the natural environment in terms of pH and oxidation-reduction potentials. J. Geol. 68:243-285. [Google Scholar]
- 2.Balser, T. C., and M. K. Firestone. 2005. Linking microbial community composition and soil processes in a California annual grassland and a mixed-conifer forest. Biogeochemistry 73:395-415.
- 3.Balser, T. C., J. W. Kirchner, and M. K. Firestone. 2002. Analytical and methodological variability in community-level physiological profiles of soil microbial communities. Soil Sci. Soc. Am. J. 66:513-517. [Google Scholar]
- 4.Blackwood, C. B., T. Marsh, S. H. Kim, and E. A. Paul. 2003. Terminal restriction fragment length polymorphism data analysis for quantitative comparison of microbial communities. Appl. Environ. Microbiol. 69:926-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borneman, J., and E. Triplett. 1997. Molecular microbial diversity in soils from eastern Amazonia. Appl. Environ. Microbiol. 63:2647-2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brodie, E., S. Edwards, and N. Clipson. 2002. Bacterial community dynamics across a floristic gradient in a temperate upland grassland ecosystem. Microb. Ecol. 44:260-270. [DOI] [PubMed] [Google Scholar]
- 7.Brune, A., P. Frenzel, and H. Cypionka. 2000. Life at the oxic-anoxic interface: microbial activities and adaptations. FEMS Microbiol. Rev. 24:691-710. [DOI] [PubMed] [Google Scholar]
- 8.Carucci, A. K. Lindrea, M. Majone, and R. Ramadori. 1999. Different mechanisms for the anaerobic storage of organic substrates and their effect on enhanced biological phosphate removal (EBPR). Water Sci. Technol. 39:21-28. [Google Scholar]
- 9.Causton, D. R. 1988. An introduction to vegetation analysis. Unwin Hyman Ltd., London, United Kingdom.
- 10.Cavigelli, M. A., and G. P. Robertson. 2001. The functional significance of denitrifier community composition in a terrestrial ecosystem. Soil Biol. Biochem. 33:297-310. [Google Scholar]
- 11.Chapelle, F. H., P. M. McMahon, N. M. Dubrovsky, R. Fujii, E. T. Oaksford, and D. A. Vroblesky. 1995. Deducing the distribution of terminal electron-accepting processes in hydrogeologically diverse groundwater systems. Water Resource Res. 31:359-371. [Google Scholar]
- 12.Cox, S. B., M. R. Willig, and F. N. Scatena. 2002. Variation in nutrient characteristics of surface soils from the Luquillo Experimental Forest of Puerto Rico: a multivariate perspective. Plant Soil 247:189-198. [Google Scholar]
- 13.Davidson, E. A., S. C. Hart, C. A. Shanks, and M. K. Firestone. 1991. Measuring gross nitrogen mineralization immobilization and nitrification by nitrogen-15 isotopic pool dilution in intact soil cores. J. Soil Sci. 42:335-350. [Google Scholar]
- 14.DeSantis, T. Z., I. Dubosarskiy, S. R. Murray, and G. L. Andersen. 2003. Comprehensive aligned sequence construction for automated design of effective probes (CASCADE-P) using 16S rDNA. Bioinformatics 19:1461-1468. [DOI] [PubMed] [Google Scholar]
- 15.Dufrêne, M., and P. Legendre. 1997. Species assemblages and indicator species: the need for a flexible asymmetrical approach. Ecol. Monogr. 67:345-366. [Google Scholar]
- 16.Eriksson, P. G., J. M. Svensson, and G. M. Carrer. 2003. Temporal changes and spatial variation of soil oxygen consumption, nitrification and denitrification rates in a tidal salt marsh of the Lagoon of Venice, Italy. Estuar. Coast. Shelf Sci. 58:861-871. [Google Scholar]
- 17.Fenchel, T., and B. J. Finlay. 1995. Ecology and evolution in anoxic worlds. Oxford University Press, Oxford, United Kingdom.
- 18.Fierer, N., and J. P. Schimel. 2002. Effects of drying-rewetting frequency on soil carbon and nitrogen transformations. Soil Biol. Biochem. 34:777-787. [Google Scholar]
- 19.Frangi, J. L. 1983. Las tierras pantanosas de la montana puertorriquena, 233-247. In A. E. Lugo (ed.), Los Bosques de Puerto Rico. USDA/Forest Service IITF and PR DNR, San Juan, Puerto Rico.
- 20.Garrity, G. M. (ed.). 2001-2003. Bergey's manual of systematic bacteriology, 2nd ed., vol I to V. Springer-Verlag, New York, N.Y.
- 21.Glimm, E., H. Heuer, B. Engelen, K. Smalla, and H. Backhaus. 1997. Statistical comparisons of community catabolic profiles. J. Microbiol. Methods 30:71-80. [Google Scholar]
- 22.Hart, S. C., J. M. Stark, E. A. Davison, and M. K. Firestone. 1994. Methods of soil analysis, part 2. Microbiological and biochemical properties, p. 985-1017. Soil Science Society of America, Madison, Wis.
- 23.Hill, Tom, C. J., K. A. Walsh, J. A. Harris, and B. F. Moffett. 2003. Using ecological diversity measures with bacterial communities. FEMS Microbiol. Ecol. 43:1-11. [DOI] [PubMed] [Google Scholar]
- 24.Hobbie, S. 1996. Temperature and plant species control over litter decomposition in Alaskan tundra. Ecol. Monogr. 66:503-522. [Google Scholar]
- 25.Imlay, J. A. 2002. How oxygen damages microbes: oxygen tolerance and obligate anaerobiosis. Adv. Microb. Physiol. 41:111-153. [DOI] [PubMed] [Google Scholar]
- 26.Jean, D., V. Briolat, and G. Reysset. 2004. Oxidative stress response in Clostridium perfringens. Microbiology 150:1649-1659. [DOI] [PubMed] [Google Scholar]
- 27.Keith-Roach, M. J., N. D. Bryan, R. D. Bardgett, and F. R. Livens. 2002. Seasonal changes in the microbial community of a salt marsh, measured by phospholipid fatty acid analysis. Biogeochemistry 60:77-96. [Google Scholar]
- 28.Keller, M., W. A. Kaplan, and S. C. Wofsy. 1986. Emissions of N2O, CH4, and CO2 from tropical forest soils. J. Geophys. Res. 91:11791-11802. [Google Scholar]
- 29.Kent, A. D., D. J. Smith, B. J. Benson, and E. W. Triplett. 2003. Web-based phylogenetic assignment tool for analysis of terminal restriction fragment length polymorphism profiles of microbial communities. Appl. Environ. Microbiol. 69:6768-6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kitts, C. 2001. Terminal restriction fragment patterns: a tool for comparing microbial communities and assessing community dynamics. Curr. Issues Intest. Microbiol. 2:17-25. [PubMed] [Google Scholar]
- 31.Liu, W. T., T. L. Marsh, H. Cheng, and L. J. Forney. 1997. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl. Environ. Microbiol. 63:4516-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lovley, D. R., and S. Goodwin. 1988. Hydrogen concentrations as an indicator of the predominant terminal electron-accepting reactions in aquatic sediments. Geochim. Cosmochim. Acta 52:2993-3004. [Google Scholar]
- 33.Ludemann, H., I. Arth, and W. Liesack. 2000. Spatial changes in the bacterial community structure along a vertical oxygen gradient in flooded paddy soil cores. Appl. Environ. Microbiol. 66:754-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Madigan, M. T., J. M. Martinko, and J. Parker. 2003. Brock biology of microorganisms. Prentice Hall, Upper Saddle River, N.J.
- 35.Marsh, T. L., P. Saxman, J. Cole, and J. Tiedje. 2000. Terminal restriction length polymorphism analysis program, a web-based research tool for microbial community analysis. Appl. Environ. Microbiol. 66:3616-3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Masumoto, M., M. Sakamoto, H. Hayashi, and Y. Benno. 2005. Novel phylogenetic assignment database for terminal-restriction fragment length polymorphism analysis of human colon microbiota. J. Microbiol. Methods 61:305-319. [DOI] [PubMed] [Google Scholar]
- 37.McCune, B., and J. B. Grace. 2002. Analysis of ecological communities. MJM Software Design, Gleneden Beach, Oreg.
- 38.McGroddy, M., and W. L. Silver. 2000. Variations in belowground carbon storage and soil CO2 flux rates along a wet tropical climate gradient. Biotropica 32:614-624. [Google Scholar]
- 39.Murphy, S. F., S. L. Brantley, A. E. Blum, A. F. White, and H. Dong. 1998. Chemical weathering in a tropical watershed, Luquillo Mountains, Puerto Rico. II. Rate and mechanism of biotite weathering. Geochim. Cosmochim. Acta 62:227-243. [Google Scholar]
- 40.Niviere, V., and M. Fontecave. 2004. Discovery of superoxide reductase: an historical perspective. J. Biol. Inorg. Chem. 9:119-123. [DOI] [PubMed] [Google Scholar]
- 41.Paul, E. A., and F. E. Clark. 1996. Soil microbiology and biochemistry. Academic Press, San Diego, Calif.
- 42.Pett-Ridge, J. 2005. Rapidly fluctuating redox regimes frame the ecology of microbial communities and their biogeochemical function in a humid tropical soil. Ph.D. thesis. Univeristy of California, Berkeley.
- 43.Picek, T., M. Simek, and H. Santruckova. 2000. Microbial responses to fluctuation of soil aeration status and redox conditions. Biol. Fertil. Soils 31:315-322. [Google Scholar]
- 44.Potter, L., P. Millington, L. Griffiths, and J. Cole. 2000. Survival of bacterial during oxygen limitation. Int. J. Food Microbiol. 55:11-18. [DOI] [PubMed] [Google Scholar]
- 45.Ricke, P. S. Kolb, and G. Braker. 2005. Application of a newly developed ARB software-intregated tool for in silico terminal restriction fragment length polymorphism analysis reveals the dominance of a novel pmoA cluster in a forest soil. Appl. Environ. Microbiol. 71:1671-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rosche, C., and H. Bothe. 2005. Improved assessment of denitrifying, N2-fixing, and total-community bacteria by terminal restriction fragment length polymorphism analysis using multiple restriction enzymes. Appl. Environ. Microbiol. 71:2026-2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Satpathy, S. N., A. K. Rath, B. Ramakrishnan, V. R. Rao, T. K. Adhya, and N. Sethunathan. 1997. Diurnal variation in methane efflux at different growth stages of tropical rice. Plant Soil 195:267-271. [Google Scholar]
- 48.Schloss, P. D., B. R. Larget, and J. Handelsman. 2004. Integration of microbial ecology and statistics: a test to compare gene libraries. Appl. Environ. Microbiol. 70:5485-5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schuler, A. J., D. Jenkins, and P. Ronen. 2001. Microbial storage products, biomass density, and settling properties of enhanced biological phosphorous removal activated sludge. Water Sci. Technol. 43:173-180. [PubMed] [Google Scholar]
- 50.Shannon, C. E., and W. Weaver. 1949. The Mathematical theory of communication. University of Illinois Press, Urbana.
- 51.Silver, W. L., A. E. Lugo, and M. Keller. 1999. Soil oxygen availability and biogeochemistry along rainfall and topographic gradients in upland wet tropical forest soils. Biogeochemistry 44:301-328. [Google Scholar]
- 52.Silver, W. L., D. J. Herman, and M. K. Firestone. 2001. Dissimilatory nitrate reduction to ammonium in upland tropical forest soils. Ecology 82:2410-2416. [Google Scholar]
- 53.Singleton, D. R., M. A. Fulong, S. L. Rathburn, and W. B. Whitman. 2001. Quantitative comparison of 16S rRNA gene sequence libraries from environmental samples. Appl. Environ. Microbiol. 67:4374-4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sposito, G. 1989. The chemistry of soils. Oxford University Press, New York, N.Y.
- 55.Tabachnick, B. G., and L. S. Fidell. 1996. Using multivariate statistics, 3rd ed. Harper Collins College Publishing, New York, N.Y.
- 56.Tanji, K. K., S. Gao, S. C. Scardaci, and A. Chow. 1999. Terminal electron-accepting processes (TEAPs) in paddy rice soils with incorporated straw: phase II, p. 153-165. In A. Zabel and G. Sposito (ed.) Soil quality in the CA environment: annual report of research projects 1998-1999. Kearney Foundation of Soil Science, Berkeley, Calif.
- 57.Teichert, A., J. Bottcher, and W. H. M. Duijnisveld. 2000. Redox measurement as a qualitative indicator of spatial and temporal variability of redox state in a sandy forest soil, p. 95-110. In J. Schuring, H. D. Schulz, W. R. Fischer, J. Bottcher, and W. H. M. Duijnisveld (ed.), Redox: fundamentals, processes and applications. Spinger-Verlag, New York, N.Y.
- 58.Tiedje, J. M., A. J. Sexstone, T. B. Parkin, N. P. Prevsbech, and D. R. Shelton. 1984. Anaerobic processes in soil. Plant Soil 76:197-212. [Google Scholar]
- 59.Updegraff, K., S. D. Bridgham, J. Pastor, and P. Weishampel. 1998. Hysteresis in the temperature response of carbon dioxide and methane production in peat soils. Biogeochemistry 43:253-272. [Google Scholar]
- 60.Waldrop, M. P., T. C. Balser, and M. K. Firestone. 2000. Linking microbial community composition to function in a tropical soil. Soil Biol. Biochem. 32:1837-1846. [Google Scholar]
- 61.Wang, H., C. A. S. Hall, J. D. Cornell, and M. H. P. Hall. 2002. Spatial dependence and the relationship of soil organic carbon and soil moisture in the Luquillo Experimental Forest, Puerto Rico. Landscape Ecol. 17:671-684. [Google Scholar]
- 62.Weaver, P. L., and P. G. Murphy. 1990. Forest structure and productivity in Puerto Rico's Luquillo mountains. Biotropica 22:69-82. [Google Scholar]
- 63.Weaver, P. L. 1994. Bano de Oro Natural Area, Luquillo Mountains, Puerto Rico. General technical report, SO-111. U.S. Department of Agriculture, Forest Service, Southern Forest Experiment Station, New Orleans, La.
- 64.Wedin, D., and D. Tilman. 1990. Species effects on N cycling: a test with perennial grasses. Oecologia 84:433-441. [DOI] [PubMed] [Google Scholar]
- 65.White, A. F., A. E. Blum, M. S. Schulz, D. V. Vivit, D. A. Stonestrom, M. Larsen, S. F. Murphy, and D. Eberl. 1998. Chemical weathering in a tropical watershed, Luquillo Mountains, Puerto Rico. I. Long-term versus short-term weathering fluxes. Geochim. Cosmochim. Acta 62:209-226. [Google Scholar]
- 66.Yu, C. P., R. Ahuja, G. Sayler, and K. H. Chu. 2005. Quantitative molecular assay for fingerprinting microbial communities of wastewater and estrogen-degrading consortia. Appl. Environ. Microbiol. 71:1433-1444. [DOI] [PMC free article] [PubMed] [Google Scholar]