Abstract
The quality and market price of truffles vary with the species and, traditionally, the place of origin. The premium species Tuber magnatum produces white truffles and has a patchy distribution restricted to Italy and some Balkan areas. We used polymorphic microsatellites to evaluate 316 specimens grouped into 26 populations sampled across the species' geographic range to determine if natural populations of T. magnatum are genetically differentiated. We found that the southernmost and the northwesternmost populations were significantly differentiated from the rest of the populations. The simple sequence repeat data also could be used to make inferences about the postglacial T. magnatum expansion pattern. This study is the first to identify a genetic and phylogeographic structure in T. magnatum. The presence of a genetic structure can be of practical interest in tracing truffle populations according to their geographic origin for marketing strategies. Evidence for extensive outcrossing in field populations of T. magnatum also is provided for the first time.
Tuber spp. are hypogeous ascomycetes that grow in ectomycorrhizal symbioses with some shrub and tree species and produce ascomata, known as truffles. Some truffles are edible and appreciated worldwide. Their quality and market price depend on the species and, traditionally, the place of origin. Natural truffle production has declined dramatically over the past century (12). This decline and the flourishing truffle market have encouraged large-scale programs for growing these fungi through the planting of nursery-produced mycorrhizal trees. These programs have been widely developed for some species, e.g., Tuber melanosporum Vittad. and Tuber uncinatum Chatin, and most of the commercial demand for these truffles is satisfied by these artificial plantations rather than natural field collections (12). How the deliberate introduction of foreign strains into native populations has affected truffle production and biodiversity remains unknown.
The complexity of the truffle life cycle, the difficulties of growing these fungi under controlled conditions, and the lack of reliable phenotypic markers to differentiate morphologically similar species have been major obstacles to understanding the distribution, propagation, and fructification of these hypogeous fungi. Molecular markers have been developed to type most of the economically important truffle species (2, 10, 15, 19, 23, 24, 25, 33, 34), but studies of the environmental and molecular determinants for different portions of the life cycle generally are lacking. The spatial distribution and ecological requirements of these symbiotic fungi vary by species. Some species are widely distributed and have pronounced morphological and molecular variability (9, 11, 22, 25, 27, 32), while others have a more restricted distribution and little intraspecific polymorphism in either morphological or genetic traits (3, 4, 8, 9, 26). Tuber magnatum Pico, which produces white ascomata, is harvested only in Italy and some countries on the Balkan Peninsula.
In the related species T. melanosporum, there is little or no genetic differentiation between the sampled populations, and the species is regarded as one that self-fertilizes for sexual reproduction and that has an ascocarp consisting primarily of diploid (dikaryotic) hyphae (3, 4, 21). The few studies of genetic structure and mode of reproduction of T. magnatum found very limited intraspecific allozyme variation and a complete absence of heterozygous individuals (8). Whether T. magnatum is normally dikaryotic and reproduces homothallically or by self-fertilization or if it is an outbreeding ascomycete remains to be determined.
Our objectives in this study were to use microsatellite markers to determine (i) if T. magnatum populations are genetically homogeneous, (ii) if the fungus is dikaryotic, and (iii) the degree of outbreeding in field populations. Understanding the life cycle and genetic differences between truffle populations is important for commercial truffle growth management and marketing strategies.
MATERIALS AND METHODS
Sample source.
Ascomata of T. magnatum were sampled from natural truffle grounds in the best known and most productive areas of Italy and from the Istrian peninsula (Croatia and Slovenia) with the help of local pickers. When multiple samples were provided by the same individual, but evidence for collection from physically different locations was lacking, the samples were screened for clones with simple sequence repeats (SSR), and strains with identical multilocus genotypes were reduced to a single representative for the statistical analyses. Thus, only 316 out of 351 ascocarps sampled were used in the statistical analysis. These truffles were grouped into 26 populations and then these populations were clustered into 13 groups based on geographic criteria (Table 1). Ascocarps also were harvested from single-truffle grounds (Ascoli, Certaldo, and Pietralunga) and analyzed to determine the amount of variation within a local population (see Table SA1 in the supplemental material).
TABLE 1.
Name and location of the studied populations and estimated diversity parameters
| Geographic group | Population no. and name | Region | No. of samples | Mean no. of allelesa | Allelic richnessa |
|---|---|---|---|---|---|
| 1 | 1. Istria | Croatia-Slovenia | 17 | 3.3 (0.61) | 2.7 (0.48) |
| 2 | 2. Pavia | Lombardia | 11 | 2.6 (0.43) | 2.4 (0.44) |
| 3. Langhe | Piemonte | 15 | 3.3 (0.81) | 2.9 (0.62) | |
| 4. Asti | Piemonte | 9 | 2.7 (0.42) | 2.6 (0.37) | |
| 5. Alba | Piemonte | 7 | 3.0 (0.49) | 2.9 (0.47) | |
| 3 | 6. Pianoro | Emilia Romagna | 9 | 2.7 (0.57) | 2.6 (0.52) |
| 7. Val di Zena | Emilia Romagna | 10 | 3.1 (0.59) | 2.8 (0.48) | |
| 4 | 8. Firenze | Toscana | 32 | 4.4 (0.97) | 3.0 (0.46) |
| 5 | 9. Pietralunga | Umbria | 7 | 2.6 (0.43) | 2.5 (0.43) |
| 10. Montemaggiore | Umbria | 12 | 2.4 (0.57) | 2.2 (0.53) | |
| 11. Città di Castello | Umbria | 12 | 3.1 (0.74) | 2.7 (0.60) | |
| 12. Gubbio | Umbria | 25 | 4.3 (0.84) | 2.9 (0.46) | |
| 13. Appennino Umbro | Umbria | 9 | 2.6 (0.53) | 2.4 (0.50) | |
| 6 | 14. Fabro | Umbria | 11 | 2.9 (0.46) | 2.6 (0.39) |
| 15. Montecastrilli | Umbria | 9 | 3.0 (0.62) | 2.8 (0.57) | |
| 7 | 16. Ascoli | Marche | 10 | 3.0 (0.58) | 2.7 (0.55) |
| 17. Chieti | Abruzzo | 5 | 2.4 (0.43) | 2.4 (0.46) | |
| 8 | 18. Valle Roveto | Abruzzo | 7 | 2.4 (0.43) | 2.4 (0.44) |
| 9 | 19. Quadri, Rosello | Abruzzo | 12 | 3.1 (0.59) | 2.8 (0.42) |
| 20. R. del Sangro, M. del. Sannio | Abruzzo-Molise | 10 | 3.1 (0.46) | 2.8 (0.36) | |
| 21. Ateleta, S.P. Avellana | Abruzzo-Molise | 18 | 3.9 (0.59) | 3.0 (0.50) | |
| 22. Castel del Giudice | Molise | 16 | 3.1 (0.46) | 2.7 (0.37) | |
| 10 | 23. Agnone | Molise | 10 | 3.7 (0.75) | 3.3 (0.65) |
| 11 | 24. Isernia, Campobasso | Molise | 16 | 3.6 (0.57) | 3.1 (0.42) |
| 12 | 25. Benevento, Avellino | Campania | 5 | 2.4 (0.37) | 2.4 (0.40) |
| 13 | 26. Potenza | Basilicata | 12 | 1.9 (0.34) | 1.7 (0.26) |
Standard errors of the means are shown in parentheses.
DNA isolation and SSR characterization.
Sample preparation and DNA isolation from all ascocarps (24) and amplification of the microsatellite loci (35) were carried out as previously described. The MA2 and MA5 loci are genetically unlinked, but each contain an (AC)n-(TC)n compound microsatellite motif. The total length of these compound loci has been previously determined following PCR with a locus-specific primer pair designed on the flanking regions of MA2 and MA5 (35). To reduce the bias due to size homoplasy, selective amplification of only one of the two motifs (MA2-1 and MA5-1) within each of the two compound loci was made with a new primer set, consisting of a nested primer (5′-AGAGAGAGAGTGTGTGTG-3′) designed at the junction between the two repeated motifs, in combination with the locus-specific flanking primers 5′-CGCAGTGCAAAAAGGAAGCATCA-3′ and 5′-CGTCTATTCCCCGCGAGATAACAAC-3′ for MA2 and MA5, respectively. PCR conditions were as previously described (35). The resulting DNA fragments were resolved with an ABI 310 genetic analyzer (Applied Biosystems, Foster City, CA) to estimate allele size for information for statistical analyses. The 15 most common alleles of MA2-1 and MA5-1 and a representative of each of the alleles of the other microsatellite loci, MA4, MA7, MA12, MA14, and MA19 (35), were sequenced. Sequencing reactions were performed directly on purified PCR products (Jet Quick spin kit; Genomed, Bad Oeynhausen, Germany) with a Big Dye terminator sequencing kit (Applied Biosystems). Estimated allele sizes were used to determine the number of repeated units.
Data analysis.
The mean number of alleles and the allelic richness, over all loci, were calculated for each population. Allelic richness, computed for each population, was weighted to the smallest sample size (n = 5) using a rarefaction method (29).
Two-locus linkage disequilibrium analysis was performed with GENEPOP, version 3.2a (30), within each population and each geographic group and across all populations. The index of association (IA) used to test for multilocus linkage disequilibrium was calculated as described by Maynard Smith et al. (18) with the program MULTILOCUS, version 1.2 (1). The same software was used to determine if IA differed significantly from zero, by performing 1,000 randomizations of the data set. The randomization procedure was first performed by shuffling alleles among all individuals. To determine if linkage disequilibrium results from the reproductive isolation of geographically distant populations, randomizations by shuffling alleles among individuals within populations and within geographical groups also were performed (17).
To test the effects of sample size reduction on linkage disequilibrium (LD) values, two locus and multilocus analyses also were performed by considering a data set of 275 individuals pooled as a single population and obtained by excluding the southernmost and northwesternmost populations (3, 4, 25, and 26).
The extent of population subdivision was evaluated by calculating Wright's FST (39) and the related RST (37) indexes, with the latter taking explicit account of the mutation process that can occur at microsatellite loci. These two indexes were estimated across populations and between pairs of populations, according to Weir and Cockerham (38) and Michalakis and Excoffier (20), by using the program SPAGEDI, version 1.1 (14).
A genetic distance matrix of pairwise FST values also was used to perform a hierarchical analysis of molecular variance (AMOVA) (7) with the ARLEQUIN program (version 2.0; Genetics and Biometry Laboratory, University of Geneva, Switzerland [http://lgb.unige.ch/arlequin/]).
The possible presence of geographic structure of genetic variation in nuclear microsatellites in truffles was evaluated with four tests. First, we tested for the presence of phylogeographic structure by comparing RST estimates to RST values computed after 10,000 random permutations of allele types among alleles (13) were calculated using the SPAGEDI program. If RST is >RST (permuted), then there is phylogeographic structure, i.e., on average, phylogenetically similar alleles are found in the same population more often than are randomly chosen alleles.
Second, we tested for a pattern of isolation by distance (31). A Mantel test with 1,000 random permutations was performed with the matrix of pairwise genetic differentiation between populations, using RST/(1 − RST), and a matrix of the ln(geographic distance) with the software GENALEX (version 5.1; Australian National University, Canberra, Australia [http://anu.edu.au/BoZo/GenAlEx/]).
Third, a spatial autocorrelation analysis was performed with SPAGEDI. This analysis used RST and evaluated 18 spatial distance classes with similar sample sizes. The 95% confidence intervals were estimated for each distance class by 10,000 random permutations.
Finally, a simulated annealing procedure implemented in the spatial analysis of molecular variance (SAMOVA) algorithm (6) was used to define groups of populations that are geographically homogeneous and maximally differentiated from each other. The program repeatedly seeks the composition of a user-defined number, K, of groups of geographically adjacent populations that maximizes FCT, the proportion of total genetic variance due to differences among groups of populations. The program was run for 10,000 iterations for K ∈ {2, …, 12} from each of 250 random initial conditions. The simulated annealing process was repeated 250 times to ensure that the final configuration of the K groups is not affected by a given initial configuration (6). For each K, the configuration with the largest FCT values after the 250 independent simulated annealing processes were completed was retained as the best grouping of populations.
RESULTS
Linkage disequilibrium analysis.
When all 316 truffles were included, the two-locus analysis had significant linkage disequilibrium between many pairs of loci (15 out of the 21 two-locus combinations).
There also was a significant departure from linkage equilibrium (IA = 0.109, P < 0.001) in the multilocus analysis. Linkage disequilibrium can result from reduced gene flow due to geographic separation. To assess the potential geographic bias in our combined analysis, these tests also were performed with samples in smaller geographic groups. When the two-locus LD was calculated for each of the 13 geographic groups, only one of the two-locus combinations had a significant LD, and no significant two-locus LD was detected when the 26 local populations were considered. Similarly, IA was not significant when the randomizations were performed by shuffling alleles among samples within the 13 geographical groups and within the 26 local populations (P values of 0.065 and 0.119, respectively).
To evaluate the effect of sample size on LD values, the two-locus LD and IA tests also were calculated for a data set of 275 individuals obtained by excluding from the whole data set the populations 3, 4, 25, and 26 that SAMOVA proved to be genetically differentiated (see below) and thus were expected to contribute largely to LD. In this situation, only 5 out of 21 two-locus combinations resulted in significant LD (P = 0.05), and the IA was not significant (IA = 0.036, P = 0.062), suggesting that the decrease in LD values when populations are analyzed separately is not due to the reduced sample size.
Patterns within and among population variability.
The seven SSR loci had 2 to 18 alleles per locus. Within each of the 351 truffles analyzed, only a single allele per locus was detected. Each multilocus genotype was then treated as a haplotype on the assumption that T. magnatum is a haploid fungus (see Discussion). The 26 populations had a mean number of alleles of 3.03 ± 0.12, ranging from 1.86 ± 0.34 to 4.43 ± 0.97, while the average allelic richness was 2.67 ± 0.06, ranging from 1.74 ± 0.26 to 3.30 ± 0.65 (Table 1).
Based on AMOVA, most of the genetic variability is located at the within-population level, which accounts for about 84% of the total molecular variance, while 6% and 10% of the genetic variation were attributable to differences among populations within geographical groups and among the 13 geographic groups, respectively (Table 2). Overall, 263 haplotypes were detected within the 316 samples. Some haplotypes were found at multiple sites in the same region, and some were found in multiple populations or groups (see supplemental Table SA1).
TABLE 2.
AMOVA based on 316 truffles using seven SSR loci
| Source of variation | df | Sum of squares | Variance component | % of total | P |
|---|---|---|---|---|---|
| Among regional groups | 11 | 95 | 0.216 | 10 | <0.01 |
| Among populations within groups | 14 | 42 | 0.112 | 5.4 | <0.01 |
| Within populations | 290 | 509 | 1.75 | 84 | <0.01 |
| Total | 315 | 646 | 2.08 |
Each population contains a relatively large number of haplotypes. Within an individual truffle ground, the genetic variation decreased slightly, because some truffles probably originated from the same vegetative mycelium. For example, of the nine ascocarps harvested in Certaldo, only three haplotypes were found. However, from the 16 and 10 ascocarps from Ascoli and Pietralunga, 10 and 7 haplotypes were identified, respectively.
Most of the alleles were not unique to a population, but some alleles could be used to characterize truffles according to their geographic origin because of their differential frequencies in some groups of populations. For example, the frequency of allele 17 at the MA7 locus was high in southern populations 23, 24, 25, and 26 (0.2, 0.4, 0.4, and 1.0, respectively), while alleles 14, 24, 34, and 35 of MA4 were found only in individuals from populations 3, 4, and 5 from Piemonte, the northwesternmost area of sampling.
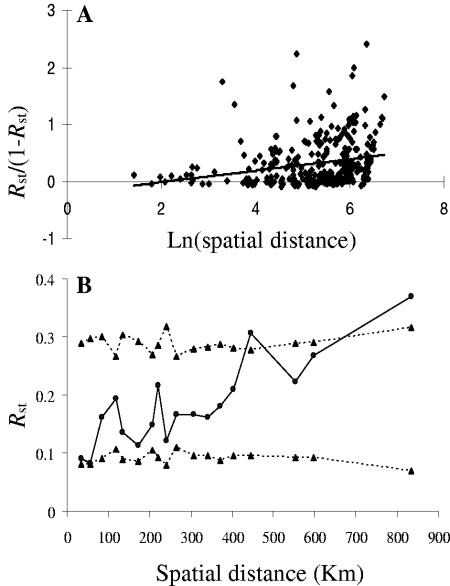
A marked and highly significant difference between populations was suggested by the FST and RST values, which were 0.15 and 0.22, respectively. A phylogeographic signal also was detected with permutation procedures in the total sample [RST of 0.22 versus RST(permuted) of 0.12, P = 0.0005]. The test of isolation by distance showed that among-population differentiation increased significantly with ln(geographical distance) (Mantel test; P < 0.006), although the regression explains only 5.4% of the total variance (R2 = 0.054) (Fig. 1A).
FIG. 1.
(A) Correlation between genetic and geographical pairwise distances. (B) Average genetic distance (RST) for 18 spatial distance classes. Broken lines indicate the boundaries of the 95% confidence intervals estimated by permutation.
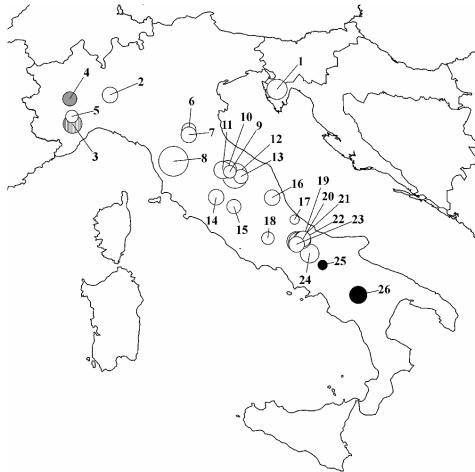
The spatial autocorrelation analysis allowed us to examine the extent of association in different geographic distance classes. This analysis confirmed the geographic pattern of genetic variation, with populations separated by >450 km being significantly further apart genetically than would be expected by chance (Fig. 1B). The results of SAMOVA confirmed the existence of geographic structure in the T. magnatum populations. In particular, when RST was used, southern populations 25 and 26 and northwestern populations 3 and 4 were clearly distinguishable from the others (Fig. 2).
FIG. 2.
Sampling localities and geographic distribution of genetic variability. Numbers refer to populations listed in Table 1. Diameters of the circles are proportional to the sample sizes. Differently shaded circles show genetically different population groups according to SAMOVA based on RST index and a K of 4.
DISCUSSION
Although a decline in production has triggered research into truffle cultivation, many basic aspects of truffle biology, such as the reproductive mode and the amount and distribution of population genetic variation, are not well understood. The limited saprophytic capabilities of truffle mycelia and the inability to obtain monosporic cultures or to cross mycelial strains under controlled conditions have slowed the direct assessment of the reproductive mode in these organisms.
We used codominant markers (polymorphic microsatellite loci) and strains sampled across the entire known range of the species to infer the genetic structure, population dynamics, and reproductive mode of T. magnatum, the most prestigious of the edible species in the genus Tuber. We found only a single allele per locus in each of the 351 truffles analyzed. These results are consistent with those of Bertault et al. (3) for T. melanosporum. They previously interpreted the absence of heterozygotes to mean that these fungi are functionally diploid with a strictly self-fertilizing reproductive system. We interpret these data to mean that the ascocarps of T. magnatum are haploid maternal tissue, as found for many ascomycetes. Two-locus and multilocus linkage disequilibrium analyses indicate that within both the T. magnatum geographical groups and the populations, extensive genetic exchange occurs. The relatively high number of multilocus genotypes within populations suggests that these fungi are not highly inbred. Within the same truffle grounds, differences between haplotypes usually occurred at multiple loci rather than at a single locus (see supplemental Table SA1), a pattern consistent with recombination in addition to mutation as a source for the variation observed in the haplotypes.
The truffle ascocarps are presumed to be primarily maternal tissue and thus would be haploid. Paternal DNA might not be as easily recoverable since it would be present only in the ascospores, and these spores are not usually broken during the DNA extraction process that we use. Thus, haploid maternal tissue will be the dominant component of the truffle and presumably represents the genotypes we analyzed in this study.
Significant linkage disequilibrium was found only when the entire truffle data set was evaluated as a whole and could be explained if there was significant geographic population subdivision. The FST and RST statistics and the correlation between genetic and geographic distance are consistent with genetic differentiation of populations, while the results of the RST permutation analysis are consistent with phylogeographic structure. If individual populations are removed from the analysis, then removal of the southernmost and two northwesternmost populations allows the rest of the populations to be grouped together.
The geographic distribution of T. magnatum tracks the postglacial expansion of the species with which this fungus can establish a mutualistic symbiosis. Refugia for some T. magnatum host species (e.g., Quercus, Corylus, Tilia, and Carpinus) were located in central and southern Italy and spread northward as the ice receded (5, 28). A refugium for T. magnatum in central Italy, from which the northernmost and southernmost populations originated, could explain the results observed.
The genetic variation and geographic distribution of the SSR markers could enable the tracing and identification of T. magnatum populations. The geographic origin of truffles often is important to associations of truffle harvesters and local governments which are promoting economic and social development of rural and marginal areas. The genetic structure of populations of T. magnatum also has important ecological implications. Artificial fungal plantations often are established in naturally productive areas to counterbalance a sharp decline in wild truffle harvests. Potential problems related to microbial competition are largely unexplored with respect to truffle cultivation, and the consequences of the deliberate introduction of nonnative strains into an indigenous population have been serious for other edible mushrooms (16, 36). Hence, the profitable commercial practice of artificial fungal plantations may cause a loss of fungal biodiversity, and local strains with unique commercial characteristics might become extinct. Thus, when white truffle cultivation is planned in naturally productive areas, ecologically adapted truffle strains that are similar to those already present in the region should be used. Similar cautions also apply to T. melanosporum (21). The SSR markers that we used could be used to type strains of T. magnatum that are inoculated on commercially produced seedlings as well as for assessing the impact the introduced strains might have on native ectomycorrhizal populations.
The presence of genetic structure in populations of both of the most prestigious truffle species argues against the hypothesis that phenotypic and organoleptic differences among truffles of different origin are due solely to environmental factors (3). If the SSR loci examined are linked to the genes controlling these commercially important traits, then the SSR markers could be used to select strains with desirable characteristics for future commercial use. As our data suggest that truffles are not strictly self-fertilizing, the possibility of improving these mushrooms through conventional breeding protocols needs to be evaluated with more care and in greater detail. This study provides the first evidence suggesting that truffles are not strictly self-fertilizing organisms and should result in a significant reevaluation of the life cycle and biology of T. magnatum and other Tuber spp.
Supplementary Material
Acknowledgments
We thank Angelo Rambelli for critically reading the manuscript.
This research was supported in part by a grant from the Regione Umbria/Comunità Montane Umbre/CNR-IGV Sezione di Perugia, contribution no. 58 from the Institute of Plant Genetics, Division of Perugia.
Andrea Rubini and Francesco Paolocci contributed equally to this work.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Agapow, P. M., and A. Burt. 2001. Indices of multilocus linkage disequilibrium. Mol. Ecol. Notes 1:101-102. [Google Scholar]
- 2.Amicucci, A., A. Zambonelli, G. Giomaro, L. Potenza, and V. Stocchi. 1998. Identification of ectomycorrhizal fungi of the genus Tuber by species-specific ITS primers. Mol. Ecol. 7:273-277. [Google Scholar]
- 3.Bertault, G., M. Raymond, A. Berthomieu, G. Callot, and D. Fernandez. 1998. Trifling variation in truffles. Nature 394:734. [Google Scholar]
- 4.Bertault, G., F. Rousset, D. Fernandez, A. Berthomieu, M. E. Hochberg, G. Callot, and M. Raymond. 2001. Population genetics and dynamics of the black truffle in a man-made truffle field. Heredity 86:451-458. [DOI] [PubMed] [Google Scholar]
- 5.Brewer, S. 2002. Recolonisation postglaciaire de quelques taxons tempérés en Europe: une approche spatiale et temporelle. Ph.D. thesis. Institut Méditerranéen d'Ecologie et de Paléoécologie, Marseille, France.
- 6.Dupanloup, I., S. Schneider, and L. Excoffier. 2002. A simulated annealing approach to define genetic structure of populations. Mol. Ecol. 11:2571-2581. [DOI] [PubMed] [Google Scholar]
- 7.Excoffier, L., P. Smouse, and J. Quattro. 1992. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frizzi, G., G. Lalli, M. Miranda, and G. Pacioni. 2001. Intraspecific isozyme variability in Italian populations of the white truffle Tuber magnatum. Mycol. Res. 105:365-369. [Google Scholar]
- 9.Gandeboeuf, D., C. Dupré, P. Roeckel-Drévet, P. Nicolas, and G. Chevalier. 1997. Grouping and identification of Tuber species using RAPD markers. Can. J. Bot. 75:36-45. [Google Scholar]
- 10.Gandeboeuf, D., C. Dupré, P. Roeckel-Drévet, P. Nicolas, and G. Chevalier. 1997. Typing Tuber ectomycorrhizae by polymerase chain amplification of the internal transcribed spacer of rDNA and the sequence characterized amplified region markers. Can. J. Microbiol. 43:723-728. [DOI] [PubMed] [Google Scholar]
- 11.Guillemaud, T., M. Raymond, G. Callot, J. C. Cleyet-Marel, and D. Fernandez. 1996. Variability of nuclear and mitochondrial ribosomal DNA of a truffle species (Tuber aestivum). Mycol. Res. 100:547-550. [Google Scholar]
- 12.Hall, I. R., W. Yun, and A. Amicucci. 2003. Cultivation of edible ectomycorrhizal mushrooms. Trends Biotechnol. 21:433-438. [DOI] [PubMed] [Google Scholar]
- 13.Hardy, O. J., N. Charbonnel, H. Fréville, and M. Heuertz. 2003. Microsatellite allele size: a simple test to assess their significance in genetic differentiation. Genetics 163:1467-1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hardy, O. J., and X. Vekemans. 2002. SPAGEDI: a versatile computer program to analyze spatial genetic structure at the individual or population level. Mol. Ecol. Notes 2:618-620. [Google Scholar]
- 15.Henrion, B., G. Chevalier, and F. Martin. 1994. Typing truffle species by PCR amplification of the ribosomal DNA spacers. Mycol. Res. 98:37-43. [Google Scholar]
- 16.Kerrigan, R. W., D. B. Carvalho, P. A. Horgen, and J. B. Anderson. 1998. The indigenous coastal Californian population of the mushroom Agaricus bisporus, a cultivated species, may be at risk of extinction. Mol. Ecol. 7:35-45. [Google Scholar]
- 17.Koufopanou, V., A. Burt, and J. W. Taylor. 1997. Concordance of gene genealogies reveals reproductive isolation in the pathogenic fungus Coccidioides immitis. Proc. Natl. Acad. Sci. USA 94:5478-5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maynard Smith, J., N. H. Smith, M. O'Rourke, and B. G. Spratt. 1993. How clonal are bacteria? Proc. Natl. Acad. Sci. USA 90:4384-4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mello, A., L. Garnero, and P. Bonfante. 1999. Specific PCR-primers as a reliable tool for the detection of white truffles in mycorrhizal roots. New Phytol. 141:511-516. [Google Scholar]
- 20.Michalakis, Y., and L. Excoffier. 1996. A generic estimation of population subdivision using distances between alleles with special reference for microsatellite loci. Genetics 142:1061-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murat, C., J. Díez, P. Luis, C. Delaruelle, C. Dupré, G. Chevalier, P. Bonfante, and F. Martin. 2004. Polymorphism at the ribosomal DNA ITS and its relation to postglacial re-colonization routes of the Périgord truffle Tuber melanosporum. New Phytol. 164:401-411. [DOI] [PubMed] [Google Scholar]
- 22.Pacioni, G., and G. Pomponi. 1991. Genotypic patterns of some Italian populations of the Tuber aestivum-T. mesentericum complex. Mycotaxon 42:171-179. [Google Scholar]
- 23.Paolocci, F., P. Angelini, E. Cristofari, B. Granetti, and S. Arcioni. 1995. Identification of Tuber spp. and corresponding ectomycorrhizae through molecular markers. J. Sci. Food Agric. 69:511-517. [Google Scholar]
- 24.Paolocci, F., A. Rubini, B. Granetti, and S. Arcioni. 1999. Rapid molecular approach for a reliable identification of Tuber spp. ectomycorrhizae. FEMS Microbiol. Ecol. 28:23-30. [Google Scholar]
- 25.Paolocci, F., A. Rubini, B. Granetti, and S. Arcioni. 1997. Typing Tuber melanosporum and Chinese black truffle species by molecular markers. FEMS Microbiol. Lett. 153:255-260. [DOI] [PubMed] [Google Scholar]
- 26.Paolocci, F., A. Rubini, C. Riccioni, B. Granetti, and S. Arcioni. 2000. Cloning and characterization of two repeated sequences in the symbiotic fungus Tuber melanosporum Vitt. FEMS Microbiol. Ecol. 34:139-146. [DOI] [PubMed] [Google Scholar]
- 27.Paolocci, F., A. Rubini, C. Riccioni, F. Topini, and S. Arcioni. 2004. Tuber aestivum and Tuber uncinatum: two morphotypes or two species? FEMS Microbiol. Lett. 235:109-115. [DOI] [PubMed] [Google Scholar]
- 28.Petit, R. J., I. Aguinagalde, J. L. de Beaulieu, C. Bittkau, S. Brewer, R. Cheddadi, R. Ennos, S. Fineschi, D. Grivet, M. Lascoux, A. Mohanty, G. Muller-Starck, B. Demesure-Musch, A. Palme, J. P. Martin, S. Rendell, and G. G. Vendramin. 2003. Glacial refugia: hotspots but not melting pots of genetic diversity. Science 300:1563-1565. [DOI] [PubMed] [Google Scholar]
- 29.Petit, R. J., A. El Mousadik, and O. Pons. 1998. Identifying populations for conservation on the basis of genetic markers. Conserv. Biol. 12:844-855. [Google Scholar]
- 30.Raymond, M., and G. Rousset. 1995. GENEPOP (version 1.2): population genetics software for exact test and ecumenicism. J. Hered. 86:248-249. [Google Scholar]
- 31.Rousset, F. 1997. Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics 145:1219-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rubini, A., F. Paolocci, S. Arcioni, and B. Granetti. 2001. Analisi morfologica comparativa delle spore e caratterizzazione molecolare di Tuber indicum Cooke e Massee e Tuber melanosporum Vitt., p 94-101. In J. C. Savignac (ed.), Actes du Ve Congrès International “Science et culture de la truffe et des autres champignons hypogés comestibles.” Fédération française des trufficulteurs, Aix en Provence, France.
- 33.Rubini, A., F. Paolocci, B. Granetti, and S. Arcioni. 2001. Morphological characterization of molecular-typed Tuber magnatum ectomycorrhizae. Mycorrhiza 11:179-185. [Google Scholar]
- 34.Rubini, A., F. Paolocci, B. Granetti, and S. Arcioni. 1998. Single step molecular characterization of morphologically similar black truffle species. FEMS Microbiol. Lett. 164:7-12. [Google Scholar]
- 35.Rubini, A., F. Topini, C. Riccioni, F. Paolocci, and S. Arcioni. 2004. Isolation and characterization of polymorphic microsatellite loci in white truffle (Tuber magnatum). Mol. Ecol. Notes 4:116-118. [Google Scholar]
- 36.Selosse, M. A., F. Martin, D. Bouchard, and F. Le Tacon. 1999. Structure and dynamics of experimentally introduced and naturally occurring Laccaria sp. discrete genotypes in a Douglas fir plantation. Appl. Environ. Microbiol. 65:2006-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Slatkin, M. 1995. A measure of population subdivision based on microsatellite allele frequencies. Genetics 139:457-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weir, B. S., and C. C. Cockerham. 1984. Estimating F-statistics for the analysis of population structure. Evolution 38:1358-1370. [DOI] [PubMed] [Google Scholar]
- 39.Wright, S. 1951. The genetical structure of populations. Ann. Eugen. 15:323-354. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.