Abstract
The sulfur present in both agricultural and uncultivated soils is largely in the form of sulfonates and sulfate esters and not as free, bioavailable inorganic sulfate. Desulfurization of the former compounds in vitro has previously been studied in Pseudomonas putida, a common rhizosphere inhabitant. Survival of P. putida strains was now investigated in three sulfur-deficient Danish soils which were found to contain 60 to 70% of their sulfur in sulfonate or sulfate ester form, as determined by X-ray near-edge spectroscopy. The soil fitness of P. putida S-313 was compared with that of isogenic strains with mutations in the sftR and asfA genes (required for in vitro desulfurization of sulfate esters and arylsulfonates, respectively) and in the ssu locus (required in vitro for the desulfurization of both sulfonates and sulfate esters). asfA or sftR mutants showed significantly reduced survival compared to the parent strain in bulk soil that had been enriched with carbon and nitrogen to mimic rhizosphere conditions, but this reduced survival was not observed in the absence of these additives. In a tomato rhizosphere grown in compost, survival of sftR and ssu mutants was reduced relative to the parent strain. The results demonstrate that the ability to desulfurize sulfonates and sulfate esters is critical for survival of bacteria in the rhizosphere but less so in bulk soils outside the influence of plant roots, where carbon is the limiting nutrient for growth.
The search for bacterial traits that help determine bacterial fitness to survive in soil and rhizosphere environments has been driven by the hope of utilizing free-living bacterial populations for biofertilization, biocontrol of plant pathogens, or biodegradation of toxic compounds. Traits which are important in bacterial fitness to survive in soil include the ability to adhere to soil particles and to the rhizoplane (4, 5), synthesis of antibiotics (32), and motility and prototrophy (6). However, the soil environment has been described as “grossly oligotrophic” (34) and microbial populations inhabiting it must possess a diverse metabolism in order to fulfill their nutritional requirements. The main driver of microbial diversity in bulk soil is therefore the availability of mineral and organic nutrients, with available carbon being the limiting nutrient (24). Within the plant rhizosphere, by contrast, rhizodeposition provides a broad range of organic and amino acids, sugars, and oligosaccharides (3) and these supply most of the carbon and nitrogen requirements to support a rich and diverse microbial community (10, 30).
Indeed, the very diversity of carbon sources plays an important role in generating niche competition between microorganisms. Soil pseudomonads can utilize a wide variety of carbon sources (28), partly explaining their ready adaptation to a variety of plant and soil types. Nitrogen can be assimilated from amino acids contained in root exudates (9, 17) or utilized in the form of nitrate (29). Micronutrients like iron and phosphorus are also essential for bacterial metabolism and can be mobilized by particular groups of bacteria. For example, sequestered iron in the soil may be accessed through siderophore biosynthesis by fluorescent pseudomonads (31) whereas insoluble phosphorus (either precipitated mineral phosphate or organic phosphates such as phytate and other phosphate esters) can be mobilized by many soil bacteria (13), especially Pseudomonas, Bacillus, and Rhizobium species (36).
The preferred source of sulfur for microorganisms is inorganic sulfate (21), but more than 95% of the soil sulfur is organically bound as a heterogeneous mixture of forms, including sulfate esters and carbon-bonded sulfur (sulfonates or peptide residues) (37). Radiochemical studies have shown that these forms of sulfur are rapidly interconverted in the soil, but it is not yet clear which organisms are responsible for these transformations or what direct effect this has on the supply of sulfur for plant uptake (22).
In previous work, we identified three main gene loci that are involved in the utilization of organosulfur (sulfonates and sulfate esters) by Pseudomonas putida S-313 during growth in laboratory media (19, 20, 23). These include the asf locus, which is required for arylsulfonate-sulfur utilization (48), the ats/sft locus, which is required for the desulfurization of sulfate esters (19), and the ssu locus, which is involved in the utilization of both sulfonates and sulfate esters (20). Since P. putida is a common soil pseudomonad, we hypothesized that these specific mineralization activities could play a key role in the fitness of P. putida in the soil environment. In this report, we use wild type/mutant comparisons to assess the importance of arylsulfonate, alkylsulfonate and sulfate ester utilization pathways on the survival of P. putida S-313 under various soil conditions and in a tomato rhizosphere.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used in this study are listed in Table 1. Pseudomonas putida strains were grown aerobically at 30°C on Luria-Bertani (LB) medium, and rifampin (100 μg/ml) and kanamycin (25 μg/ml) were added as required. For the preparation of soil inocula, the P. putida strains were grown for 24 h as a bacterial lawn on LB medium containing the appropriate antibiotic and then harvested, washed twice with 0.1 M MgCl2, and resuspended in 0.1 M MgCl2 before application to soil microcosms. For enumeration of bacteria recovered from the soil, cycloheximide (100 μg/ml) was added to the medium to inhibit fungal growth.
TABLE 1.
Bacterial strains used in this study
| Pseudomonas putida strain | Description/genotype | Reference |
|---|---|---|
| S-313 | Prototroph; utilizes arylsulfonates as sulfur source | 50 |
| S-313R | Rifampin-resistant derivative of strain S-313 | 19 |
| SN36 | asfA::miniTn5Km derivative of strain S-313; unable to utilize arylsulfonates as sulfur source | 48 |
| PH18 | sftR::miniTn5Km derivative of strain S-313; unable to utilize sulfate esters as sulfur source | 19 |
| SN34 | ssuE::miniTn5Km derivative of strain S-313; unable to utilize sulfonates or sulfate esters as sulfur source | 20 |
Plant growth conditions.
Licopersicon esculentum var. Ailsa Craig was used as a model plant in rhizosphere colonization assays. Seeds were sterilized in a 10% (vol/vol) solution of NaOCl for 20 min, washed three times with sterile distilled water, placed on sulfur-free Murashige-Skoog-agarose plates, and incubated for 5 days in a growth chamber on a cycle of 16 h light (25°C) and 8 h dark (22°C). Plantlets of L. esculentum were transferred into propagators set up with five pots per soil condition.
Soils and nutrient enrichment.
The effect of soil organic content and nutrient enrichment conditions was studied in three sandy soils collected in the area of Karup, Denmark. These were collected from an agricultural site (A), a forest site (F), and an area of natural grassland (G). The soils were washed with distilled water, air dried, and sieved (<4 mm). Full characterizations of the soils are shown in Table 2. Viking MH compost (Taylors Organex, United Kingdom) was used to assess bacterial survival in the rhizosphere of tomato. Soil nutrient enrichment was achieved by adding dilution buffer (1% [wt/vol] tryptone-0.1% [wt/vol] sodium pyrophosphate at 5 ml per 15-g microcosm) to the soil together with the bacterial inoculum. The solution contained no detectable sulfate (<0.1 μM), as measured by suppressed ion chromatography (see below).
TABLE 2.
Characteristics of soils used in this study
| Soil type | Physical properties
|
Chemical properties
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sand (%) | Silt (%) | Clay (%) | pH (H2O) | Organic carbon (g/kg) | P (mg/kg) | K (mg/kg) | Ca (mg/kg) | Mg (mg/kg) | S (mg/kg) | |
| Agricultural | 75.8 | 16.9 | 7.3 | 6.12 | 16.9 | 330 | 225 | 1,120 | 66 | 176 |
| Forest | 78.2 | 13.9 | 7.9 | 4.94 | 44.8 | 147 | 162 | 1,180 | 146 | 177 |
| Grassland | 89.6 | 6.1 | 4.3 | 5.28 | 19.8 | 328 | 103 | 610 | 68 | 82 |
Measurement of bacterial population dynamics.
For population dynamics studies, the bacterial inocula were added to obtain a bacterial density of 106, 107, or 108 CFU g−1 dry soil, using separate propagators for each strain. For rhizosphere experiments, inoculated propagators were incubated for 3 days before introduction of the germinated seeds. To measure population dynamics, four samples of bulk and/or rhizosphere soil were collected from independent replicate pots at suitable time intervals (usually 7, 14, 21, 28, and 33 days). Each soil sample was suspended in 5 ml sterile dilution buffer (1% [wt/vol] tryptone-0.1% [wt/vol] sodium pyrophosphate) (43) and mixed for 60 s with a vortex shaker. The soil suspensions were diluted in the dilution buffer and plated on LB medium containing the appropriate antibiotics. After enumeration of colonies, the bacterial concentrations [B] were expressed as CFU per gram of dry soil (CFU g−1 dry soil). The kinetics of survival were approximated by linear regression of the log [B] values against time. Linear regression analysis was performed according to the method of Gardner and Altman (12), using a 95% confidence interval.
Characterization of soil sulfur composition.
X-ray absorption near-edge spectroscopy (XANES) measurements at the sulfur K-edge were used to characterize the sulfur oxidation states present in the soils studied. The measurements were made at the Daresbury Synchrotron radiation source (2-GeV storage ring, operating typically between 250 and 180 mA) on beamline 3.4. This beamline has a planar premirror which functions as a high-energy cutoff and a toroidal focusing mirror. For these experiments, Si(III) crystals were used in the monochromator and the toroidal mirror was set to provide collimated light through the monochromator; energy resolution in this geometry is approximately 0.3 eV. Incident intensity (Io) was monitored by placing a 0.75-μm Al foil sheet between the monochromator and the sample. Measurements were performed within a chamber pumped down to high vacuum (<1 × 10−7 Torr). Fluorescence yield intensity measurements reported here were made using a gas microstrip detector (39); fluorescence intensities were optimized by adjusting the angle of incidence onto the sample. The detector was at 90o relative to the incident beam and located approximately 25 mm from the sample surface. XANES scans were subdivided into several separate energy regions spanning the region from approximately 2,430 to 2,550 eV. Extensive standardization with reference compounds was completed as discussed below, using the theoretical value for the K-shell absorption edge of elemental sulfur (2,472 eV) as reference. Samples were prepared as pressed 13-mm-diameter pellets diluted approximately 50% with either pure carbon or boron nitride. Gaussian curves were fitted to the XANES spectra of the standard compounds used, representing the range of sulfur oxidation states expected in aerobic soils. The relative amount of each oxidation state within a soil sample was then calculated by fitting the experimental XANES spectra for each soil to a linear combination of the standard spectra, using the least-squares fitting routine SOLVER supplied in Microsoft Excel.
Total inorganic sulfate in the soils was quantified by extraction and ion chromatography. Soil samples (5 g) were extracted with 16 mM KH2PO4 (25 ml) for 30 min (7) and filtered. The sulfate concentration in the filtrate was determined by suppressed anion chromatography using an AG14/AS14 anion-exchange column (Dionex), and conductimetric detection (eluent, 3.5 mM Na2CO3-1.0 mM NaHCO3; flow rate, 1.2 ml/min).
RESULTS
Soil management and nutrient availability affect the fitness of bacteria to survive in soil.
We compared the bacterial population dynamics of four P. putida strains (S-313R, SN36, PH18, SN34) (Table 1) in three soils (agricultural, grassland, and forest soils from central Denmark) and two nutrient enrichment conditions (enriched [E] and nonenriched [NE]). For the purposes of this experiment, we considered the plant rhizosphere to represent a general source of nutrients for soil microbial communities, and this was modeled by the nutrient enrichment procedure used. After addition of a bacterial inoculum to the soil, a reproducible pattern of bacterial population dynamics was observed, with a rapid increase in population for 1 day, followed by a decrease over time. These bacterial population dynamics were influenced in two main ways by the addition of nutrients: (i) initial bacterial densities (day 1, i.e., 1 day after inoculation) were significantly higher in the enriched soil (8.80 ± 0.17 log CFU g−1 dry soil) than in the nonenriched soil (7.24 ± 0.06 log CFU g−1 dry soil), and (ii) initial decreases in bacterial densities (day 1 to day 8) were lower in enriched soils (0.2 day−1) than in nonenriched soils (0.3 day−1). However, from day 8 to day 32, the log-transformed bacterial densities decreased linearly. These linear decreases represent the equilibrium between the growth and death rates of bacterial populations and are referred to here as survival or survival rate.
Overall, P. putida populations were maintained at a higher density in enriched soil than in nonenriched soil. In the forest soil, enrichment also improved the bacterial survival rate whereas it did not affect the survival in agricultural and grassland soils. This suggests that the enrichment provides nutrients that limit bacterial survival in the forest soil but not in the agricultural or grassland soils, probably phosphorus, since this is present at much lower levels in the forest soil than in the other two (Table 2).
Pathways for utilizing organic sulfur sources are involved in soil bacterial fitness.
To evaluate the importance of bacterial organosulfur metabolism in soil fitness, we measured the survival rates in each soil for the parent strain P. putida S-313R and the mutant strains PH18, SN34, and SN36 under enriched conditions (Table 3). In the agricultural soil, the survival of strain PH18 was significantly reduced compared to the other strains. In the grassland soil, mutants PH18 and SN36 showed similar reduced survival rates compared to both the wild type and mutant strain SN34. All strains showed similar survival in the forest soil. The poor survival of mutant strains PH18 and SN36 in enriched soil indicates that the abilities to desulfurize sulfate esters (defective in strain PH18) and aromatic sulfonates (defective in strain SN36) are important for P. putida S-313R to compete for nutrients.
TABLE 3.
Survival of P. putida strains in agricultural, forest, and grassland soils under enriched conditions
| Soil | Survival of inoculated strain
|
|||
|---|---|---|---|---|
| S-313R | PH18 | SN34 | SN36 | |
| Agricultural | 0.083a ± 0.017b,1 | 0.122 ± 0.0202 | 0.075 ± 0.0101 | 0.084 ± 0.0191 |
| Forest | 0.096 ± 0.0161,2 | 0.114 ± 0.0201,2 | 0.102 ± 0.0132 | 0.116 ± 0.0251,2 |
| Grassland | 0.083 ± 0.0281,2 | 0.161 ± 0.0163 | 0.106 ± 0.0122 | 0.146 ± 0.0163 |
First-order decay rate of the bacterial population (day−1).
Confidence interval for the slope of the regression line (P = 0.05). Values with the same superscript number are not significantly different for P = 0.05.
Although the sulfonatase and sulfatase genes were important for P. putida survival in enriched soil, in unenriched soils, the situation was different (Table 4). In the agricultural soil, the survival rates of all strains were similar. In the grassland soil, the survival rates of strains SN34 and SN36 were significantly higher than that of strain PH18, though SN34 and SN36 did not survive significantly better than the wild type. In the forest soil, strain SN34 had a significantly higher survival rate than either strain S-313R or mutant strains PH18 and SN36. These results imply that in unenriched soils, the survival of P. putida S-313R is not reduced by the various mutations affecting the ability to assimilate organically bound sulfur but that other factors are limiting bacterial survival in native soils. These factors may be different for different soils—forest soil contained much higher levels of organic carbon than the other two soils but also a much lower phosphorus level and had the lowest pH (Table 2). Interestingly, the better survival of the ssu mutant strain SN34 in grassland and forest soils, compared to strain S-313R, would seem to indicate a negative effect of expression of the ssu genes on survival of P. putida, but this has not yet been explored further.
TABLE 4.
Survival of P. putida strains in agricultural, forest, and grassland soils under nonenriched conditions
| Soil | Survival of inoculated strain
|
|||
|---|---|---|---|---|
| S-313R | PH18 | SN34 | SN36 | |
| Agricultural | 0.083a ± 0.048b,1,2,3 | 0.068 ± 0.0281 | 0.067 ± 0.0101 | 0.055 ± 0.0181 |
| Forest | 0.142 ± 0.0353,4 | 0.128 ± 0.0303,4 | 0.099 ± 0.0152 | 0.147 ± 0.0214 |
| Grassland | 0.136 ± 0.0413,4 | 0.152 ± 0.0304 | 0.108 ± 0.0242,3 | 0.114 ± 0.0202,3 |
First-order decay rate of the bacterial population (day−1).
Confidence interval for the slope of the regression line (P = 0.05). Values with the same superscript number are not significantly different for P = 0.05.
Determination of soil sulfur composition using XANES.
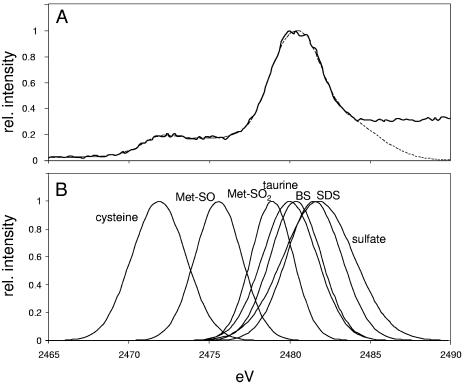
Differential soil competence of P. putida strains with differing ability to utilize sulfonates and sulfate esters (Table 3) suggests that the soils studied may differ in their sulfur compositions. Sulfur compositions of the soils were measured by XANES. K-edge spectra were obtained for each of the soil samples studied, and these were then fitted with standard spectra obtained for cysteine, methionine sulfoxide, methionine sulfone, a sulfate ester (sodium dodecyl sulfate), two sulfonates [taurine (2-aminoethanesulfonate) and benzenesulfonate], and sodium sulfate (Fig. 1). Two approaches were used for the fitting, which gave convergent results. In the first of these, the standard spectra were fitted to each total soil spectrum in order to determine the proportion of each sulfur state in the soil. In the second approach, the soil spectra were deconvoluted into a series of Gaussian curves, which could then be assigned to the standard spectra. The concentration of each sulfur species in each soil sample was then calculated from the total sulfur content of the soil (Table 2).
FIG. 1.
X-ray near-edge spectroscopy (XANES) to determine sulfur composition of soil. Sulfur K-edge spectra were measured for a number of standard compounds and fitted to Gaussian curves (B). These were then used to model the XANES spectrum of the studied Danish agricultural soil (A, unbroken line), as described in Materials and Methods. The optimized model spectrum (A, dashed line) gave the sulfur species composition given in Table 5. BS, benzenesulfonate; Met-SO, methionine sulfoxide; Met-SO2, methionine sulfone.
Overall, the grassland soil showed much lower levels of sulfur than either agricultural or forest soils (Table 2). All the soils contained very low levels of inorganic sulfate (0.41 to 2.2% of total sulfur), with the agricultural soil poorest in sulfate (Table 5). Grassland soil was dominated by sulfonate sulfur, but it was not possible to determine from the spectra whether this sulfonate sulfur was bound mainly to aliphatic or aromatic carbon. The agricultural soil contained approximately equal portions of sulfonate and sulfate ester sulfur, whereas the forest soil was dominated by sulfate ester sulfur. The proportion of amino acid sulfur was fairly uniform for the three soils (27 to 38% of total sulfur) (Table 5), which is significant because this sulfur source is expected to be available to all the strains examined.
TABLE 5.
Sulfur species composition of the soils under study
| Soil | Sulfur species (mg S/kg soil)a
|
|||||
|---|---|---|---|---|---|---|
| Sulfate | Sulfonates | Sulfate esters | Met sulfoxide | Met sulfone | Cysteine | |
| Agricultural | 0.72 (0.4) | 61.4 (34.9) | 55.4 (31.5) | 19.9 (11.3) | 13.4 (7.6) | 25.2 (14.3) |
| Forest | 2.6 (1.5) | 41.4 (23.4) | 64.6 (36.5) | 25.6 (14.5) | 17.7 (9.9) | 25.2 (14.2) |
| Grassland | 1.8 (2.2) | 40.0 (48.7) | 18.0 (21.9) | 8.4 (10.2) | 3.7 (4.5) | 10.2 (12.5) |
Values in parentheses indicate percent total sulfur for each soil.
Utilization of organic sulfur sources is important in rhizosphere competence.
The above experiments explored the importance of sulfatases and sulfonatases for bacterial survival in soils, with nutrient enrichment used to model a rhizosphere effect. These results were now extended by examining the survival of mutant strains SN36, PH18, and SN34 in the rhizosphere of tomato plants. To reduce the variables in the system and obtain consistent plant growth, the plants were cultivated in compost—the compost used contained high levels of inorganic sulfate (0.7 g S/kg), but in the immediate vicinity of the plant root, where bacterial colonization occurs, we anticipated that sulfate levels were reduced due to plant uptake. Each bacterial strain was inoculated at three initial densities in order to measure density effects on survival, but the initial differences of bacterial concentration were found to be maintained over the course of the experiment, resulting in similar survival rates regardless of initial density for all strains except for strain S-313 at low cell density (Table 6). Overall regression analysis indicated that the survival rates in the tomato rhizosphere varied according to the inoculated strain. Compared to the wild-type strain S-313R, the survival of the asf mutant strain SN36 was lower, whereas survival rates of both the sftR mutant strain PH18 and the ssu mutant strain SN34 were greatly reduced. These differences highlight the importance of the sulfonate and sulfate ester assimilation pathways for the fitness of P. putida in the rhizosphere of tomato cultivated in compost. XANES measurements of sulfur composition of the compost were unsuccessful because of the high content of organic carbon and the fibrous nature of the sample. However, the low survival of strains PH18 and SN34 suggests that sulfate esters are likely to be the primary source of organic sulfur in the compost used.
TABLE 6.
Survival of P. putida strains in the rhizosphere of L. esculentum cultivated in compost
| Inoculum (CFU/g soil) | Survival of inoculated strain
|
|||
|---|---|---|---|---|
| S-313R | PH18 | SN34 | SN36 | |
| 108 | 0.191a ± 0.036b,2 | 0.279 ± 0.0523 | 0.291 ± 0.0523 | 0.207 ± 0.0462,3 |
| 107 | 0.197 ± 0.0242 | 0.318 ± 0.0443 | 0.347 ± 0.0473 | 0.283 ± 0.0503 |
| 106 | 0.146 ± 0.0171 | 0.336 ± 0.0373 | 0.326 ± 0.0383 | 0.267 ± 0.0413 |
| Mean survival | 0.170 ± 0.0281 | 0.299 ± 0.0423 | 0.312 ± 0.0473 | 0.243 ± 0.0412,3 |
First-order decay rate of the bacterial population (day−1).
Confidence interval for the slope of the regression line (P = 0.05). Values with the same superscript number are not significantly different for P = 0.05.
DISCUSSION
Deficiency in inorganic sulfate levels in soils has been of concern in agriculture for some years now, especially in areas with sandy soils (37, 38). Sulfur deficiency in crop plants affects both the quality and the quantity of crop yield and has other effects, including chlorosis and stunted growth (37). However, although many soils are low in inorganic sulfate, they often contain sufficient amounts of sulfur as carbon-bound sulfur, largely as sulfonates and sulfate esters. In this report, we show that these organically bound sulfur forms are important for the survival of bacteria, both in the bulk soil and in a tomato rhizosphere. Mutants of P. putida S-313 with mutations in the asf, ssu, and ats/sft gene loci showed impaired soil and rhizosphere competence compared to the wild type, with the highest effects seen in the soil that was lowest in total sulfur. Since rhizosphere bacteria often have a very close and mutually protective relationship with their host plants, these findings also have important implications for plant sulfur metabolism.
In unamended soils, survival profiles and first-order decay rates for P. putida S-313 were comparable with those previously observed for pseudomonads (first-order decay rates between 0.02 and 0.11 day−1 [45]). We found that bacterial fitness to survive in all three soils was very similar for the four strains studied and was therefore largely independent of the ability to use sulfonates or sulfate esters (Table 4). However, this is not an unexpected result in bulk soil, since previous studies have shown that carbon supply, and not sulfur supply, is the main factor limiting bacterial growth (measured either by thymidine incorporation [1] or using reporter systems [26, 47]). Carbon supply to soil microbes comes largely from plant roots, and the microbial environment in the rhizosphere may be envisaged as a gradient of carbon limitation that increases dramatically with increasing distance from the rhizoplane (41). By contrast, in bulk soil, nitrogen and phosphate are not limiting nutrients (15, 27), but on the rhizoplane, they may become so, due to competition with plants for nutrient uptake (27). Although it might be anticipated that starving a bacterial population for carbon before inoculating it into the soil might increase adaptation and increase the bacterial survival rate, this is not in fact the case (14, 46). However, increased survival was observed for a Pseudomonas fluorescens R2f that had been starved of multiple nutrients (C, N, P, S) before soil inoculation, but this was not found for another soil strain, P. fluorescens CHA0 (14), and the role of adaptation to nutrient deprivation in the soil is therefore still uncertain. In this work, we supplemented the soils with a source of carbon, nitrogen, and phosphorus in order to create conditions where total sulfur concentrations might limit bacterial growth. Under these conditions, the mutant strains that were defective in the ability to utilize sulfonates or sulfate esters often survived more poorly than the wild type, depending on the soil used (Table 3). This effect was seen most strongly in the grassland soil, corresponding to the lowest value for total sulfur of the soils studied (Table 2). A similar effect was seen previously for nitrogen limitation, where an N-regulated lux reporter in P. fluorescens DF57 was not expressed in the soil unless carbon and phosphorus supplementation was applied (15). We conclude that sulfonates and sulfate esters are important sources of sulfur for bacteria residing in bulk soil but are not usually limiting for growth.
Supplementation of the bulk soil with carbon and nitrogen constituted a crude model for the rhizosphere itself, where these additional nutrients are provided by rhizodeposition. However, it is an inadequate model, since it ignores many other factors, including spatial variability, predation, and microbial community effects (42). We therefore also carried out the fitness studies in a tomato rhizosphere and found that P. putida strains that are unable to use sulfate esters or sulfonates in vitro showed greatly reduced survival rates compared to the wild-type strain (Table 6). This is a very significant result, suggesting that the competition for sulfur in the rhizosphere is very strong (both between microorganisms and possibly between plants and microbes) and may play an important part in bacterial colonization and survival. This competition may well also have an impact on the utilization of other nutrients. Previous studies with a phosphate limitation reporter (27), for example, have shown that phosphate is a limiting nutrient for bacteria in a gnotobiotic barley rhizosphere, but not in a natural rhizosphere. It was suggested that phosphate limitation might be reduced in the natural system by protozoan grazing or by phosphate mobilization within the total community (phosphate release by bacteria has been reviewed [36]). However, the effect could in principle also be caused if increased competition for sulfur within the natural system reduced the net requirement for phosphorus compared with the gnotobiotic one. This is currently being explored in more detail in our laboratory, using reporter strains for the genes of interest.
Carbon is present within the soil as low-molecular-weight compounds (especially within root exudates), but it is also present as highly polymeric compounds, especially as degradation products from plants (24). The same is also true for organically bound soil sulfur, and many of the XANES studies of soil sulfur have in fact been carried out with the high-molecular-weight humic fraction (33, 40, 49). Mobilization of sulfur from such high-molecular-weight polymers requires extracellular enzymes to break down the polymers before sulfur assimilation can occur. Interestingly, the sulfonatase and sulfatase enzymes encoded by the ssu, asf, and ats gene loci all require cofactors and are intracellularly located: the alkanesulfonatase and arylsulfonatases of P. putida S-313 require a reduced flavin mononucleotide pool (20, 48), and the main alkylsulfatase of P. putida S-313 is an α-ketoglutarate-dependent dioxygenase (18). In addition, all three gene loci studied contain specific transporter components that are coregulated with the oxygenase genes. We postulate that sulfur-containing polymers are cleaved either by extracellular fungal and actinomycetal enzymes or by uncharacterized bacterial activities present in the native rhizosphere, providing low-molecular-weight intermediates that can then be taken up and processed intracellularly. Extracellular sulfatase enzymes are common in soil (25) (though not produced by strain S-313 [19]), but their role remains uncertain since the addition of sulfatase to two different soils had no effect on the measured rate of sulfate release (11).
Two methods can be used for the determination of soil sulfur composition. Different sulfur species show differential reactivities to reducing agents, and traditionally, this property has been used to determine soil sulfur composition. Sulfate esters can be quantified by hydrogen sulfide released after hydriodic acid treatment, amino acid sulfur can be measured similarly after Raney-Ni reduction, and carbon-bound sulfur can then be calculated from the residual sulfur after subtraction of the inorganic sulfate present (8). More recently, sulfur species have often been determined by sulfur K-edge XANES, the method used in this work, using either whole soils and sediments (16, 35) or extracted humic substances (40, 44, 49). The method is nondestructive and requires little material and allows the determination of soil content of a range of sulfur oxidation states (Fig. 1). The soils used in this study were from a small area of central Denmark, with similar texture but taken from sites with different land use histories. All these soils showed low levels of inorganic sulfate (Table 5), but they differed in their content of other sulfur species. In particular, the agricultural soil showed higher levels of sulfonate-sulfur, while grassland soil was poor in sulfate esters. XANES analysis of all three soils showed that 27 to 30% of total sulfur was present as amino acid sulfur (Table 5). This is higher than the average values obtained in previous studies (e.g., 7 to 25% of total sulfur found as amino acids in a range of forest soils [2]) probably because we obtained XANES spectra from whole soils rather than from extracted humic materials. This method gives a broader picture, since it includes both inorganic and organic sulfur, but it is likely that the reduced sulfur present in bulk soil contains uncharacterized inorganic compounds which could bias the measured amino acid sulfur pool.
Initially, we attempted to correlate bacterial survival rates in the three soils studied with the soil properties, including both sulfur composition and other characteristics (Table 2). We anticipated that bacteria deficient in sulfonatase activity, for example, would survive significantly less well in a soil containing a high proportion of sulfonate-sulfur. However, although these effects can be seen for individual comparisons (e.g., the sulfonatase mutant strain SN36 survived poorly in the grassland soil compared to the agricultural soil [Table 4]), the small number of soils studied and the multifactorial nature of bacterial soil fitness made it hard to reach statistically significant generalizations. In particular, although bacteria showed significantly lower survival in the grassland soil, this was presumably not because of specific changes in sulfur composition but because the overall sulfur content was lower than for the other two soils. However, a few other correlations were noted; hence, the grassland soil contained reduced levels of silt and clay compared with the forest and agricultural soils, and this correlated with a decreased content of sulfate ester sulfur, confirming previous conclusions that clay particles are enriched in sulfate ester sulfur (40).
Acknowledgments
This work was supported by the Biotechnology and Biological Sciences Research Council and the Natural Environment Research Council.
We are grateful to Lars Elsgaard for assistance and advice in obtaining and preparing soils and to Marie Goldrick for help in the measurement of bacterial population dynamics.
REFERENCES
- 1.Alden, L., F. Demoling, and E. Baath. 2001. Rapid method of determining factors limiting bacterial growth in soil. Appl. Environ. Microbiol. 67:1830-1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Autry, A. R., and J. W. Fitzgerald. 1990. Sulfonate S—a major form of forest soil organic sulfur. Biol. Fertil. Soils 10:50-56. [Google Scholar]
- 3.Bertin, C., X. H. Yang, and L. A. Weston. 2003. The role of root exudates and allelochemicals in the rhizosphere. Plant Soil 256:67-83. [Google Scholar]
- 4.Boelens, J., D. Zoutman, J. Campbell, W. Verstraete, and W. Paranchych. 1993. The use of bioluminescence as a reporter to study the adherence of the plant-growth promoting rhizopseudomonads 7NSK2 and ANP15 to canola roots. Can. J. Microbiol. 39:329-334. [Google Scholar]
- 5.DeFlaun, M. F., B. M. Marshall, E. P. Kulle, and S. B. Levy. 1994. Tn5 insertion mutants of Pseudomonas fluorescens defective in adhesion to soil and seeds. Appl. Environ. Microbiol. 60:2637-2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Weger, L. A., I. Kuiper, A. J. van der Bij, and B. J. J. Lugtenberg. 1997. Use of a lux-based procedure to rapidly visualize root colonisation by Pseudomonas fluorescens in the wheat rhizosphere. Antonie Leeuwenhoek 72:365-372. [DOI] [PubMed] [Google Scholar]
- 7.Eriksen, J. 1997. Sulphur cycling in Danish agricultural soils: inorganic sulphate dynamics and plant uptake. Soil Biol. Biochem. 29:1379-1385. [Google Scholar]
- 8.Freney, J. R., G. E. Melville, and C. H. Williams. 1975. Soil organic matter fractions as sources of plant-available sulfur. Soil Biol. Biochem. 7:217-221. [Google Scholar]
- 9.Futamata, H., M. Sakai, H. Ozawa, Y. Urashima, T. Sueguchi, and T. Matsuguchi. 1998. Chemotactic response to amino acids of fluorescent pseudomonads isolated from spinach roots grown in soils with different salinity levels. Soil Sci. Plant Nutr. 44:1-7. [Google Scholar]
- 10.Gamliel, A., and J. Katan. 1992. Influence of seed and root exudates on fluorescent pseudomonads and fungi in solarized soil. Phytopathology 82:320-327. [Google Scholar]
- 11.Ganeshamurthy, A. N., and N. E. Nielsen. 1990. Arylsulfatase and the biochemical mineralization of soil organic sulfur. Soil Biol. Biochem. 22: 1163-1165. [Google Scholar]
- 12.Gardner, M. J., and D. G. Altman. 1989. Calculating confidence intervals for regression and correlation, p. 34-49. In M. J. Gardner and D. G. Altman (ed.), Statistics with confidence. British Medical Journal, London, United Kingdom. [DOI] [PMC free article] [PubMed]
- 13.Gyaneshwar, P., G. N. Kumar, L. J. Parekh, and P. S. Poole. 2002. Role of soil microorganisms in improving P nutrition of plants. Plant Soil 245:83-93. [Google Scholar]
- 14.Hase, C., F. Mascher, Y. Moenne-Loccoz, and G. Defago. 1999. Nutrient deprivation and the subsequent survival of biocontrol Pseudomonas fluorescens CHA0 in soil. Soil Biol. Biochem. 31:1181-1188. [Google Scholar]
- 15.Jensen, L. E., L. Kragelund, and O. Nybroe. 1998. Expression of a nitrogen regulated lux gene fusion in Pseudomonas fluorescens DF57 studied in pure culture and in soil. FEMS Microbiol. Ecol. 25:23-32. [Google Scholar]
- 16.Jokic, A., J. N. Cutler, E. Ponomarenko, G. van der Kamp, and D. W. Anderson. 2003. Organic carbon and sulphur compounds in wetland soils: insights on structure and transformation processes using K-edge XANES and NMR spectroscopy. Geochim. Cosmochim. Acta 67:2585-2597. [Google Scholar]
- 17.Jones, D. L., A. C. Edwards, K. Donachie, and P. R. Darrah. 1994. Role of proteinaceous amino acids released in root exudates in nutrient acquisition from the rhizosphere. Plant Soil 158:183-192. [Google Scholar]
- 18.Kahnert, A., and M. A. Kertesz. 2000. Characterization of a sulfur-regulated oxygenative alkylsulfatase from Pseudomonas putida S-313. J. Biol. Chem. 275:31661-31667. [DOI] [PubMed] [Google Scholar]
- 19.Kahnert, A., P. Mirleau, R. Wait, and M. A. Kertesz. 2002. The LysR-type regulator SftR is involved in soil survival and sulphate ester metabolism in Pseudomonas putida. Environ. Microbiol. 4:225-237. [DOI] [PubMed] [Google Scholar]
- 20.Kahnert, A., P. Vermeij, C. Wietek, P. James, T. Leisinger, and M. A. Kertesz. 2000. The ssu locus plays a key role in organosulfur metabolism in Pseudomonas putida S-313. J. Bacteriol. 182:2869-2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kertesz, M. A. 1999. Riding the sulfur cycle—metabolism of sulfonates and sulfate esters in gram-negative bacteria. FEMS Microbiol. Rev. 24:135-175. [DOI] [PubMed] [Google Scholar]
- 22.Kertesz, M. A., and P. Mirleau. 2004. The role of soil microbes in plant sulfur nutrition. J. Exp. Bot. 55:1939-1945. [DOI] [PubMed] [Google Scholar]
- 23.Kertesz, M. A., and C. Wietek. 2001. Desulfurization and desulfonation. Sulfur-controlled gene expression in bacteria. Appl. Microbiol. Biotechnol. 57:460-466. [DOI] [PubMed] [Google Scholar]
- 24.Killham, K., and C. Yeomans. 2001. Rhizosphere carbon flow measurement and implications: from isotopes to reporter genes. Plant Soil 232:91-96. [Google Scholar]
- 25.Klose, S., and M. A. Tabatabai. 1999. Arylsulfatase activity of microbial biomass in soils. Soil Sci. Soc. Am. J. 63:569-574. [Google Scholar]
- 26.Koch, B., J. Worm, L. E. Jensen, O. Hojberg, and O. Nybroe. 2001. Carbon limitation induces σS-dependent gene expression in Pseudomonas fluorescens in soil. Appl. Environ. Microbiol. 67:3363-3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kragelund, L., C. Hosbond, and O. Nybroe. 1997. Distribution of metabolic activity and phosphate starvation response of lux-tagged Pseudomonas fluorescens reporter bacteria in the barley rhizosphere. Appl. Environ. Microbiol. 63:4920-4928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Latour, X., T. S. Corberand, G. Laguerre, F. Allard, and P. Lemanceau. 1996. The composition of fluorescent pseudomonad populations associated with roots is influenced by plant and soil type. Appl. Environ. Microbiol. 62:2449-2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin, J. T., and V. Stewart. 1998. Nitrate assimilation by bacteria. Adv. Microb. Physiol. 39:1-30. [DOI] [PubMed] [Google Scholar]
- 30.Lynch, J. M., and J. M. Whipps. 1990. Substrate flow in the rhizosphere. Plant Soil 129:1-10. [Google Scholar]
- 31.Mirleau, P., S. Delorme, L. Philippot, J. M. Meyer, S. Mazurier, and P. Lemanceau. 2000. Fitness in soil and rhizosphere of Pseudomonas fluorescens C7R12 compared with a C7R12 mutant affected in pyoverdine synthesis and uptake. FEMS Microbiol. Ecol. 34:35-44. [DOI] [PubMed] [Google Scholar]
- 32.Natsch, A., C. Keel, H. A. Pfirter, D. Haas, and G. Defago. 1994. Contribution of the global regulator gene gacA to persistence and dissemination of Pseudomonas fluorescens biocontrol strain CHA0 introduced into soil microcosms. Appl. Environ. Microbiol. 60:2553-2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pickering, T. J., R. C. Prince, T. Divers, and G. N. George. 1998. Sulfur K-edge X-ray absorption spectroscopy for determining the chemical speciation of sulfur in biological systems. FEBS Lett. 441:11-14. [DOI] [PubMed] [Google Scholar]
- 34.Poindexter, J. S. 1981. Oligotrophy. Adv. Microb. Ecol. 5:63-89. [Google Scholar]
- 35.Prietzel, J., J. Thieme, U. Neuhausler, J. Susini, and I. Kogel-Knabner. 2003. Speciation of sulphur in soils and soil particles by X-ray spectromicroscopy. Eur. J. Soil Sci. 54:423-433. [Google Scholar]
- 36.Rodriguez, H., and R. Fraga. 1999. Phosphate solubilizing bacteria and their role in plant growth promotion. Biotechnol. Adv. 17:319-339. [DOI] [PubMed] [Google Scholar]
- 37.Scherer, H. W. 2001. Sulphur in crop production. Eur. J. Agron. 14:81-111. [Google Scholar]
- 38.Schnug, E., and S. Haneklaus. 1998. Diagnosis of sulphur nutrition, p. 1-38. In E. Schnug (ed.), Sulphur in agroecosystems. Kluwer, Dordrecht, The Netherlands.
- 39.Smith, A. D., J. E. Bateman, G. E. Derbyshire, D. M. Duxbury, J. Lipp, E. J. Spill, and R. Stephenson. 2001. A gas microstrip X-ray detector for soft energy fluorescence EXAFS. Nucl. Instrum. Methods Phys. Res. A 467:1136-1139. [Google Scholar]
- 40.Solomon, D., J. Lehmann, and C. E. Martinez. 2003. Sulfur K-edge XANES spectroscopy as a tool for understanding sulfur dynamics in soil organic matter. Soil Sci. Soc. Am. J. 67:1721-1731. [Google Scholar]
- 41.Standing, D., A. A. Meharg, and K. Killham. 2003. A tripartite microbial reporter gene system for real-time assays of soil nutrient status. FEMS Microbiol. Lett. 220:35-39. [DOI] [PubMed] [Google Scholar]
- 42.Toal, M. E., C. Yeomans, K. Killham, and A. A. Meharg. 2000. A review of rhizosphere carbon flow modelling. Plant Soil 222:263-281. [Google Scholar]
- 43.Trevors, J. T., and S. Cook. 1992. A comparison of plating media and diluents for enumeration of aerobic bacteria in a loam soil. J. Microbiol. Methods 14:271-275. [Google Scholar]
- 44.Vairavamurthy, M. A., D. Maletic, S. K. Wang, B. Manowitz, T. Eglinton, and T. Lyons. 1997. Characterization of sulfur-containing functional groups in sedimentary humic substances by X-ray absorption near-edge structure spectroscopy. Energy Fuels 11:546-553. [Google Scholar]
- 45.van Elsas, J. D., and L. S. van Overbeek. 1993. Bacterial responses to soil stimuli, p. 55-79. In S. Kjelleberg (ed.), Starvation in bacteria. Plenum Press, New York, N.Y.
- 46.van Overbeek, L. S., L. Eberl, M. Givskov, S. Molin, and J. D. van Elsas. 1995. Survival of, and induced stress resistance in, carbon-starved Pseudomonas fluorescens cells residing in soil. Appl. Environ. Microbiol. 61:4202-4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Overbeek, L. S., J. D. van Elsas, and J. A. van Veen. 1997. Pseudomonas fluorescens Tn5-B20 mutant RA92 responds to carbon limitation in soil. FEMS Microbiol. Ecol. 24:57-71. [Google Scholar]
- 48.Vermeij, P., C. Wietek, A. Kahnert, T. Wüest, and M. A. Kertesz. 1999. Genetic organization of sulfur-controlled aryl desulfonation in Pseudomonas putida S-313. Mol. Microbiol. 32:913-926. [DOI] [PubMed] [Google Scholar]
- 49.Xia, K., F. Weesner, W. F. Bleam, P. R. Bloom, U. L. Skyllberg, and P. A. Helmke. 1998. XANES studies of oxidation states of sulfur in aquatic and soil humic substances. Soil Sci. Soc. Am. J. 62:1240-1246. [Google Scholar]
- 50.Zürrer, D., A. M. Cook, and T. Leisinger. 1987. Microbial desulfonation of substituted naphthalenesulfonic acids and benzenesulfonic acids. Appl. Environ. Microbiol. 53:1459-1463. [DOI] [PMC free article] [PubMed] [Google Scholar]