Abstract
Babesia and Hepatozoon infections of dogs in a village of eastern Sudan were analyzed by using a single PCR and sequencing. Among 78 dogs, 5 were infected with Babesia canis rossi and 2 others were infected with B. canis vogeli. Thirty-three dogs were positive for Hepatozoon. Hepatozoon canis was detected by sequence analysis.
Both Babesia and Hepatozoon infections are important tick-borne protozoal diseases of dogs (2, 8). The diagnosis of infections with these protozoa is usually based on the detection of pathogens in peripheral blood under a microscope. However, such morphology-based methods are labor- and time-consuming because of their low sensitivities. Recently, molecular techniques, including PCR and sequence analysis, have been used for the diagnosis and epidemiological studies of canine Babesia and Hepatozoon infections (1, 3, 10, 11, 21, 22). The advantages of the molecular methods over other techniques are their higher sensitivities and specificities for the detection of the target pathogens in peripheral blood. Babesia canis is divided into three subspecies, B. canis canis, B. canis vogeli, and B. canis rossi (7, 14, 19). By using these molecular methods, the diagnosis of Babesia infection is easily performed at the subspecies level. Hepatozoon has also been analyzed by using molecular technologies to identify two species, Hepatozoon canis and H. americanum (1, 2, 20). Because most epidemiological studies of protozoal infections in African countries are performed based on morphology, little information is available on canine Babesia and Hepatozoon infections in Africa (5,9, 17). Thus, the objective of this study was to clarify the infection rates and subspecies of Babesia and Hepatozoon in dogs in Barbar el Fugara, a village in eastern Sudan, by using a combination of screening PCR and the sequencing methodology. We used a screening PCR to detect both Babesia and Hepatozoon simultaneously, followed by sequencing to identify the organisms to the species or the subspecies level.
Peripheral blood was obtained from 78 randomly selected dogs in the village during May 1997, May 1998, April 1999, and January 2000 (6). As these dogs were all free roaming around the village, the ages and histories of dogs were unknown. The sex and health status of the dogs were not recorded. Ticks were recovered from 61 dogs for identification. Rhipicephalus sanguineus was the most dominant tick species: it was recovered from 44 of 61 dogs (72.1%), in agreement with the findings presented in a previous report (12), followed by Rhipicephalus evertsi evertsi (3 of 61 dogs [4.9%]) and Amblyomma lepidum (4of 61 dogs [6.6%]). Total DNA was extracted from each sample of canine blood with a QIAamp DNA Mini kit (QIAGEN GmbH, Hilden, Germany), adjusted to 200 μl with TE (Tris-EDTA) buffer, and stored at −20°C until it was used. Detection of DNA fragments of Babesia and Hepatozoon was attempted by PCR with primers of Babesia-F (GTG-AAA- CTG-CGA-ATG-GCT-CA) and Babesia-R (CCA-TGC-TGA- AGT-ATT-CAA-GAC). This primer set was previously reported to be specific for the genus Babesia (11), but it could amplify both Babesia and Hepatozoon simultaneously in our preliminary experiments. To confirm the results of PCR and to identify the infectious agents at the species or subspecies level, selected products of the PCR were purified with a QIAPCR purification kit (QIAGEN) or QIAquick gel extraction kit (QIAGEN) for direct sequence analysis. A fluorescence-labeled dideoxynucleotide technology was used for the DNA sequencing reactions (Perkin- Elmer, Applied Biosystems Division, Foster City, CA). The samples were then sequenced by using a Perkin-Elmer ABI Prism 377 automated DNA sequencer at the DNA Core Facility of the Center for Gene Research, Yamaguchi University. The sequences of the agent determined were analyzed for phylogenetic relationships with other sequences registered in GenBank. Multiple-sequence alignment analysis, the determination of pairwise percent identities of the sequences, distance matrix calculations, and the construction of phylogenetic trees were all performed with the ClustalW program (18), version 1.8, in the DNA data bank ofJapan (DDBJ; Mishima, Japan [http://www.ddbj.nig.ac.jp/htmls/E-mail/clustalw-e.html]), as described in a previous report (11). The distance matrices for the aligned sequences with all gaps ignored were calculated by using the Kimura two-parameter method (13), and the neighbor-joining method was used to construct a phylogenetic tree (16). The stability of the tree obtained was estimated by bootstrap analysis for 100 replications by using the same program. Tree figures were generated by using the Tree View program, version 1.61 (15). The GenBank accession numbers of the 18S rRNA gene sequences of other species used to analyze the data are as follows: Babesia divergense, GenBank accession no. U16370; Babesia odocoilei, GenBank accession no. U16369; Babesia gibsoni Asia-1, GenBank accession no. AF175300; B. gibsoni Asia-2, GenBank accession no. AF175301; B. canis vogeli, GenBank accession no. AY072925; B. canis canis, GenBank accession no. AY072926; Babesia caballi, GenBank accession no. Z15104; Babesia bigemina, GenBank accession no. X59607; Babesia bovis, GenBank accession no. L19078; Theileria sergenti, GenBank accession no. AB000271; Hepatozoon canis Japan, GenBank accession no. AF418558; Hepatozoon canis Italia, GenBank accession no. AF176835; Hepatozoon americanum, GenBank accession no. AF176836; Hepatozoon catesbianae, GenBank accession no. AF176837; and Neosporum caninum, GenBank accession no. U03069.
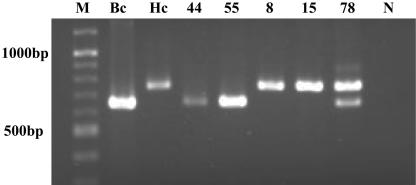
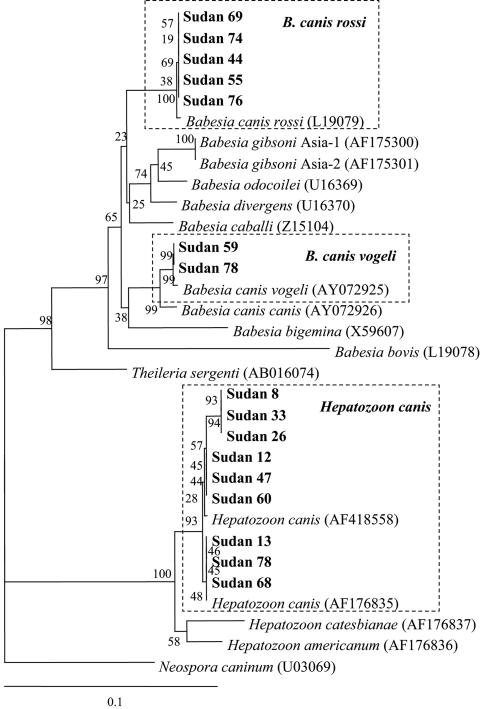
Among the 78 dogs examined, 7 (9.0%) dogs (dogs 44, 55, 59, 69, 74, 76, and 78) showed a band positive for Babesia at about 645 bp. A total of 33 (42.3%) dogs were positive for Hepatozoon with a band of about 780 bp. Among these, three dogs (dogs 59, 74, and 78) showed dual positivity for bands at both 645 and 780 bp (Fig. 1). By analyzing the seven sequences of the Babesia 645-bp PCR products, excluding the primer region, five were identified as B. canis rossi (GenBank accession no. L19079) with percent identities of 99.7 to 99.8% (Fig.2). The other two were very similar to B. canis rossi (GenBank accession no. AY072925), with percent identities of 99.8% (Fig. 2). Nine PCR products were randomly selected from among 33 Hepatozoon-positive PCR products for sequence analysis. All nine samples examined showed higher similarities with H. canis (GenBank accession no. AF176835), with percent identities of 99.1 to 100% (Fig. 2).
FIG. 1.
Results of PCR for five positive samples. Screening by PCR produced a 645-bp fragment for Babesia (lanes Bc, 44, and 55) and a 780-bp fragment for Hepatozoon (lanes Hc, 8, and 15). Isolate 78 showed dual positivity for Babesia and Hepatozoon, with 645- and 780-bp fragments. Lane M, molecular size marker; lane N, negative control.
FIG. 2.
Phylogenetic relationships between Babesia and Hepatozoon spp. in Sudan detected in this study and sequences registered in GenBank based on partial nucleotide sequences of the 18S rRNA gene. The numbers at the nodes are the proportions of 100 bootstrap resamplings that support the topology shown. The scale bar represents 10% divergence.
B. canis has three subspecies: B. canis canis, B. canis rossi, and B. canis vogeli. Each subspecies has a different vector and has a different pathogenesis in canine hosts. B. canis rossi is known to be the most pathogenic among the three subspecies and is transmitted by Hemaphysalis leachi (7). The pathogenesis of B. canis vogeli is comparatively weaker than those of the other two subspecies, and it is transmitted by Rhipicephalus sanguineus (7). In the present study, the predominant tick species recovered from dogs was R. sanguineus, and H. leachi was not detected. Babesia canis rossi may also be transmitted by ticks, such as R. sanguineus, R. evertsi evertsi, or A. lepidum, which were recovered from dogs in this study. Although the clinical symptoms of the infected dogs were not recorded in this study, infection with B. canis rossi might cause clinical disease in the canine host. The findings reported here are the first evidence of infection with B. canis rossi and B. canis vogeli in dogs in Sudan.
Our findings are also the first evidence of Hepatozoon canis infection in dogs in Sudan. H. canis is also known to be transmitted by R. sanguineus (4), which was the most common tick found in the present study. The rate of infection with H. canis was higher than that with B. canis in the present study. The weak pathogenesis of H. canis infection in canine hosts might contribute to the higher infection rate in this group, although the clinical symptoms of the infected dogs were not recorded.
Infections with B. canis rossi, B. canis vogeli, and H. canis in dogs may have a clinical impact on the quality of dogs' lives in this area. Dogs may also be reservoirs for continued propagation or may be the cause of increased infection rates. Furthermore, R. sanguineus may play an important role in the transmission of Babesia and Hepatozoon in Sudan.
In the present study, a single PCR was successfully used to detect Babesia and Hepatozoon simultaneously in canine blood samples. This provided an easy screening method for the detection of both Babesia and Hepatozoon in a single PCR. In combination with subsequent sequence analysis, this PCR assay may provide accurate information about the infectious agents. There were no difficulties in determining the subspecies of Babesia or the species of Hepatozoon in the sequence analysis in the present study. A dog might be infected with more than one subspecies of Babesia or more than one species of Hepatozoon at the same time. In such a case, the results of subsequent sequence analysis would be more difficult to interpret, because the results of the direct sequencing of the PCR products could not be read accurately. A subspecies-specific PCR for Babesia canis and a species-specific PCR for Hepatozoon would be required to evaluate the infection rate with more accuracy in those cases.
Nucleotide sequence accession number.
The nucleotide sequences of the 18S rRNA genes of the following Babesia and Hepatozoon isolates detected from dogs in this study have been deposited in the GenBank database under the indicated accession numbers: Babesia canis rossi Sudan-44, GenBank accession no. DQ111760; Babesia canis rossi Sudan-55, GenBank accession no. DQ111761; Babesia canis rossi Sudan-69, GenBank accession no. DQ111762; Babesia canis rossi Sudan-74, GenBank accession no. DQ111763; and Babesia canis rossi Sudan-76, GenBank accession no. DQ111764; Babesia canis vogeli Sudan-59, GenBank accession no. DQ111765; Babesia canis vogeli Sudan-78, GenBank accession no. DQ111766; Hepatozoon canis Sudan-8, GenBank accession no. DQ111751; Hepatozoon canis Sudan-12, GenBank accession no. DQ111752; Hepatozoon canis Sudan-13, GenBank accession no. DQ111753; Hepatozoon canis Sudan-26, GenBank accession no. DQ111754; Hepatozoon canis Sudan-33, GenBank accession no. DQ111755; Hepatozoon canis Sudan-47, GenBank accession no. DQ111756; Hepatozoon canis Sudan-60, GenBank accession no. DQ111758; Hepatozoon canis Sudan-68, GenBank accession no. DQ111759; and Hepatozoon canis Sudan-78, GenBank accession no. DQ111757.
Acknowledgments
We acknowledge the technical expertise of the DNA Core Facility of the Center for Gene Research, Yamaguchi University.
Our study was supported by grants from the Ministry of Education, Science, Sports and Culture of Japan, the Institut National de la Sante et de la Recherche Medicale, and the Japan Society for the Promotion of Science.
REFERENCES
- 1.Baneth, G., J. R. Barta, V. Shkap, D. S. Martin, D. K. Macintire, and N. Vincent-Johnson. 2000. Genetic and antigenic evidence supports the separation of Hepatozoon canis and Hepatozoon americanum at the species level. J. Clin. Microbiol. 38:1298-1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baneth, G., J. S. Mathew, V. Shkap, D. K. Macintire, J. R. Barta, and S. A. Ewing. 2003. Canine hepatozoonosis: two disease syndromes caused by separate Hepatozoon spp. Trends. Parasitol. 19:27-31. [DOI] [PubMed] [Google Scholar]
- 3.Caccio, S. M., B. Antunovic, A. Moretti, V. Mangili, A. Marinculic, R. R. Baric, S. B. Slemenda, and N. J. Pieniazek. 2002. Molecular characterization of Babesia canis canis and Babesia canis vogeli from naturally infected European dogs. Vet. Parasitol. 106:285-292. [DOI] [PubMed] [Google Scholar]
- 4.Christophers, S. R. 1907. The sexual life cycle of Leucocytozoon canis in the tick. Sci. Mem. Off. Med. Sanit. Dep. Gov. India 28:1-11. [Google Scholar]
- 5.Collett, M. G. 2000. Survey of canine babesiosis in South Africa. J. S. Afr. Vet. Assoc. 71:180-186. [DOI] [PubMed] [Google Scholar]
- 6.Dereure, J., S. H. El-Safi, B. Bucheton, M. Boni, M. M. Kheir, B. Davoust, F. Pratlong, E. Feugier, M. Lambert, A. Dessein, and J. P. Dedet. 2003. Visceral leishmaniasis in eastern Sudan: parasite identification in humans and dogs; host-parasite relationships. Microbes Infect. 5:1103-1108. [DOI] [PubMed] [Google Scholar]
- 7.Hauschild, S., and E. Schein. 1996. The subspecies specificity of Babesia canis. Berl. Munch. Tierarztl. Wochenschr. 109:216-219. [PubMed] [Google Scholar]
- 8.Homer, M. J., I. Aguilar-Delfin, S. R. Telford III, P. J. Krause, and D. H. Persing. 2000. Babesiosis. Clin. Microbiol. Rev. 13:451-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ibrahim, N. D., P. M. Rahamathulla, and C. O. Njoku. 1989. Neutrophil myeloperoxidase deficiency associated with canine hepatozoonosis. Int. J. Parasitol. 19:915-918. [DOI] [PubMed] [Google Scholar]
- 10.Inokuma, H., Y. Yoshizaki, K. Matsumoto, M. Okuda, T. Onishi, K. Nakagome, R. Kosugi, and M. Hirakawa. 2004. Molecular survey of Babesia infection in dogs in Okinawa, Japan. Vet. Parasitol. 121:341-346. [DOI] [PubMed] [Google Scholar]
- 11.Inokuma, H., Y. Yoshizaki, Y. Shimada, Y. Sakata, M. Okuda, and T. Onishi. 2003. Epidemiological survey of Babesia species in Japan performed with specimens from ticks collected from dogs and detection of new Babesia DNA closely related to Babesia odocoilei. J. Clin. Microbiol. 41:3494-3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jongejan, F., D. Zivkovic, R. G. Pegram, R. J. Tatchell, T. Fison, A. A. Latif, and G. Paine. 1987. Ticks (Acari:Ixodidae) of the Blue and White Nile ecosystems in the Sudan with particular reference to the Rhipicephalus sanguineus group. Exp. Appl. Acarol. 3:331-346. [DOI] [PubMed] [Google Scholar]
- 13.Kimura, M. 1980. A simple method for estimating evolutional rates of base substitutions through comparative studies of nucleotide sequence. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 14.Kjemtrup, A. M., A. A. Kocan, L. Whiteworth, J. Meinkoth, A. J. Birkenheuer, J. Cummings, M. K. Boudreaux, S. L. Stockham, A. Irizarry-Rovira, and P. A. Conrad. 2000. There are at least three genetically distinct small piroplasms from dogs. Int. J. Parasitol. 30:1501-1505. [DOI] [PubMed] [Google Scholar]
- 15.Page, R. D. M. 1996. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 16.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Med. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 17.Shakespeare, A. S. 1995. The incidence of canine babesiosis amongst sick dogs presented to the Onderstepoort Veterinary Academic Hospital. J. S. Afr. Vet. Assoc. 66:247-250. [PubMed] [Google Scholar]
- 18.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uilenberg, G., F. F. Franssen, N. M. Perie, and A. A. Spanjer. 1989. Three groups of Babesia canis distinguished and a proposal for nomenclature. Vet. Q. 11:33-40. [DOI] [PubMed] [Google Scholar]
- 20.Vincent-Johnson, N. A., D. K. Macintire, D. S. Lindsay, S. D. Lenz, G. Baneth, V. Shkap, and B. L. Blagburn. 1997. A new Hepatozoon species from dogs: description of the causative agent of canine hepatozoonosis in North America. J. Parasitol. 83:1165-1172. [PubMed] [Google Scholar]
- 21.Zahler, M., H. Rinder, E. Schein, and R. Gothe. 2000. Detection of a new pathogenic Babesia microti-like species in dogs. Vet. Parasitol. 89:241-248. [DOI] [PubMed] [Google Scholar]
- 22.Zahler, M., H. Rinder, E. Zweygarth, T. Fukata, Y. Maede, E. Schein, and R. Gothe. 2000. ‘Babesia gibsoni ’ of dogs from North America and Asia belong to different species. Parasitology 120:365-369. [DOI] [PubMed] [Google Scholar]