Abstract
Bordetella avium is the etiologic agent of turkey coryza or bordetellosis, a respiratory disease responsible for substantial economic losses to the turkey industry. At present, identification of this bacterium relies on isolation and biochemical testing. Although a PCR for the detection of B. avium was proposed a number of years ago (P. H. Savelkoul, L. E. de Groot, C. Boersma, I. Livey, C. J. Duggleby, B. A. van der Zeijst, and W. Gaastra, Microb. Pathog. 15:207-215, 1993), lack of analytical verification precludes its use as a diagnostic tool. Furthermore, a number of details pertaining to the reaction conditions used are missing or unclear. In the present study we have identified an optimal set of PCR conditions for use with the previously described primer pair and determined the limit of detection under these conditions to be approximately 20 pg. Assay sensitivity is 100%, based on an analysis of 72 B. avium isolates from diverse geographic locations and covering a time span of at least 25 years. Evaluation of a separate group of 87 bacterial isolates from poultry, comprising both gram-positive and gram-negative commensals and pathogens representing 11 genera, demonstrated an assay specificity of 98.8%. Reproducibility is 100% using either purified genomic DNA or boiled cell lysates less than 3 days old. Sequence analysis of the B. avium PCR amplicons identified only three occasional sequence polymorphisms. These data indicate the B. avium PCR assay can provide clinically significant results.
Bordetella avium is the etiologic agent of turkey coryza or bordetellosis, a respiratory disease responsible for substantial economic losses to the turkey industry. Although mortality associated with uncomplicated bordetellosis is low, morbidity often approaches 100% (10) and infected turkeys are particularly susceptible to secondary colisepticemia. B. avium is highly transmissible and may be spread either through exposure to contaminated litter or water or by direct contact with infected birds (20). Rigorous biosecurity measures are required to prevent inadvertent spread from infected to clean flocks.
Despite the increasing use and superior performance of PCR-based diagnostic assays in clinical laboratories, identification of B. avium still depends upon isolation and biochemical testing. This approach is complicated by the slow growth of B. avium, which may result in the masking of pinpoint colonies by other fast-growing organisms that typically contaminate samples. B. avium must also be differentiated from Bordetella hinzii, a newly recognized and closely related bacterium not currently believed to cause clinical disease in poultry (10).
A review of the literature reveals only a single report describing the use of PCR for the detection of B. avium (19). While the assay shows promise as a diagnostic tool, several critical points remain to be addressed before it can be implemented with confidence. The reported primer set was shown to amplify a fragment of the predicted size from “several” B. avium isolates. However, the exact number and identities of the isolates included were not stated. Accordingly, it is unclear whether the targeted DNA sequence is present in strains from diverse geographic locations and whether it has been stably maintained over time. The report contains a discrepancy in the sequence of the forward primer, which must be resolved. Although the specificity of the primer set was tested on several other bacterial genera, most isolates were not derived from poultry. Additionally, many bacterial genera commonly found in the turkey respiratory tract, including both commensals and respiratory pathogens, were not tested. It is also unclear whether the targeted amplicon is present in B. hinzii, potentially resulting in a false-positive result, since this bacterium was not recognized as a distinct species at the time the work was carried out. Furthermore, the original report does not address the limit of detection, and the exact composition of the PCR mix is not provided. Finally, no obvious attempt was made to optimize the assay, and at least some of the stated conditions do not intuitively seem optimal (e.g., a 2-min extension step for a 500-bp amplicon).
The goal of the present study is to define and optimize conditions for PCR identification of B. avium using the previously proposed primer set and to determine the sensitivity, specificity, reproducibility, and limit of detection of the assay.
MATERIALS AND METHODS
Bacterial isolates.
Bacterial isolates included in this study are listed in Table 1. All were obtained from commercially raised turkeys, except where indicated. Pasteurella multocida isolates (kindly provided by Mark Wilson at the USDA National Veterinary Services Laboratories, Ames, IA) originated from western, Midwestern, and southern states and represent capsular types A, D, and F. Other isolates obtained in 2003 or 2004 were collected during farm visits by swabbing the upper third of the trachea with BBL CultureSwabs (Becton Dickinson and Company, Sparks, MD). Apparently healthy birds as well as some with clinical signs of bordetellosis were sampled. Swabs were streaked on the day of collection onto duplicate 5% sheep's blood agar plates, which were incubated at 37°C for 24 to 48 h with or without 5% CO2. In order to select a wide range of commensal organisms for testing the specificity of the B. avium primers, a representative of every distinguishable colony type was selected for identification. Isolates were evaluated with Gram's stain and then identified using the MicroLog System (BioLog, Inc., Hayward, CA). Briefly, the results of 95 carbon source utilization tests were matched against a database of either gram-positive or gram-negative bacteria, as appropriate. Bacterial isolates obtained prior to 2003, most of which were provided by collaborators, were selected from the National Animal Disease Center collection. Identification was based on standard biochemical testing and colony morphology.
TABLE 1.
Bacterial isolates included in this study
| Isolate | Yr of isolation | Geographic origin | No. of isolates |
|---|---|---|---|
| B. avium | 1979 or prior | North Carolina | 4 |
| 1979 or prior | Minnesota | 2 | |
| 1997 | Minnesota | 6 | |
| 1979 or prior | Iowa | 1 | |
| 1981 | Iowa | 7 | |
| 2001 | Iowa | 1 | |
| 2004 | Iowa | 13 | |
| 1979 or prior | Ohio | 17 | |
| 1983 | Ohio | 1 | |
| 1990 | California | 2 | |
| 1998 | New Jersey | 9a | |
| 1979 or prior | Germany | 6b | |
| 1981 | South Africa | 3 | |
| B. hinzii | 1979 or prior | Minnesota | 5 |
| 1981 or prior | Minnesota | 5 | |
| 1991 | Minnesota | 1 | |
| 1989 | New York | 1 | |
| 1979 or prior | Ohio | 5 | |
| 1987 | Ohio | 11 | |
| 1982 or prior | Iowa | 4 | |
| 1990 | California | 3 | |
| 1994 | Washington | 1 | |
| 1985 or prior | United States | 1 | |
| 1996 | United States | 1 | |
| 1992 | Switzerland | 1 | |
| 1993 | Switzerland | 1 | |
| 1999 | Spain | 1 | |
| 1981 or prior | South Africa | 1 | |
| UKc | Belgium | 1 | |
| UK | Australia | 1b | |
| UK | UK | 3 | |
| Alcaligenes faecalis | 1979 or prior | North Carolina | 1 |
| 1982 or prior | UK | 1 | |
| Acinetobacter baumannii | 1979 or prior | Minnesota | 2 |
| Bacillus pumilus | 2004 | Iowa | 1 |
| Enterococcus cecorum | 2004 | Iowa | 2 |
| Enterococcus sulfureus | 2003 | Iowa | 1 |
| Escherichia coli | 2004 | Iowa | 5 |
| 2004 | Arkansas | 1 | |
| Ornithobacterium rhinotracheale | UK | Arkansas | 1 |
| 1996 | UK | 1 | |
| 1997 | North Carolina | 1 | |
| 2004 | Iowa | 1 | |
| 1988 | United Kingdom | 1b | |
| Pasteurella multocida | 1996 | Minnesota | 1 |
| 2001-2002 | United States | 11d | |
| Macrococcus caseolyticus | 2003 | Iowa | 1 |
| Micrococcus luteus | 2003 | Iowa | 1 |
| Staphylococcus aureus | 2003 | Iowa | 1 |
| 2004 | Iowa | 1 | |
| Staphylococcus epidermis | 2003 | Iowa | 1 |
| Staphylococcus hyicus | 2003 | Iowa | 1 |
| 2004 | Iowa | 1 | |
| Streptococcus spp. | 2004 | Iowa | 1 |
| Streptococcus hyointestinales | 2004 | Iowa | 1 |
Seven were from wild ducks, one was from a wild turkey, and one was from a wild goose.
Includes the ATCC type strain.
UK, unknown.
Five were from chickens.
PCR.
PCR was carried out in an Applied Biosystems 9700 thermal cycler. Whole-cell lysates used as templates were prepared by suspending a colony, ∼1.5 mm in diameter or the equivalent, in 10 μl water. The mixture was boiled for 10 min, placed on ice until chilled, and centrifuged at 16,000 × g for 1 min to pellet cell debris. Supernatant (2.5 μl) was used as the template in each PCR. Lysates were stored at 4°C for up to 1 week. Chromosomal DNA purified using a commercially available kit (Gentra Systems, Minneapolis, MN) was used to assess the limit of detection. DNA was quantitated with PicoGreen (Molecular Probes, Eugene, OR), a highly sensitive fluorescent stain specific for double-stranded DNA.
B. avium-specific primer sequences were obtained from the previous report of Savelkoul et al. (19). However, it was necessary to unequivocally establish the sequence of the forward primer, N-avium. This sequence appears twice in the original report, as a 25-mer in the text of the paper but as a 24-mer (missing a C at an internal position) in one of the figures. Comparison of the two sequences with the GenBank entry (accession no. X74117) suggests that the 24-mer is the correct sequence. This was confirmed by communication with one of the authors (P. Savelkoul, personal communication). The sequence of the reverse primer, C-avium, is as described previously (19). A DNA fragment of 500 bp was reported to be generated with this primer set.
Since the reaction component concentrations previously used with the N-avium/C-avium primer set are not known, the following arbitrarily selected conditions were used, except where otherwise indicated: 1 U AmpliTaq polymerase (Applied Biosystems, Foster City, CA), 2.5 μl 10× buffer II (100 mM Tris-HCl, pH 8.3, 500 mM KCl), 2.5 μl dimethyl sulfoxide (DMSO), 1.0 mM MgCl2, 0.5 μM primers, and 200 μM deoxynucleoside triphosphates, in a final volume of 25 μl. The cycling conditions were originally described as 35 cycles of 1 min at 95°C, 2 min at 50°C, and 2 min at 72°C. The modified conditions used in the present study for initial testing were 5 min at 95°C, 35 cycles of 1 min at 95°C, 30 s at 50°C, and 30 s at 72°C, followed by a final extension step of 7 min at 72°C. Alterations to these biochemical and cycling conditions were tested as detailed in Results.
As a positive control, a 16S rRNA-specific PCR was carried out on all samples determined to be negative with the N-avium/C-avium primer set. The forward primer (5′-AGAGTTTGATCCTGGCTCAG-3′), designated univ16S-3, is homologous to a highly conserved sequence from the 5′ end of the 16S rRNA gene and has been previously described (5, 23). The reverse primer (5′-GCGGCTGCTGGCACG-3′), designated univ16S-4, was selected from highly conserved sequence between the third and fourth variable regions of the 16S rRNA gene. This primer set generates an amplicon of approximately 520 bp from multiple isolates of the 20 species of bacteria with which it has so far been tested (K. B. Register, unpublished data). Reactions were carried out in a volume of 25 μl and contained 1 U AmpliTaq polymerase, 200 μM deoxynucleoside triphosphates, 0.5 μM each primer, 1.5 mM MgCl2, and 2.5 μl of 10× buffer II. An initial denaturation step of 1 min at 95°C was followed by 35 cycles of 1 min at 95°C, 30 s at 53°C, and 30 s at 72°C, with a final extension step of 5 min at 72°C.
Ten microliters of each PCR was analyzed by agarose gel electrophoresis in 3:1 NuSieve (Cambrex BioScience Rockland, Inc., Rockland, ME) containing 0.5 μg/ml ethidium bromide.
Performance characteristics.
PCR performance characteristics were established based on standardized guidelines (8, 14). Assay sensitivity was defined as follows: true positives/(true positives + false negatives) × 100. Assay specificity was defined as follows: true negatives/(true negatives + false positives) × 100. The limit of detection per mass of DNA was determined, and results were converted to genome number based on data from the Sanger Institute's B. avium genome sequencing project (http://www.sanger.ac.uk/Projects/B_avium).
DNA sequencing.
PCR products were purified with spin columns (QIAGEN, Valencia, CA) and sequenced directly by fluorescence-based cycle sequencing with AmpliTaq and BigDye Terminators on an ABI 377 sequencer at the National Animal Disease Center Genomics Unit. Sequences were analyzed using Vector NTI Suite software (Invitrogen, Carlsbad, CA). Final sequences are the results of a minimum of three reactions with at least one sequence from each strand.
Nucleotide sequence accession numbers.
GenBank accession numbers for sequences reported here are AY925018 to AY925089.
RESULTS
Optimization of reaction conditions.
In initial experiments, results obtained with boiled lysates from six randomly chosen B. avium strains were compared using either the original cycling conditions or the more typical modified cycling conditions detailed above. Amplicons of the predicted mobility were detected from all samples; however, in most cases the modified cycling conditions resulted in higher yields. Accordingly, the modified cycling program (henceforth designated program A) was adopted as a starting point for further testing.
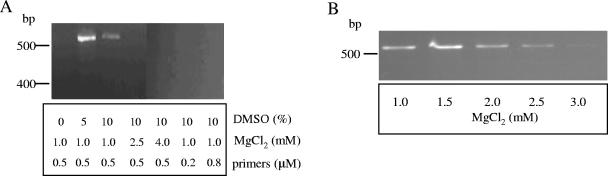
The limit of detection of program A with various amounts of purified B. avium chromosomal DNA, ranging from 25 pg to 1,300 pg, was estimated to be between 25 pg and 260 pg (K. B. Register, unpublished data). The effects of altering the concentration of DMSO, MgCl2, or primers on product yield of PCRs containing template amounts within this range were next compared. Optimal concentrations were identified as 5% DMSO, 0.5 μM primers, and 1.5 mM MgCl2 (Fig. 1).
FIG. 1.
Effect of DMSO, MgCl2, and primer concentrations on the yield of the N-avium/C-avium amplicon. A) One hundred picograms of B. avium chromosomal DNA was amplified in the presence of the DMSO, MgCl2, and primer concentrations indicated. B) One hundred twenty picograms of B. avium chromosomal DNA was amplified in the presence of 5% DMSO, 0.5 μM primers, and the MgCl2 concentration indicated. Other conditions were as described in the text.
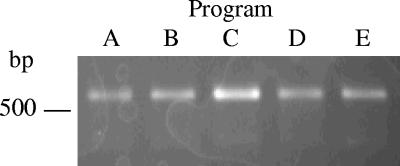
In subsequent experiments, the effects of altering individual steps of program A on product yield were compared in a similar manner. The cycling conditions evaluated are outlined in Table 2. Each program was tested with 65 pg, 100 pg, 120 pg, or 260 pg of the template, and the presence and intensities of the resulting amplicons were compared. Results are summarized in Table 3. When 65 pg of template was used, no detectable product was generated with any program. Program D, designed to test the effect of the originally reported annealing time on yield, was the only program that failed to result in detectable product with 100 pg of template. The replacement of program A with the originally reported extension time (program E) did not appear to affect the results. Similarly, no discernible effect was noted when the initial denaturation time was shortened from 5 min to 2 min (program B). Only program C, which shortened the cycle denaturation time from 1 min to 30 s, resulted in a consistent increase in product intensity with all template amounts tested. Representative results obtained with 260 pg of template are shown in Fig. 2. Based on these data, program C was selected for standard use.
TABLE 2.
Programs tested with the N-avium/C-avium primer pair
| Step | Time, temp (°C) ina:
|
||||
|---|---|---|---|---|---|
| Program A | Program B | Program C | Program D | Program E | |
| Initial denaturation | 5 min, 95 | 2 min, 95 | 5 min, 95 | 5 min, 95 | 5 min, 95 |
| 35 cycles: | |||||
| Denaturation | 1 min, 95 | 1 min, 95 | 30 s, 95 | 1 min, 95 | 1 min, 95 |
| Annealing | 30 s, 50 | 30 s, 50 | 30 s, 50 | 2 min, 50 | 30 s, 50 |
| Extension | 30 s, 72 | 30 s, 72 | 30 s, 72 | 30 s, 72 | 2 min, 72 |
| Final extension | 5 min, 72 | 5 min, 72 | 5 min, 72 | 5 min, 72 | 5 min, 72 |
Steps in bold indicate alterations from program A.
TABLE 3.
Effect of cycling program modifications on product yield
| Amt of template (pg) | Fluorescence intensity of ampliconsa in program:
|
||||
|---|---|---|---|---|---|
| A | B | C | D | E | |
| 65 | − | − | − | − | − |
| 100 | ++ | ++ | +++ | − | ++ |
| 120 | ++ | ++ | +++ | + | ++ |
| 260 | ++ | ++ | +++ | ++ | ++ |
The fluorescence intensity of amplicons resulting from program A was arbitrarily designated (++) for all template amounts tested. Relative intensities of amplicons from other PCRs carried out with the same amount of template are indicated as increased (+++) or decreased (+). −, indicates no fluorescence.
FIG. 2.
Effect of different cycling conditions on the yield of the N-avium/C-avium amplicon. Two hundred sixty picograms of B. avium chromosomal DNA was amplified using the program indicated, as outlined in Table 2. Other conditions were as described in the text.
The originally reported size of the N-avium/C-avium amplicon is 500 bp (19). However, it was noted that the mobility of the amplicon from the B. avium isolates included in our study was consistent with a slightly larger fragment. Examination of the DNA sequence from the original GenBank entry revealed that the actual length of the amplicon is 528 bp.
Performance characteristics.
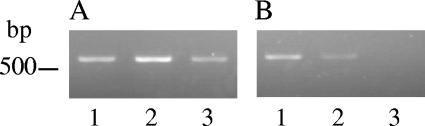
Under the optimized cycling and biochemical conditions identified above, a preliminary estimate of 80 pg to 100 pg was obtained for the limit of detection with the N-avium/C-avium primer set (K. B. Register, unpublished data). A single B. avium genome is predicted to have a mass of 0.004 pg based on the reported genome size of 3,732,255 bp, found at the Sanger Institute website (http://www.sanger.ac.uk/Projects/B_avium/). This corresponds to a relatively high limit of detection of roughly 20,000 to 24,000 genomes. A BLAST search of the B. avium genome assembly using the N-avium and C-avium primer sequences identified numerous potential secondary binding sites that include stretches of 10 to 14 consecutive bases with 100% homology to the primers. These sites do not result in the generation of secondary amplification products but may negatively affect the limit of detection by decreasing the number of primer molecules available for binding to the target gene. It was hypothesized that increasing the annealing temperature of the PCR might lower the limit of detection by preventing at least a portion of these illegitimate primer binding events. The product yield from PCRs in which the annealing temperature was raised by 2°C increments indicated that 52°C is optimal over a range of template concentrations; results obtained using 100 pg are shown in Fig. 3A. Under these conditions, the limit of detection is between 15 pg and 20 pg (Fig. 3B), corresponding to 3,750 to 5,000 genomes.
FIG. 3.
Limit of detection for the N-avium/C-avium amplicon. A) One hundred picograms of purified B. avium DNA was used as the template in optimized PCRs as described in the text, with an annealing temperature of 50°C (lane 1), 52°C (lane 2), or 54°C (lane 3). B) Optimized PCRs using an annealing temperature of 52°C and 40 pg (lane 1), 20 pg (lane 2), or 15 pg (lane 3) of purified B. avium DNA.
Assay sensitivity was determined by testing lysates from 72 B. avium isolates, obtained from a variety of geographic locations and over a span of at least 25 years (Table 1). A single band of the predicted size was generated from all isolates. Thus, the assay sensitivity was defined as 100%.
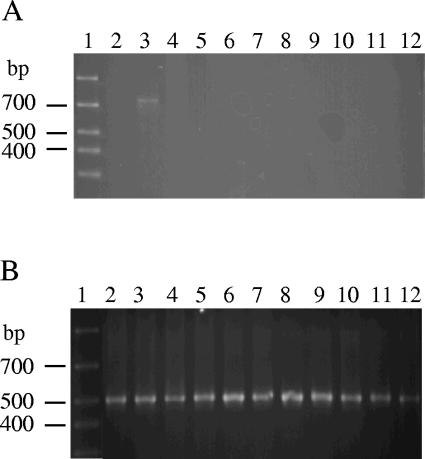
The specificity of the N-avium/C-avium primer set was assessed with a variety of avian bacterial isolates, including both pathogens and the normal flora isolated from tracheal swabs (Table 1). In particular, it was important to determine whether the primer set amplifies DNA from B. hinzii, since many of those isolates had been misidentified as B. avium or B. avium-like prior to the recent establishment of B. hinzii as a unique species. None of the 47 B. hinzii strains tested with the N-avium/C-avium primer pair displayed a product, although a 16S rRNA-specific amplicon was apparent for all when the univ16S-3/univ16S-4 primer set was used as a positive control. All but 1 of the additional 40 bacterial isolates evaluated were likewise negative with the N-avium/C-avium primer set and positive with the univ16S-3/univ16S-4 primer set. Lysate from one of the two Staphylococcus hyicus isolates tested resulted in a weak but reproducible band of approximately 750 bp, easily distinguishable from the smaller B. avium-specific band. The outcome was identical when new lysates from both strains were retested. Attempts to eliminate the product by increasing the annealing temperature were not successful. Representative results are shown in Fig. 4. Based on these data, the specificity of the N-avium/C-avium PCR is 98.8%.
FIG. 4.
Specificity of the N-avium/C-avium primer set. Representative results obtained from PCRs with the N-avium/C-avium primer set (A) or the univ16S-3/univ16S-4 primer set (B). Lanes: 1, marker; 2, S. hyicus 14-4A; 3, S. hyicus 1-C; 4. Staphylococcus aureus; 5, Staphylococcus epidermis; 6, Enterococcus cecorum; 7, Micrococcus luteus; 8, Escherichia coli; 9, Acinetobacter baumannii; 10, P. multocida; 11. Ornithobacterium rhinotracheale; 12, B. hinzii.
The reproducibility of the PCR was evaluated by comparing the results using genomic DNA preparations, derived from several different B. avium isolates, multiple times over a period of several weeks. Each set of PCRs included a range of template amounts, from 20 pg to 260 pg. Both positive and negative results were 100% identical each time the assays were carried out. Reproducibility was also determined using cell lysates from approximately one-third of the B. avium isolates as well as lysates from roughly half of the other genera and species included in this study. There was 100% concordance among reaction mixtures prepared from replicate lysates of the same isolate. Since lysates contain supernatant sufficient for at least three PCRs, reproducibility using a single lysate in individual PCRs set up on consecutive days was also determined. Results were identical with lysates up to 2 days old; lysates from B. avium isolates stored for 3 days or longer prior to use occasionally gave rise to false-negative results with the N-avium/C-avium primer set. In most cases, false-negative results were also seen with the 16S rRNA primer pair.
DNA sequence analysis of N-avium/C-avium amplicons.
No alterations in the mobility of the N-avium/C-avium amplicon were noted among the 72 B. avium strains evaluated. However, the DNA sequence of each amplicon was determined in order to identify any polymorphisms that might be present. Three single-base polymorphisms were noted, which combine to result in four unique sequence variants differing by one or two base substitutions (Fig. 5). None of the sequences is identical to the originally reported sequence, which contains two substitutions and two single-base deletions not noted in the other four variants.
FIG. 5.
DNA sequence variants identified in the region amplified by the N-avium/C-avium primer pair. Dashes indicate conserved bases, asterisks indicate deleted bases, and substitutions are indicated by the appropriate letter, compared to the sequence defined as variant A. The sequence originally reported for this amplicon by Savelkoul et al. (19) was derived from GenBank accession no. X74117.
The sequence designated variant A was found in 49 of the isolates evaluated (68.0%). These include isolates acquired from all the geographic locations listed in Table 1 and from all years of isolation represented in this study. Variant B occurred in only one isolate (1.4%), obtained at least 25 years ago in North Carolina. Four isolates (5.6%) share the sequence designated variant C; all originated in Germany at least 25 years ago. Variant D was found in 18 isolates (25.0%). Seven were acquired in 1998 from wild ducks sampled in New Jersey, while the remaining 11 were cultured in 2004 from turkeys on four different Iowa farms. The minimal sequence variation noted among all isolates indicates that the region amplified by the N-avium/C-avium primer set is sufficiently stable to be a reliable target for PCR.
DISCUSSION
In the present study, we sought to optimize and determine the performance characteristics of a PCR for the identification of B. avium. Because the original reaction conditions used with the N-avium/C-avium primers were not clearly stated (19), a standard set was arbitrarily chosen for our initial evaluation and used as a starting point for optimization. Comparisons were carried out with template amounts approaching the limit of detection, since small differences in yield were more readily apparent when the target was not present in great excess. It was expected that annealing and extension times shorter than those originally used (2 min each) would decrease the limit of detection because of a reduction in thermal inactivation of the DNA polymerase. However, only a shorter annealing step had this effect. Nonetheless, there was no obvious detriment to also limiting the duration of the extension step. Thirty seconds approximates the general rule of allowing 1 min of extension time/kb synthesized (6), and this permits the PCR to be completed in a shorter period of time. Accordingly, 30 seconds was adopted for both the annealing and extension steps.
Assay verification and validation are essential when implementing a new diagnostic procedure. Analytical verification is defined as the process of establishing performance characteristics, such as limit of detection, sensitivity, specificity, and reproducibility (8, 14). Data reported here indicate that the N-avium/C-avium PCR can be expected to provide clinically significant results with a high degree of sensitivity, specificity, and reproducibility compared to those of the gold standard of culture and biochemical testing. The ability to evaluate suspect colonies without prior purification of DNA is advantageous, delivering results more quickly and with less cost.
Even under optimal cycling conditions, the limit of detection found with the N-avium/C-avium primer set is somewhat disappointing. Although the optimized PCR described is useful for testing single colonies, identification of B. avium directly from clinical samples, without previous culture, cannot be recommended at this time since swabs from infected poultry may not always contain a sufficiently high number of organisms. Given that amplification efficiency is inversely proportional to amplicon size (18), it may be possible to lower the limit of detection by replacement of one or both primers such that a smaller region of the target is amplified. Based on a search ofthe recently completed B. avium genome sequence, this target is in a single-copy gene (http://www.sanger.ac.uk/Projects/B_avium/private/). Through additional analysis of the genome, it may be possible to identify novel B. avium-specific gene sequences that are present in multiple copies, permitting the development of an alternative PCR with a lower limit of detection.
B. hinzii, referred to as B. avium-like or as Alcaligenes faecalis type II prior to 1995 (22), is commonly acquired from the respiratory tracts of diseased poultry but has not been demonstrated to be pathogenic for birds (2, 9, 11). Consequently, differentiating between B. avium and B. hinzii is of importance for veterinary diagnostic laboratories. Only a few phenotypic tests have been shown to delineate these species, and they do not correctly identify all strains (22). Moreover, results of some may vary depending on inoculum size, culture conditions, and the procedure used (1, 3, 7, 12, 22). Ribotyping and restriction enzyme analysis have recently been found to reliably distinguish between B. avium and B. hinzii (16, 17) but are not readily carried out in most diagnostic laboratories. Based on the specificity of the N-avium/C-avium PCR demonstrated in this study, the assay can be recommended as an alternative means to discriminate between these species that is more readily incorporated into the repertoire of a diagnostic laboratory. It is noteworthy that three of the B. hinzii isolates included in this study were misidentified as B. avium or B. avium-like at the time of their isolation. Ribotyping and restriction enzyme analysis correctly identified them as B. hinzii (16); negative results were obtained with these isolates using the N-avium/C-avium PCR.
In recent years sequence polymorphisms have been discovered in selected virulence genes, including fimbrial genes, from several species of Bordetella (4, 13, 15, 21). The B. avium DNA sequence immediately upstream of the region targeted by the N-avium/C-avium primers was previously reported to have 80% homology to the region just upstream of the Bordetella pertussis fimbrial gene fim-2 (19). However, the amplicon sequence itself bears little resemblance to any known Bordetella fimbrial gene, and the currently available annotation of the B. avium genome indicates some similarity to the C-terminal region of putative transcriptional regulators from other bacteria, including other species of Bordetella. Nevertheless, since target sequence stability is important for reliable PCR identification of an infectious agent, we determined the sequence of the N-avium/C-avium amplicon from all B. avium isolates included in this study. A majority of the isolates had no sequence variation, with only three point mutations identified from those remaining. Since the isolates originated from a variety of geographic locations and over an extended period of time, sequence polymorphisms are unlikely to interfere with the reliability of the N-avium/C-avium PCR.
Acknowledgments
We gratefully acknowledge the technical assistance of Michael Mullins, Gwen Nordholm, and Pamala Beery. We are indebted to David Alt and the National Animal Disease Center Genomics Unit for DNA sequence data. Genome sequence data for B. avium was generated by the Pathogen Sequencing Unit (PSU) at the Sanger Institute and is available from http://www.sanger.ac.uk/Projects/B_avium. We are grateful to the PSU of the Sanger Institute for access to unpublished annotation data.
REFERENCES
- 1.Berkhoff, H. A., and G. D. Riddle. 1984. Differentiation of Alcaligenes-like bacteria of avian origin and comparison with Alcaligenes spp. reference strains. J. Clin. Microbiol. 19:477-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blackall, P. J., and C. M. Doheny. 1987. Isolation and characterisation of Bordetella avium and related species and an evaluation of their role in respiratory disease in poultry. Aust. Vet. J. 64:235-239. [DOI] [PubMed] [Google Scholar]
- 3.Blackall, P. J., and J. G. Farrah. 1986. An evaluation of two methods of substrate alkalinization for the identification of Bordetella avium and other similar organisms. Vet. Microbiol. 11:301-306. [DOI] [PubMed] [Google Scholar]
- 4.Boursaux-Eude, C., and N. Guiso. 2000. Polymorphism of repeated regions of pertactin in Bordetella pertussis, Bordetella parapertussis, and Bordetella bronchiseptica. Infect. Immun. 68:4815-4817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cai, H., M. Archambault, and J. F. Prescott. 2003. 16S ribosomal RNA sequence-based identification of veterinary clinical bacteria. J. Vet. Diagn. Investig. 15:465-469. [DOI] [PubMed] [Google Scholar]
- 6.Grunenwald, H. 2003. Optimization of polymerase chain reactions, p. 89-99. In J. M. Bartlett and D. Stirling (ed.), PCR protocols, 2nd ed. Humana Press, Totowa, N.J.
- 7.Hinz, K.-H., and G. Glünder. 1986. Identification of Bordetella avium sp. nov. by the API 20NE system. Avian Pathol. 15:611-614. [DOI] [PubMed] [Google Scholar]
- 8.Hoorfar, J., and N. Cook. 2003. Critical aspects of standardization of PCR, p. 51-64. In K. Sachse and J. Frey (ed.), PCR detection of microbial pathogens. Humana Press, Totowa, N.J. [DOI] [PubMed]
- 9.Jackwood, M. W., S. M. McCarter, and T. P. Brown. 1995. Bordetella avium: an opportunistic pathogen in Leghorn chickens. Avian Dis. 39:360-367. [PubMed] [Google Scholar]
- 10.Jackwood, M. W., and Y. M. Saif. 2003. Bordetellosis (turkey coryza), p. 705-718. In Y. M. Saif (ed.), Diseases of poultry, 11th ed. Iowa State Press, Ames, Iowa.
- 11.Jackwood, M. W., Y. M. Saif, P. D. Moorhead, and R. N. Dearth. 1985. Further characterization of the agent causing coryza in turkeys. Avian Dis. 29:690-705. [PubMed] [Google Scholar]
- 12.Kattar, M. M., J. F. Chavez, A. P. Limaye, S. L. Rassoulian-Barrett, S. L. Yarfitz, L. C. Carlson, Y. Houze, S. Swanzy, B. L. Wood, and B. T. Cookson. 2000. Application of 16S rRNA gene sequencing to identify Bordetella hinzii as the causative agent of fatal septicemia. J. Clin. Microbiol. 38:789-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mooi, F. R., H. van Oirschot, K. Heuvelman, H. G. J. van der Heide, W. Gaastra, and R. J. Willems. 1998. Polymorphism in the Bordetella pertussis virulence factors P.69/pertactin and pertussis toxin in The Netherlands: temporal trends and evidence for vaccine-driven evolution. Infect. Immun. 66:670-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nolte, F. S., and A. M. Caliendo. 2003. Molecular detection and identification of microorganisms, p. 234-256. In P. R. Murray, E. J. Baron, J. H. Jorgensen, M. A. Pfaller, and R. H. Yolken (ed.), Manual of clinical microbiology, 8th ed. American Society for Microbiology, Washington, D.C.
- 15.Register, K. B. 2001. Novel genetic and phenotypic heterogeneity in Bordetella bronchiseptica pertactin. Infect. Immun. 69:1917-1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Register, K. B., R. E. Sacco, and G. E. Nordholm. 2003. Comparison of ribotyping and restriction enzyme analysis for inter- and intraspecies discrimination of Bordetella avium and Bordetella hinzii. J. Clin. Microbiol. 41:1512-1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sacco, R. E., K. B. Register, and G. E. Nordholm. 2000. Restriction enzyme analysis and ribotyping distinguish Bordetella avium and Bordetella hinzii isolates. Epidemiol. Infect. 124:83-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sachse, K. 2003. Specificity and performance of diagnostic PCR assays, p. 3-29. In K. Sachse and J. Frey (ed.), PCR detection of microbial pathogens. Humana Press, Totowa, N.J. [DOI] [PubMed]
- 19.Savelkoul, P. H., L. E. de Groot, C. Boersma, I. Livey, C. J. Duggleby, B. A. van der Zeijst, and W. Gaastra. 1993. Identification of Bordetella avium using the polymerase chain reaction. Microb. Pathog. 15:207-215. [DOI] [PubMed] [Google Scholar]
- 20.Simmons, D. G., and J. G. Gray. 1979. Transmission of acute respiratory disease (rhinotracheitis) of turkeys. Avian Dis. 23:132-138. [PubMed] [Google Scholar]
- 21.Tsang, R. S., A. K. Lau, M. L. Sill, S. A. Halperin, P. Van Caeseele, F. Jamieson, and I. E. Martin. 2004. Polymorphisms of the fimbria fim3 gene of Bordetella pertussis strains isolated in Canada. J. Clin. Microbiol. 42:5364-5367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vandamme, P., J. Hommez, M. Vancanneyt, M. Monsieurs, B. Hoste, B. Cookson, C. H. Wirsing von Konig, K. Kersters, and P. J. Blackall. 1995. Bordetella hinzii sp. nov., isolated from poultry and humans. Int. J. Syst. Bacteriol. 45:37-45. [DOI] [PubMed] [Google Scholar]
- 23.Wilmotte, A., G. Van der Auwera, and R. De Wachter. 1993. Structure of the 16 S ribosomal RNA of the thermophilic cyanobacterium Chlorogloeopsis HTF (′Mastigocladus laminosus HTF') strain PCC7518, and phylogenetic analysis. FEBS Lett. 317:96-100. [DOI] [PubMed] [Google Scholar]