Abstract
Currently available methods for the diagnosis of cutaneous leishmaniasis (CL) have low sensitivities or are unable to quantify the number of viable parasites. This constitutes a major obstacle for the diagnosis of the disease and for the study of the effectiveness of treatment schedules and urges the development of improved detection methods. In this study, quantitative nucleic acid sequence-based amplification (QT-NASBA) technology was used to detect and quantify Leishmania parasites in skin biopsy samples from CL patients. The assay is based on the detection of a small subunit rRNA (18S rRNA), which may allow for the detection of viable parasites. The QT-NASBA assay was evaluated using in vitro-cultured promastigotes and amastigotes and 2-mm skin biopsy samples from Old and New World CL patients. The study demonstrated that the lower detection limit of the QT-NASBA was two parasites per biopsy sample. Parasites could be quantified in a range of 2 to 11,300,000 parasites per biopsy sample. The QT-NASBA could detect levels of parasites 100-fold lower than those detected by conventional PCR. Test evaluation revealed that the QT-NASBA had a sensitivity of 97.5% and a specificity of 100% in the present study. The QT-NASBA is a highly sensitive and specific method that allows quantification of both Old and New World Leishmania parasites in skin biopsy samples and may provide an important tool for diagnosis as well as for monitoring the therapy of CL patients.
Leishmania comprises a genus of flagellate protozoan parasites with a worldwide distribution and with more than 20 species that are pathogenic in humans. The parasites are transmitted by the bite of infected female sand flies of the genera Phlebotomus and Lutzomyia. Leishmaniasis prevalence is estimated to be 12 million people, and, in developing countries, approximately 350 million people, mainly poor, are at risk of contracting the disease (33). The infection can cause a wide range of clinical manifestations and is considered to be one of the most threatening parasitic diseases for humans. Nowadays, more and more nonimmune humans are entering areas where leishmaniasis is endemic (for example, refugees in countries with civil unrest or forces on military missions) (2), and there is an evident increase of patients with human immunodeficiency virus type 1 being coinfected with Leishmania parasites (1).
Cutaneous leishmaniasis (CL) is the clinical manifestation in which the parasite causes one or more slow-healing ulcers on the skin. Geographically, it can be divided into Old World (southern Europe, Africa, the Middle East, and west and central Asia) and New World (southern United States to northern Argentina) leishmaniases (27). The Old World Leishmania species cause benign and often self-limiting ulcers. The New World species cause manifestations of greater variety, of which mucocutaneous leishmaniasis (MCL) is the most severe. MCL is most likely the result of the spreading of parasites from cutaneous lesions to the mucosal membranes, where they can progressively destroy connective tissue, such as cartilaginous structures, in the nasopharyngeal tract (9).
Early and accurate diagnosis and effective treatment is important to avoid the development of long-lasting chronic disease, disfiguring scars, and, in the case of MCL, extensive damage to the nasopharynx. Traditionally, the diagnosis of CL or MCL is based on microscopic demonstration or culturing of Leishmania parasites from skin biopsy samples or aspirates from lesions. However, these methods are rather insensitive, and culturing of parasites is time-consuming and laborious (21). In areas of endemicity, CL is most frequently diagnosed by epidemiological and clinical evaluation, because most conventional diagnostic methods are available only in reference laboratories. In contrast to its usefulness for the diagnosis of visceral leishmaniasis, which is the systemic manifestation of leishmaniasis, serology is not very useful for the diagnosis of CL, because antibodies tend to be undetectable or present in low titers (8, 18).
In the last decades, several PCR assays have been developed for the detection of Leishmania parasites. These assays often have high sensitivities (96% to 100%) (6, 7). In countries where PCR is available, it has already become an important diagnostic tool (7). However, most PCRs are based on DNA amplification, which does not provide information on the viability of the pathogen. For example, bacterial DNA has been detected several weeks after the pathogens had been killed (10, 28). DNA of Leishmania parasites can be detected in scars of CL patients even months after clinical cure (14, 24).
A sensitive tool to detect viable parasites would be very useful in improving the diagnosis of leishmaniasis. Furthermore, such an assay would make it possible to monitor the effectiveness of treatment schemes. Current treatment schemes have several shortcomings due to variation in the sensitivities of Leishmania species, different clinical pictures, and emerging drug resistance (5). There is also no gold standard to decide whether a patient is cured. Up to this date, a patient is declared cured on the basis of clinical criteria, which may delay the detection of a treatment failure.
In the present study, we have employed quantitative nucleic acid sequence-based amplification (QT-NASBA) technology to develop a tool for the diagnosis of leishmaniasis and for the quantification of Leishmania parasites. QT-NASBA is based on the amplification of single-stranded RNA sequences without the interference of DNA and has been applied for several infectious diseases, such as human immunodeficiency virus, mycobacterial diseases, and malaria (20, 23, 29, 31). This technology has proven to be highly sensitive and specific and is able to detect very low target levels in clinical samples (23). Moreover, the detection of specific RNA molecules may serve as a suitable marker for pathogen viability (26, 29). The QT-NASBA assay already has been successfully applied in studies to assess the efficacy of drug therapies for Plasmodium falciparum infection and to predict treatment outcomes (16, 19).
In this study, we have developed an 18S rRNA QT-NASBA for the detection and quantification of Leishmania parasites in in vitro cultures of promastigotes and amastigotes and in skin biopsy samples from patients with CL diagnosed in areas where it is endemic and areas where it is not. We compared the 18S rRNA QT-NASBA results with those of a standard 18S rRNA PCR assay running in our laboratory (30).
MATERIALS AND METHODS
In vitro-cultured promastigotes.
Leishmania (Viannia) braziliensis guyanensis (MHOM/BR/75/M4147) and L. donovani (MHOM/SD/68/1S) were cultured at 25°C in RPMI 1640 medium (Gibco, Scotland) supplemented with 15% fetal calf serum. Parasites were subcultured weekly.
In order to determine the lower detection limit of the developed NASBA assay and the number of rRNA molecules in promastigotes for the calculation of the number of parasites in biopsy samples, promastigote culture samples were analyzed. Two samples were collected from 3-day-old cultures of L. donovani and L. (V.) braziliensis guyanensis promastigotes. The parasites were harvested, counted (using a Neubauer hemacytometer), and resuspended in phosphate-buffered saline (PBS) at a concentration of 103 parasites per μl. One hundred μl was added to 900 μl of guanidinium isothiocyanate L6 lysis buffer (3), and after RNA extraction (see “RNA/DNA extraction” below), 10-fold dilutions were made (103, 102, 101, and 1 parasites per μl). Additionally, four serial dilutions of L. donovani promastigotes were made by using blood samples with 107, 105, 104, 103, 102, and 0 parasites per ml of blood.
In vitro generation of amastigotes.
Human monocyte cell line U-937 was used for the in vitro generation of amastigotes (12). U-937 cells were maintained at 37°C and 5% CO2 in RPMI 1640 medium containing fetal calf serum (10%). After 3 days, monocytes were counted, harvested, and resuspended in RPMI medium containing 10 ng/ml phorbol myristate acetate (Sigma, St. Louis, MO). Three Falcon chamber slides (Falcon, Oxnard, CA) with four chambers each were filled with 150,000 monocytes and incubated for 48 h at 37°C and 5% CO2. Phorbol myristate acetate stimulates the monocytes to differentiate into an adherent phagocytic cell line. After 48 h, the differentiated cells were infected with 3,000,000 promastigotes of L. (V.) braziliensis guyanensis in stationary phase for 16 h at 37°C at a parasite-to-host cell ratio of 20 to 1. Next, the phagocytic cells were washed three times with PBS to remove noninvading parasites, and fresh medium was added. The cells in the three chamber slides were maintained at 37°C and 5% CO2 for 1, 4, and 8 h, respectively. After incubation, the cells were washed again three times with PBS. Cells in two of the four chambers of each slide were lysed by adding 950 μl L6 lysis buffer. The lysate was transferred to an Eppendorf tube and stored at −70°C until further analysis. Cells in the remaining two chambers of the slide were fixed in methanol and stained with Giemsa (Merck, Darmstadt, Germany), and subsequently, >400 macrophages were counted.
Skin biopsy samples.
All biopsy samples were taken from the active edge of the lesion by dermatologists using sterile disposable 2-mm-diameter skin biopsy sample punchers. In order to make sure that all biopsy samples were collected according to the same standard, particular attention was paid to the fact that the complete 4-mm length of the biopsy sample was collected rather than only part of the sample.
In total, 55 skin biopsy samples of 2-mm diameter were collected from 37 individuals diagnosed with CL at the dermatology clinic of the Academic Medical Center, Amsterdam, The Netherlands, before (n = 33) and after (n = 22) treatment. In addition, one needle aspiration sample was collected from the lymph node of a CL patient with lymph node involvement. Patient characteristics and probable areas of infection are presented in Table 1. The diagnosis of CL was based on demonstrating the presence of Leishmania parasites in skin biopsy samples by means of staining of smears, culturing, histopathological examination, and/or PCR.
TABLE 1.
Characteristics of 37 CL patients (skin biopsy samples collected before and/or after treatment) who visited the outpatient clinic of the Department of Dermatology, AMC
| Characteristic | Median (range) | No. (%) |
|---|---|---|
| Age (yr) | 32 (10-79) | |
| Male gender | 27 (73) | |
| Probable area of Leishmania infection | ||
| Central America | 14 (37.8) | |
| South America | 11 (29.8) | |
| Middle East | 6 (16.2) | |
| Southern Europe | 6 (16.2) |
Another 44 skin biopsy samples were collected from CL patients residing in countries of endemicity in the New and Old Worlds (Table 2). Twenty-five samples were collected at the Cutaneous Leishmaniasis Center, Sanliurfa City, Turkey; eight samples at the Bolan Medical College Complex Hospital, Quetta, Balochistan, Pakistan; six samples at the Academic Hospital, Paramaribo, Suriname; and five samples at the Fundação de Medicina Tropical do Amazonas (FMTAM), Manaus, Brazil. All samples were collected before any antiprotozoan drug was administered to the patient.
TABLE 2.
Skin biopsy samples collected from CL patients in areas of nonendemicity and endemicity before and after treatment (n = 99)
| Countrya | No. of CL samples collected before/after treatment
|
|||
|---|---|---|---|---|
| New World
|
Old World
|
|||
| Before | After | Before | After | |
| The Netherlands | 21 | 21 | 12 | 1 |
| Turkey | 25 | 0 | ||
| Pakistan | 8 | 0 | ||
| Brazil | 5 | 0 | ||
| Suriname | 6 | 0 | ||
Turkey, Pakistan, Brazil, and Suriname are countries of endemicity, whereas The Netherlands is a country of nonendemicity.
As negative controls, 12 skin biopsy samples were collected from individuals with normal skin undergoing reconstructive surgery, and 13 skin biopsy samples were collected from patients with other skin diseases, such as sarcoidosis (n = 1), ulcerating pyoderma (n = 5), aspecific dermatitis (n = 1), folliculitis (n = 1), folliculitis keloidalis (n = 1), dermatofibroma (n = 1), and persistent insect bites (n = 3). All the skin biopsy samples were individually collected in tubes filled with 950 μl L6 lysis buffer and stored at −20°C until the RNA isolation procedure was performed.
RNA/DNA extraction.
RNA and DNA were extracted from the samples according to the method of Boom et al. (3). Briefly, each 2-mm skin biopsy sample was added to 950 μl of L6 lysis buffer. To each sample, 106 molecules of quantification RNA (Q-RNA) were added before nucleic acid isolation (see “Cloning and in vitro transcription” below). To disrupt the tissue, one 5-mm-diameter stainless steel bead (Retsch) was added to a tube containing a skin biopsy sample, which was subsequently placed in an MM 301 mixer mill (Retsch GmbH & Co. KG) and shaken for 300 seconds at a frequency of 30 Hz. Next, the supernatant was transferred to a new tube, and 30 μl of silica suspension was added. After washing and drying of the silica, the nucleic acids were eluted in 50 μl nuclease-free water and stored at −20°C. For every five skin biopsy samples, negative controls (water or negative blood) were included in order to assess carryover contamination.
(i) Cloning and in vitro transcription.
Primers R174 and R798, designed by Van Eys et al. (30), were used to amplify a 600-bp region of the Leishmania 18S rRNA gene (Table 3). The amplified product was cloned directly into plasmid pCR2.1-TOPO (Invitrogen, Carlsbad, Calif.) according to the manufacturer's instructions. After purification, the plasmids were cut with the restriction enzyme PvuII, and a large amount of in vitro wild-type RNA (WT-RNA) was synthesized with an SP6/T7 transcription kit (Boehringer Mannheim).
TABLE 3.
Primers and probes used for the amplification and detection of Leishmania parasites
| Primer or probe | Sequence |
|---|---|
| Primers | |
| R174 | 5′-GGTTCCTTTCCTGATTTACG-3′ |
| R798 | 5′-GGCCGGTAAAGGCCGAATAG-3′ |
| Fp1 (forward primer 1)a | 5′-GATGCA AGG TCG CAT ATG AGC CAA AGT GTG GAG ATC GAA G-3′ |
| Rp2 (reverse primer 2)b | 5′-AAT TCT AAT ACG ACT CAC TAT AGG GAG AAG GGC CGG TAA AGG CCG AAT AG-3′ |
| Capture probes | |
| 18S WT | 5′ biotin-GAC CAT TGT AGT CCA CAC TG-3′ |
| 18S Q | 5′ biotin-CTT AGG TCC ACT AAG GTA CC-3′ |
The region of Fp1 which is presented in bold is for generic ECL detection.
The region of Rp2 which is underlined is a T7 promotor sequence.
For quantification purposes and to serve as an internal control, Q-RNA was constructed by site-directed mutagenesis according to the method of Schoone et al. (23). Before each RNA/DNA extraction, 106 molecules of Q-RNA were added to each sample. This standard allowed for accurate quantification of the parasites (see “Calculations and statistical analysis” below) and served as an internal control for monitoring the efficiency of RNA extraction and the presence of NASBA-inhibiting substances.
(ii) NASBA development.
Primers and probes were based on the target sequence of the 18S rRNA gene identified by Van Eys et al. (30). The sequences of the primers Fp1 and Rp2 and of the WT and Q probes are shown in Table 3. The primers amplify the rRNAs from all Leishmania species, because the 170-bp regions of the rRNA sequences are homologous, according to the published sequence data (30).
The NASBA reaction was performed using a NucliSense basic kit for amplification according to the manufacturer's instructions (bioMérieux) in a 10-μl total reaction volume at a final KCl concentration of 70 mM. The reaction mixture was incubated with 2.5 μl RNA extract at 65°C for 5 min and subsequently at 41°C for 5 min. The isothermal amplification took place for 90 min at 41°C.
(iii) ECL detection of WT- and Q-RNA amplicons.
For quantification, WT-RNA was coamplified with a fixed amount of Q-RNA (internal standard) as a competitor. Amplification products were hybridized (30 min, 41°C) to a ruthenium-labeled generic probe, and the respective capture probes were bound to streptavidin-coated magnetic beads (Table 3). This was followed by electrochemiluminescence (ECL) detection in a NucliSense ECL reader (bioMérieux). Emitted light is detected by a photoelectric cell, offering a precise measurement of amplification products.
Calculations and statistical analysis.
Quantification was achieved by competitive amplification of WT-RNA and Q-RNA molecules in one reaction using the same primers. The ECL provides for each sample a WT-RNA and a Q-RNA signal, and the ratio between these signals is directly correlated to the number of parasites in a sample.
In each experiment, a standard curve of blood samples spiked with in vitro WT-RNA (109, 107, 105, 104, and 0 molecules per 50 μl blood), each mixed with 106 molecules of Q-RNA, was included. Final ECL results were calculated by best-fit regression analysis as described by Schoone et al. (23).
The sensitivities and specificities of the QT-NASBA and PCR in the present study were calculated as follows: sensitivity = TP/(TP + FN) × 100%, and specificity = TN/(TN + FP) × 100%, where TN represents true negative, TP true positive, FN false negative, and FP false positive. The performances of QT-NASBA and PCR were statistically compared using Fisher's exact test.
PCR.
PCR was also performed on the same extractions of the clinical samples that were analyzed with QT-NASBA. The PCR is based on the amplification of a 600-bp region of the 18S rRNA gene (30), and the reaction was performed according to the method of Osman et al. (17). Briefly, 2.5 μl of DNA was amplified in a final volume of 25 μl with primers R174 and R798 (Table 3) for 40 cycles. Amplification conditions were 50°C for 5 min and 94°C for 10 min, followed by 40 cycles of 94°C for 75 s, 60°C for 60 s, and 72°C for 120 s, with a final extension of 72°C for 50 min. In each PCR run, positive and negative samples were included. The PCR products were detected by UV light on a 2% agarose gel stained with ethidium bromide. A 100-bp DNA ladder (Amersham Biosciences, Buckinghamshire, United Kingdom) was used as a marker, and DNA from the reference L. donovani strain, MHOM/SD/68/1S, was used as a positive control.
RESULTS
Sensitivity.
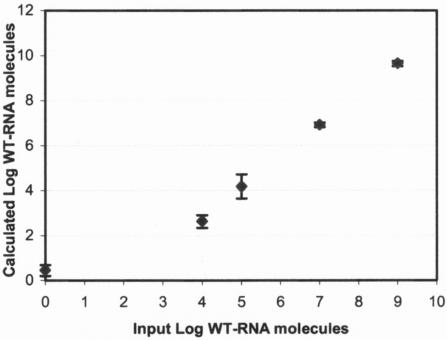
The sensitivity of the QT-NASBA procedure was assessed by performing the assay on serial dilutions of in vitro-generated WT-RNA (samples containing 109, 107, 105, 104, and 0 molecules of WT-RNA per 50 μl blood). A good correlation was obtained between the input of WT-RNA molecules and the number of WT-RNA molecules calculated on the basis of the QT-NASBA (Fig. 1). The correlation coefficient was calculated to be 0.938 (P < 0.001) on the basis of the mean of five independent assays per sample. The interassay variation coefficients were 10.6, 12.9, 1.5, and 1.2% for 104, 105, 107, and 109 molecules/50 μl, respectively. This confirmed the high efficacies of the RNA extraction and the NASBA procedure. The lower detection limit for reliable quantification was at least 104 molecules of RNA per 50 μl blood. The in vitro WT-RNA serial dilutions were used as the standard curve for the quantification of parasites in biopsy samples.
FIG. 1.
Quantification of Leishmania in vitro WT-RNA molecules by QT-NASBA. Serial dilutions of WT-RNA were added to 50 μl blood, and RNA was extracted, which was followed by QT-NASBA. The figure is based on five independent NASBA runs. Error bars indicate standard errors.
Analysis of four serial dilutions of L. donovani promastigotes (samples containing 107, 105, 104, 103, 102, and 0 parasites/ml blood) revealed that the lower detection limit for quantification was at least 102 parasites/ml, which corresponds to 0.1 parasite per μl (data not shown). The relation between the quantification result by QT-NASBA and the log number of microscopically counted promastigotes was significant, with an r2 of 0.959 (P < 0.001). The samples were also tested in conventional PCR and showed a lower detection limit of 104 parasites/ml. This corresponds with 10 parasites per μl (data not shown).
Comparison of promastigotes and amastigotes.
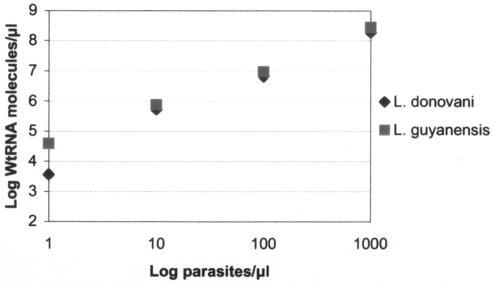
To quantify the number of Leishmania parasites in skin biopsy samples, the number of rRNA copies per parasite had to be calculated. This was achieved by quantitative analysis of serial dilutions of in vitro WT-RNA and promastigotes of L. donovani and L. (V.) braziliensis guyanensis. Comparisons were made between QT-NASBA results for 103, 102, 101, and 1 parasites per μl. Figure 2 shows that the average log difference between the log number of promastigotes and that of WT-RNA was 4.82 (standard deviation = 0.56), corresponding to 63,000 rRNA copies per parasite. The number of parasites in skin biopsy samples was calculated on the basis of this result.
FIG. 2.
Comparison of the numbers of rRNA molecules of in vitro-cultured promastigotes of L. donovani and L. (V.) braziliensis guyanensis with those of a serial dilution of wild-type in vitro rRNA molecules. The average log number of rRNA molecules per parasite was 4.8.
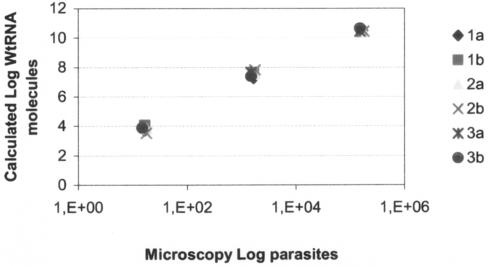
Duplicate samples (n = 3) of in vitro-cultured amastigotes were also analyzed with QT-NASBA. After RNA extraction from the amastigotes, two dilutions were made of each sample (1:100 and 1:1,000). A standard curve with in vitro WT-RNA was included in the analysis. The QT-NASBA results were plotted against the different dilutions and showed that quantification was linear on a 4-log range (Fig. 3). The duplicate curves obtained from the three different samples had similar slopes, and correlation coefficients were greater than 0.998.
FIG. 3.
Dilution of three duplicate samples of in vitro-cultured amastigotes of L. (V.) braziliensis guyanensis in the human cell line U-937. After the extraction procedure, Leishmania RNA was quantified. The number of amastigotes was determined by microscopy.
QT-NASBA results were also compared with amastigote counts obtained by microscopic examination of two Giemsa-stained chambers on each of the three Falcon chamber slides. QT-NASBA counts were compared with the number of amastigotes counted, and an average log difference of 4.78 (standard deviation = 0.47), which corresponds with 60,000 copies of rRNA per parasite, was found.
QT-NASBA and PCR analysis of clinical samples.
Skin biopsy samples collected from untreated (n = 33) and treated (n = 22) individuals with imported CL who visited the outpatient clinic of the Department of Dermatology (Academic Medical Center, The Netherlands) and 44 skin biopsy samples from untreated CL patients from areas of endemicity were analyzed with QT-NASBA and PCR. In addition, a needle aspiration sample collected from the lymph node of a CL patient was also tested. Furthermore, 13 skin biopsy samples from patients with other skin diseases and 12 from healthy individuals were also tested. The lower detection limit of QT-NASBA was 0.1 parasite/μl, and therefore 2 parasites/biopsy sample is considered to be a reliable threshold of quantification in 2-mm skin biopsy samples (2-mm biopsy sample, ∼12.6 μl).
Thirty-three of the 34 samples obtained from untreated patients with imported CL were positive by QT-NASBA (Table 4). Parasite numbers ranged from 2 to 11,300,000 parasites per biopsy sample. QT-NASBA was negative for 16 out of 22 biopsy samples collected from 1 day to 24 weeks after treatment. Six skin biopsy samples were found to be positive after treatment with parasite counts between 2 to 513 parasites per sample. These patients were followed up, and four out of six of them developed relapses. These recurrences were clinically and parasitologically confirmed. Sixteen patients with negative QT-NASBA results did not develop any clinical disease after treatment.
TABLE 4.
QT-NASBA results for skin biopsy samples and one lymph node sample from CL patients and other individuals (n = 125)
| Sample type or country of endemicity | No. (%) positive/range (P/biopsy sample)a | No. (%) negative |
|---|---|---|
| Sample from patient: | ||
| Before treatment | 33 (97)/2-11,300,000 | 1 (3) |
| After treatment | 6 (27)/2-513 | 16 (73) |
| With other skin diseases | 0 (0) | 13 (100) |
| With normal skin | 0 (0) | 12 (100) |
| Country of endemicity | ||
| Turkey | 25 (100)/2-530,000 | 0 (0) |
| Pakistan | 8 (100)/94-15,000 | 0 (0) |
| Brazil | 5 (100)/325-800,000 | 0 (0) |
| Suriname | 5 (83)/193-125,000 | 1 (17) |
P, parasites.
The results of QT-NASBA analysis of skin biopsy samples collected from confirmed CL patients living in four different areas of endemicity are presented in Table 4. Forty-three out of 44 were found positive, and parasite counts ranged from 2 to 800,000 parasites per biopsy sample. One sample from Suriname was negative. QT-NASBA analysis of biopsy samples from 13 patients with skin disease other than Leishmania and from 12 individuals with normal skin found all samples to be negative (Table 4). On the basis of these results, the sensitivity of the QT-NASBA was calculated to be 97.5%, and its specificity was 100%.
All skin biopsy samples were also analyzed with a conventional PCR. The PCR results for all untreated CL patients in comparison to those from QT-NASBA are presented in Table 5. The PCR was positive in 71 out of 78 skin biopsy samples from confirmed CL patients, resulting in a sensitivity of 91.8%. The control biopsy samples (from healthy individuals and patients with other skin diseases) were all negative by PCR (specificity of 100%). Statistical analysis revealed that the sensitivity of QT-NASBA was not significantly higher than the sensitivity found for the PCR employed in the present study (Fisher's P = 0.172). However, it should be noted that five biopsy samples from CL patients negative by PCR were found to be positive with QT-NASBA. These five samples (one from the AMC, two from Turkey, and two from Pakistan) had parasite loads of 2 to 15,000 parasites/biopsy sample. Furthermore, the needle aspiration sample collected from the lymph node of a confirmed CL patient was negative with PCR but positive in QT-NASBA, albeit with a very low parasite count of two parasites. One biopsy sample was PCR positive but QT-NASBA negative. The histopathology of the lesion from which this biopsy sample was taken revealed a granulomatous reaction. Finally, one biopsy sample obtained from an imported case of CL in The Netherlands was negative in both assays. Histopathological analysis of this patient showed superficial and deep perivascular infiltrates in the dermis, consisting of lymphocytes, histiocytes, eosinophils, and few plasma cells. However, Leishmania parasites could not be found. The PCR results of the 22 treated CL patients in The Netherlands in comparison to those from QT-NASBA are presented in Table 5. QT-NASBA found six follow-up samples positive, but in contrast, only three positive PCR results were observed.
TABLE 5.
Comparison of QT-NASBA and PCR results
| QT-NASBA result | PCR result
|
|
|---|---|---|
| Positive | Negative | |
| Before treatmenta | ||
| Positive | 70 | 6 |
| Negative | 1 | 1 |
| After treatmentb | ||
| Positive | 3 | 3 |
| Negative | 0 | 16 |
Results before treatment are for 77 skin biopsy samples and 1 lymph node sample from CL patients from areas of endemicity and nonendemicity.
Results after treatment are for 22 skin biopsy samples from CL patients at the outpatient clinic of the Department of Dermatology.
DISCUSSION
The Leishmania QT-NASBA developed in this study is a highly sensitive and reproducible method for the detection and quantification of parasites in skin biopsy samples (2 to 11,300,000 parasites per sample). The assay is based on the 18S rRNA gene sequence, and this target has been found to be highly efficient for the diagnosis of leishmaniasis from human clinical material (15, 30, 34). Each parasite contains a large number (∼160) of copies of the 18S rRNA gene (11), and it is assumed that the number of rRNA copies in the cytoplasm is >104 (30). This corresponds with our observation that one Leishmania parasite contains around 104.8 copies of 18S rRNA. The difference in copy numbers of rRNA and of DNA predicts a much higher sensitivity of the QT-NASBA in comparison to the conventional PCR. This was confirmed in our experiments when L. donovani promastigote dilutions were compared in the QT-NASBA and PCR (data not shown). The Leishmania QT-NASBA could detect levels of parasites at least 100-fold lower than conventional PCR, as we found a detection limit of 0.1 parasite per amplification reaction compared to a limit of 10 parasites for the latter. No statistically significant difference was found between QT-NASBA and conventional PCR for the detection of parasites in biopsy samples from CL patients before treatment (97.5% and 91.8%, respectively). One biopsy sample, collected before treatment, was negative by both molecular methods. This biopsy sample was obtained from an individual that had a persistent lesion for over 2 years, which started after a visit to French Guiana. He was identified as a CL patient by the smear method only, while no parasites could be detected in culture or by histopathological analysis. Moreover, the histopathological result was not compatible with leishmaniasis. Therefore, the diagnosis of CL is in this case questionable. An alternative explanation may be that this individual had a long-standing CL infection, which is easily misdiagnosed due to a very low density of Leishmania parasites in these kinds of lesions, which can persist for more than 6 months (32). Another skin biopsy sample was negative by the QT-NASBA but positive by PCR. This sample was collected from an individual from Suriname with a 6-month-old lesion. The histopathology of the lesion was typical for a granulomatous reaction, a result indicative of an efficient cell-mediated immune response. It may be possible that viable parasites were not present in the sample and only persistent DNA of the parasite was detected by PCR. In contrast, 6 out of 78 samples from confirmed CL patients were negative by the PCR but positive by the QT-NASBA, showing a higher sensitivity of the assay. Two out of six samples were obtained from patients at the Department of Dermatology in Amsterdam (one skin biopsy sample and one needle aspiration sample). Histopathological results and PCR were negative for both patients. To confirm CL diagnosis in these two specific cases, needle aspiration and skin biopsy sample collection were repeated, and PCR gave a positive result in this second attempt.
Recently, several real-time PCR assays, which have detection limits comparable to that of QT-NASBA, have been developed for quantification of Leishmania parasites (4, 13, 25). Some of these real-time PCRs have the Leishmania infantum kinetoplast minicircle DNA as the target for amplification, because it has a high copy number in the parasite (∼104 copies). However, the kinetoplast minicircle DNA is in general species specific and may even differ within one species (15). This constitutes a problem for the quantification of Leishmania parasites in a clinical sample and affects the accuracy of the real-time PCR assay. In contrast, the QT-NASBA has the 18S rRNA, which does not show any variation in copy number between the L. donovani and L. (V.) braziliensis guyanensis species or even between the promastigote and amastigote stages of the parasite, as its target. The 18S rRNA target is present in all Leishmania species, ensuring high sensitivity and allowing quantification of all species in a similar manner by QT-NASBA (30).
A real-time PCR assay based on an 18S rRNA gene target has been developed by Schulz et al. (25). This method was able to quantify different Leishmania species and to discriminate between three clinically relevant groups (L. donovani complex, L. braziliensis complex, and other Leishmania spp.). However, the advantage of QT-NASBA over this assay is the detection of RNA instead of the DNA of the parasite. This is particularly the case when the assay is used to monitor the efficacy of treatment schedules in which the detection of viable parasites is crucial. Real-time PCR has the advantage over the current QT-NASBA that it is able to detect directly a signal during the amplification reaction. The establishment of a real-time QT-NASBA will add to the further improvement of the Leishmania QT-NASBA. Schneider et al. (22) compared real-time QT-NASBA with real-time PCR for Plasmodium falciparum and showed that the real-time QT-NASBA was more convenient, since the QT-NASBA uses less sample material to achieve the same sensitivity.
A sensitive and quantitative assay, such as QT-NASBA, could be of great importance for the establishment of duration and efficacy of treatment schedules and the early recognition of treatment failures. Recently, Bossolasco et al. (4) developed a real-time PCR assay for visceral leishmaniasis by quantifying Leishmania parasites following treatment and showed that persistence of parasites in the blood after treatment was associated with a high risk of relapse. So far, no follow-up studies have been performed on skin biopsy samples of CL patients. QT-NASBA may be the tool that enables the design of new clinical trials for the follow-up of treatment of CL patients. Preliminary results of our present study indicate that QT-NASBA may detect early clinical treatment failures. Six skin biopsy samples collected from 1 day to 8 weeks after treatment were positive by QT-NASBA, with parasite counts of 2 to 513 parasites/sample. Four patients of this group did develop treatment failures. In contrast, PCR was positive in three out of the six samples, two of the donors of which developed relapses. In parallel, Omar et al. (16) demonstrated that QT-NASBA can predict antimalarial treatment failures.
In conclusion, this study shows that QT-NASBA assay is a reliable, sensitive, and specific method for the detection of Leishmania parasites in small skin biopsy samples. The method provides quantitative data for all Old and New World Leishmania species tested, and this could make the test ideal for diagnostic purposes and for monitoring the effectiveness of treatment.
Acknowledgments
This work was supported by a grant from The Netherlands Foundation for the Advancement of Tropical Research (WOTRO contract 96-210).
We thank Annigje Jensema, Remco van der Ham, Adinda van Ginkel, Chantal van der Horst (AMC, Amsterdam, The Netherlands), Seray Ozensoy Toz, Fadile Yildiz Zeyrek (Faculty of Medicine, Ege University, Izmir, Turkey), and Mahrukh Fatima (Bolan Medical College, Quetta, Pakistan) for collecting skin biopsy samples. We also thank Inge Peekel and Nel Kroon (KIT Biomedical Research, Amsterdam, The Netherlands) for maintaining parasite cultures, Saskia van Scheij (AMC, Amsterdam, The Netherlands) for assisting in the establishment of the human monocyte cell line, Petra Schneider (University Medical Centre Nijmegen, The Netherlands) for her advice and suggestions, and Mirjam Bakker and Birgit van Benthem (KIT Biomedical Research, Amsterdam, The Netherlands) for assistance with the statistical analysis of the results.
REFERENCES
- 1.Alvar, J., C. Canavate, B. Gutierrez-Solar, M. Jimenez, F. Laguna, R. Lopez-Velez, R. Molina, and J. Moreno. 1997. Leishmania and human immunodeficiency virus coinfection: the first 10 years. Clin. Microbiol. Rev. 10:298-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berman, J. D. 2005. Recent developments in leishmaniasis: epidemiology, diagnosis, and treatment. Curr. Infect. Dis. Rep. 7:33-38. [DOI] [PubMed] [Google Scholar]
- 3.Boom, R., C. J. A. Sol, M. M. M. Salimans, C. L. Jansen, P. M. E. Wertheim-van Dillen, and J. van der Noordaa. 1990. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 28:495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bossolasco, S., G. Gaiera, D. Olchini, M. Gulletta, L. Martello, A. Bestetti, L. Bossi, L. Germagnoli, A. Lazzarin, C. Uberti-Foppa, and P. Cinque. 2003. Real-time PCR assay for clinical management of human immunodeficiency virus-infected patients with visceral leishmaniasis. J. Clin. Microbiol. 41:5080-5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Croft, S. L. 2001. Monitoring drug resistance in leishmaniasis. Trop. Med. Int. Health 6:899-905. [DOI] [PubMed] [Google Scholar]
- 6.de Oliveira, C. I., A. Báfica, F. Oliveira, C. B. F. Favali, T. Correa, L. A. R. Freitas, E. Nascimento, J. M. Costa, and A. Barral. 2003. Clinical utility of polymerase chain reaction-based detection of Leishmania in the diagnosis of American cutaneous leishmaniasis. Clin. Infect. Dis. 37:e149-e153. [DOI] [PubMed] [Google Scholar]
- 7.Faber, W. R., L. Oskam, T. van Gool, N. C. M. Kroon, K. J. Knegt-Junk, H. Hofwegen, A. C. van der Wal, and P. A. Kager. 2003. Value of diagnostic techniques for cutaneous leishmaniasis. J. Am. Acad. Dermatol. 49:70-74. [DOI] [PubMed] [Google Scholar]
- 8.Hepburn, N. C. 2001. Management of cutaneous leishmaniasis. Curr. Opin. Infect. Dis. 14:151-154. [DOI] [PubMed] [Google Scholar]
- 9.Herwaldt, B. L. 1999. Leishmaniasis. Lancet 354:1191-1199. [DOI] [PubMed] [Google Scholar]
- 10.Josephson, K. L., C. P. Gebra, and I. L. Pepper. 1993. Polymerase chain reaction detection of nonviable bacterial pathogens. Appl. Environ. Microbiol. 59:3513-3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leon, W., D. L. Fouts, and J. Manning. 1978. Sequence arrangement of the 16S and 26S rRNA genes in pathogenic haemoflagellate Leishmania donovani. Nucleic Acids Res. 5:491-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Looker, D. L., S. Martinez, J. M. Horton, and J. J. Marr. 1986. Growth of Leishmania donovani amastigotes in the continuous human macrophage cell line U937: studies of drug efficacy and metabolism. J. Infect. Dis. 154:323-327. [DOI] [PubMed] [Google Scholar]
- 13.Mary, C., F. Faraut, L. Lascombe, and H. Dumon. 2004. Quantification of Leishmania infantum DNA by a real-time PCR assay with high sensitivity. J. Clin. Microbiol. 42:5249-5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mendonça, M. G., M. E. F. De Brito, E. H. G. Rodrigues, V. Bandeira, M. L. Jardim, and F. G. C. Abath. 2004. Persistence of Leishmania parasites in scars after clinical cure of American cutaneous leishmaniasis: is there a sterile cure? J. Infect. Dis. 189:1018-1023. [DOI] [PubMed] [Google Scholar]
- 15.Meredith, S. E., E. E. Zijlstra, G. J. Schoone, Kroon, N. C. M., G. J. van Eys, K. U. Schaeffer, A. M. el-Hassan, and P. G. Lawyer. 1993. Development and application of the polymerase chain reaction for the detection and identification of Leishmania parasites in clinical material. Arch. Inst. Pasteur Tunis 70:419-431. [PubMed] [Google Scholar]
- 16.Omar, S. A., P. F. Mens, G. J. Schoone, A. Yusuf, J. Mwangi, S. Kaniaru, G. A. A. Omer, and H. D. F. H. Schallig. 2005. Plasmodium falciparum: evaluation of a quantitative nucleic acid sequence-based amplification assay to predict the outcome of sulfadoxine-pyrimethamine treatment of uncomplicated malaria. Exp. Parasitol. 110:73-79. [DOI] [PubMed] [Google Scholar]
- 17.Osman, O. F., L. Oskam, E. E. Zijlstra, N. C. M. Kroon, G. J. Schoone, E. A. G. Khalil, M. El-Hassan, and P. A. Kager. 1997. Evaluation of PCR for diagnosis of visceral leishmaniasis. J. Clin. Microbiol. 35:2454-2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salman, S., N. G. Rubeiz, and A. G. Kibbi. 1999. Cutaneous leishmaniasis: clinical features and diagnosis. Clin. Dermatol. 17:291-296. [DOI] [PubMed] [Google Scholar]
- 19.Schallig, H. D. F. H., G. J. Schoone, E. J. M. Lommerse, N. C. M. Kroon, P. J. de Vries, and T. van Gool. 2003. Usefulness of quantitative nucleic acid sequence based amplification for diagnosis of malaria in an academic hospital setting. Eur. J. Clin. Microbiol. Infect. Dis. 22:555-557. [DOI] [PubMed] [Google Scholar]
- 20.Schallig, H. D. F. H., G. J. Schoone, N. C. M. Kroon, A. Hailu, F. Chappuis, and H. Veeken. 2001. Development and application of “simple” diagnostic tools for visceral leishmaniasis. Med. Microbiol. Immunol. 190:69-71. [DOI] [PubMed] [Google Scholar]
- 21.Schallig, H. D. F. H., and L. Oskam. 2002. Review: molecular biological applications in the diagnosis and control of leishmaniasis and parasite identification. Trop. Med. Int. Health 7:641-651. [DOI] [PubMed] [Google Scholar]
- 22.Schneider, P., L. Wolters, G. Schoone, H. D. F. H. Schallig, R. Hermsen, P. Sillekens, and R. Sauerwein. 2004. Real-time nucleic acid sequence-based amplification is more convenient than real-time PCR for quantification of Plasmodium falciparum. J. Clin. Microbiol. 43:402-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schoone, G. J., L. Oskam, N. C. M. Kroon, H. D. F. H. Schallig, and S. A. Omar. 2000. Detection and quantification of Plasmodium falciparum in blood samples using quantitative nucleic acid sequence-based amplification. J. Clin. Microbiol. 38:4072-4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schubach, A., F. Haddad, M. P. Oliveira-Neto, W. Degrave, C. Pirmez, G. Grimaldi, Jr., and O. Fernandes. 1998. Detection of Leishmania DNA by PCR in scars of treated human patients. J. Infect. Dis. 178:911-914. [DOI] [PubMed] [Google Scholar]
- 25.Schulz, A., K. Mellenthin, G. Schönian, B. Fleischer, and C. Drosten. 2003. Detection, differentiation, and quantitation of pathogenic Leishmania organisms by a fluorescence resonance energy transfer-based real-time PCR assay. J. Clin. Microbiol. 41:1529-1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smits, H. L., G. C. Gussenhoven, W. Terpstra, R. A. F. Schukkink, B. Van Gemen, and T. Van Gool. 1997. Detection, identification and semi-quantification of malaria parasites by NASBA amplification of small subunit ribosomal RNA sequences. J. Microbiol. Methods 28:65-75. [Google Scholar]
- 27.Tavares, C. A. P., A. P. Fernandes, and M. N. Melo. 2003. Molecular diagnosis of leishmaniasis. Expert Rev. Mol. Diagn. 3:89-99. [DOI] [PubMed] [Google Scholar]
- 28.Van der Vliet, G. M. E., P. Schepers, R. A. F. Schukkink, B. van Gemen, and P. R. Klatser. 1994. Assessment of mycobacterial viability by RNA amplification. Antimicrob. Agents Chemother. 38:1959-1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van der Vliet, G. M. E., S. Cho, K. Kampirapap, J. van Leeuwen, R. A. F. Schukkink, B. van Gemen, P. K. Das, W. R. Faber, G. P. Walsh, and P. R. Klatser. 1996. Use of NASBA RNA amplification for detection of Mycobacterium leprae in skin biopsies from untreated and treated leprosy patients. Int. J. Lepr. 64:396-403. [PubMed] [Google Scholar]
- 30.Van Eys, G. J. J. M., G. J. Schoone, N. C. M. Kroon, and S. B. Ebeling. 1992. Sequence analysis of small subunit ribosomal RNA genes and its use for detection and identification of Leishmania parasites. Mol. Biochem. Parasitol. 51:133-142. [DOI] [PubMed] [Google Scholar]
- 31.Van Gemen, B., R. Van Beuningen, and A. Nabbe. 1994. A one-tube quantitative HIV-1 RNA NASBA nucleic acid amplification assay using electrochemiluminescent (ECL) labelled probes. J. Virol. Methods 49:157-167. [DOI] [PubMed] [Google Scholar]
- 32.Weigle, K. A., L. A. Labrada, C. Lozano, C. Santrich, and D. C. Barker. 2002. PCR-based diagnosis of acute and chronic cutaneous leishmaniasis caused by Leishmania (Viannia). J. Clin. Microbiol. 40:601-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.World Health Organization. 1990. The leishmaniasis. WHO Tech. Rep. Ser. 753:154. [Google Scholar]
- 34.Wortmann, G., C. Sweeney, H. S. Houng, N. Aronson, J. Stiteler, J. Jackson, and C. Ockenhouse. 2001. Rapid diagnosis of leishmaniasis by fluorogenic polymerase chain reaction. Am. J. Trop. Med. Hyg. 65:583-587. [DOI] [PubMed] [Google Scholar]