Abstract
Avian H9N2 influenza A virus has caused repeated human infections in Asia since 1998. Here we report that an H9N2 influenza virus infected a 5-year-old child in Hong Kong in 2003. To identify the possible source of the infection, the human isolate and other H9N2 influenza viruses isolated from Hong Kong poultry markets from January to October 2003 were genetically and antigenically characterized. The findings of this study show that the human H9N2 influenza virus, A/Hong Kong/2108/03, is of purely avian origin and is closely related to some viruses circulating in poultry in the markets of Hong Kong. The continued presence of H9N2 influenza viruses in poultry markets in southern China increases the likelihood of avian-to-human interspecies transmission.
Since the mid-1990s, two lineages of H9N2 influenza A viruses have become established in domestic poultry in southern China, Duck/HK/Y280/97 (Y280-like) and Quail/HK/G1/97 (G1-like) viruses, which are predominantly found in chickens and quail, respectively (2, 3). These viruses have been sporadically introduced into humans and cause a flu-like illness (4, 5, 7, 16, 21). In the meantime, H9N2 influenza Y280-like viruses were also isolated from domestic pigs in Hong Kong in 1998 (20). These findings indicate that the interspecies transmission of H9N2 influenza virus from avian to mammalian hosts continues to occur and poses a persistent threat for humans.
Systematic surveillance studies revealed that those H9N2 influenza viruses have also been regularly isolated from different types of poultry in live poultry markets in Hong Kong and mainland China (1, 14, 17). Antigenic and genetic analyses revealed that, in the recent past, some of those H9N2 influenza viruses continued to evolve and reassort with other influenza viruses of avian origin to generate multiple genotypes. Furthermore, some of the H9N2 influenza viruses currently circulating in southern China have molecular features that allow them to preferentially bind to α-2,6-NeuAcGal receptors (1, 14, 18, 19).
In November 2003, an H9N2 avian influenza was isolated from a child with a flu-like illness in Hong Kong. Although highly pathogenic avian influenza H5N1 has been a major health threat to humans in Southeast Asia since 2003 (15), this event demonstrated that H9N2 virus also has the potential to infect humans. The continuing prevalence of both H5N1 and H9N2 influenza viruses in poultry and their repeated introduction into humans highlight the importance of understanding the behavior of these viruses in Southeast Asia. This study characterizes the H9N2 virus from this human case along with H9N2 viruses isolated from poultry in Hong Kong during the same period of time to identify the possible source of infection.
CASE REPORT
In late November 2003, an H9N2 influenza virus, HK/2108/03, was isolated from the nasopharyngeal aspirate of a 5-year-old child hospitalized with an influenza-like illness in Hong Kong. The child presented with mild fever (temperature, 38.7°C), cough, and mild dehydration, with dried lips and decreased skin turgor. The neutrophil count (10.0 ×109/liter) was slightly elevated (reference range, 1.5 to ×109 8.5 ×109/liter), and the lymphocyte count (1.6 ×109/liter) was slightly below the reference range (2.0 ×109 to 8.5 ×109/liter). This may indicate that a secondary bacterial superinfection had occurred during the period of illness. Radiological examination of the chest did not reveal any abnormal findings. The child had no history of contact with domestic poultry prior to the onset of illness and was discharged from the hospital after 2 days, as the patient's body temperature had dropped to normal and the other symptoms had disappeared.
MATERIALS AND METHODS
Virus sampling and isolation.
To characterize the virus and also determine the possible source of HK/2108/03, we sequenced and analyzed 22 additional H9N2 viruses isolated from terrestrial poultry in Hong Kong markets as described previously (2, 14) (Table 1). No H9N2 viruses were isolated from aquatic poultry in Hong Kong during this period. Both the human and poultry isolates were initially grown in 10-day-old embryonated chicken eggs (2, 14).
TABLE 1.
Influenza A H9N2 viruses characterized in this study
| Virus and hostb | Sampling date (day/mo/yr) | Sample type | Genotype |
|---|---|---|---|
| HK/2108/03 | 27/11/03 | Nasopharyngeal aspirate | A |
| Gf/HK/SSP607/03 | 18/08/03 | Fecal droppings | A |
| Ph/HK/CSW1323/03 | 04/09/03 | Trachea | A |
| Gf/HK/NT184/03 | 15/04/03 | Fecal droppings | A |
| Ck/HK/NT142/03 | 20/03/03 | Fecal droppings | B |
| Ck/HK/WF126/03 | 22/04/03 | Fecal droppings | Y280a |
| SCk/HK/WF285/03 | 20/08/03 | Fecal droppings | Y280a |
| CK/HK/YU463/03 | 19/05/03 | Fecal droppings | D |
| CK/HK/YU577/03 | 16/01/03 | Fecal droppings | D+ |
| SCk/HK/YU663/03 | 21/07/03 | Fecal droppings | D+ |
| Ck/HK/CSW161/03 | 16/01/03 | Trachea | D+ |
| Gf/HK/NT101/03 | 17/02/03 | Fecal droppings | D+ |
Nonreassortant viruses that belong to the Y280 lineage.
Abbreviations: Ck, chicken; Gf, guinea fowl; Ph, pheasant; SCk, silky chicken.
Antigenic analysis.
The virus subtype was determined by standard hemagglutination inhibition (HI) and neuraminidase inhibition tests, as recommended by the World Health Organization (http://www.who.int/csr/resources/publications/en/#influenza). Antigenic analysis was performed with a panel of monoclonal antibodies (MAbs) to the hemagglutinin (HA) of H9N2 strains (1).
Phylogenetic and molecular analyses.
RNA extraction, cDNA synthesis, and PCR were carried out as described previously (2, 3). Sequencing was performed with the BigDye Terminator v3.1 cycle sequencing kit on an ABI PRISM 3700 DNA analyzer (Applied Biosystems) by following the manufacturer's instructions. Sequence fragments were assembled with Lasergene (version 6.0; DNASTAR) and then aligned by using BioEdit, version 7, and residue analysis was performed with BioEdit, version 7 (9). Phylogenetic trees were generated by neighbor-joining bootstrap analysis (1,000 replicates) by using the Tamura-Nei algorithm in MEGA, version 2.1 (13). All eight genes were sequenced for each virus isolate. Virus genotypes were defined by gene phylogeny. A distinct phylogenetic lineage with bootstrap support of ≥70% (≥50% for the M, NP, and PA genes) indicated a common origin. We followed the scheme presented by Choi et al. (1); however, nonreassortant Y280-like viruses were incorrectly described as genotype C by Choi et al. Also, genotypes D and F are identical across seven gene segments and differ only in the presence (genotype D) or absence (genotype F) of a 3-amino-acid deletion in the neuramidase (NA) molecule. As such, we have renamed genotype F genotype D+ to better reflect their similarity.
Nucleotide sequence accession numbers.
The sequence data generated in this study have been submitted to GenBank and have been given accession numbers DQ226095 to DQ226182.
RESULTS
Antigenic analysis.
The antigenic properties of the H9N2 viruses were investigated by using a panel of H9 MAbs raised against Ck/HK/G9/97, Qa/HK/G1/97, and Dk/HK/Y280/97 viruses by using HI assays (Table 2). The majority of the H9N2 viruses characterized in this study reacted well with all Ck/HK/G9/97 and Dk/HK/Y280/97 MAbs but showed little cross-reactivity with the Qa/HK/G1/97 MAbs. Human isolate HK/2108/03 and four other H9N2 viruses (Gf/HK/NT184/03, Gf/HK/SSP607/03, Ph/HK/CSW1323/03, and WDk/ST/4808/01) have distinctly different patterns of antigenic reactivity. They showed no cross-reaction (HI titer < 200) with any of the MAbs against previous H9N2 viruses except MAbs 7B10, 8C4, and 18B10 against Dk/HK/Y280/97. Compared to other genotype A viruses, WDk/ST/4808/01 showed decreased antigenicity to MAb 8C4. The consistent pattern of discrimination of these monoclonal antibodies between groups of viruses is in agreement with the results of our phylogenetic and molecular analyses (see below).
TABLE 2.
Hemagglutination inhibition titers from antigenic analysis of influenza A H9N2 virusesa
| Virus | HI titer of the following isolates to the indicated MAb:
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CK/HK/G9/97
|
Qa/HK/G1/97
|
DK/HK/Y280/97
|
|||||||||||||
| G9-6 | G9-25 | 1073-9 | 26 | 29 | 7B10 | 8C4 | 15F1 | 18G4 | 3D11 | 4G3 | 19A10 | 18B10 | 2F4 | 18B1 | |
| CK/HK/G9/97 | ≥ | ≥ | 800 | 800 | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | 6,400 | ≥ | ≥ |
| DK/HK/Y280/97 | ≥ | ≥ | 200 | 400 | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | 6,400 | ≥ | ≥ |
| HK/1073/99 | ≥ | ≥ | <200 | ≥ | ≥ | <200 | <200 | <200 | <200 | <200 | <200 | <200 | <200 | <200 | <200 |
| Qa/HK/G1/97 | ≥ | ≥ | 1600 | ≥ | ≥ | 400 | <200 | <200 | <200 | <200 | <200 | <200 | <200 | <200 | <200 |
| HK/2108/03 | <200 | <200 | <200 | <200 | <200 | 400 | ≥ | <200 | <200 | <200 | <200 | <200 | 3,200 | <200 | <200 |
| Gf/HK/SSP607/03 | <200 | <200 | <200 | <200 | <200 | <200 | ≥ | <200 | <200 | <200 | <200 | <200 | 3,200 | <200 | <200 |
| Ph/HK/CSW1323/03 | <200 | <200 | <200 | <200 | <200 | <200 | ≥ | <200 | <200 | <200 | <200 | <200 | 3,200 | <200 | <200 |
| Gf/HK/NT184/03 | <200 | <200 | <200 | <200 | <200 | 300 | ≥ | <200 | <200 | <200 | <200 | <200 | 1,600 | <200 | <200 |
| WDk/ST/4808/01 | <200 | <200 | <200 | <200 | <200 | <200 | 400 | <200 | <200 | <200 | <200 | <200 | 800 | <200 | <200 |
| CK/HK/NT142/03 | ≥ | 6400 | <200 | <200 | ≥ | ≥ | ≥ | 6,400 | ≥ | ≥ | 6,400 | ≥ | 1,600 | ≥ | ≥ |
| CK/HK/WF126/03 | ≥ | ≥ | <200 | 800 | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | 6,400 | ≥ | ≥ |
| SCk/HK/WF285/03 | ≥ | ≥ | <200 | 400 | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | 800 | ≥ | ≥ |
| CK/HK/YU463/03 | ≥ | ≥ | <200 | 800 | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | 3,200 | ≥ | ≥ |
| CK/HK/YU577/03 | ≥ | ≥ | <200 | 800 | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ |
| SCk/HK/YU663/03 | ≥ | ≥ | <200 | 800 | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ |
| Ck/HK/CSW161/03 | 12,500 | ≥ | <200 | <200 | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | 800 | ≥ | ≥ |
| GF/HK/NT101/03 | ≥ | ≥ | 800 | 800 | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ | ≥ |
Abbreviations and symbols: Ck, chicken; Ph, pheasant; Gf, guinea fowl; Qa, quail; WDk, wild duck; SCk, silky chicken; ≥, HI titer greater than 12,800.
Phylogenetic analysis.
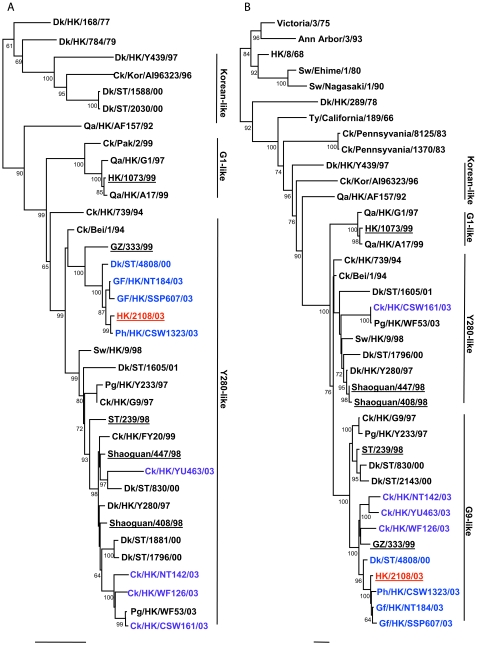
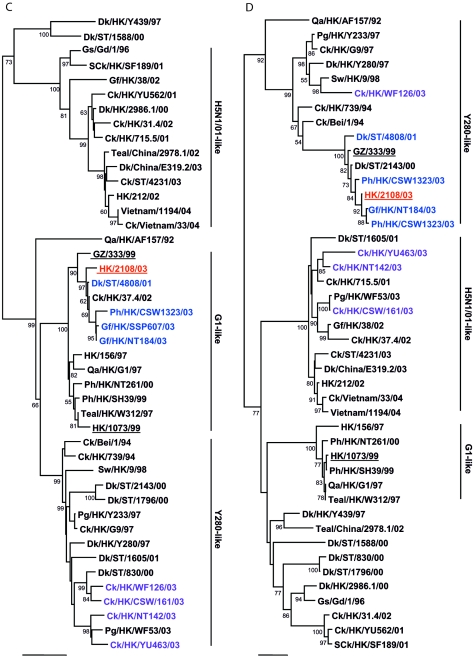
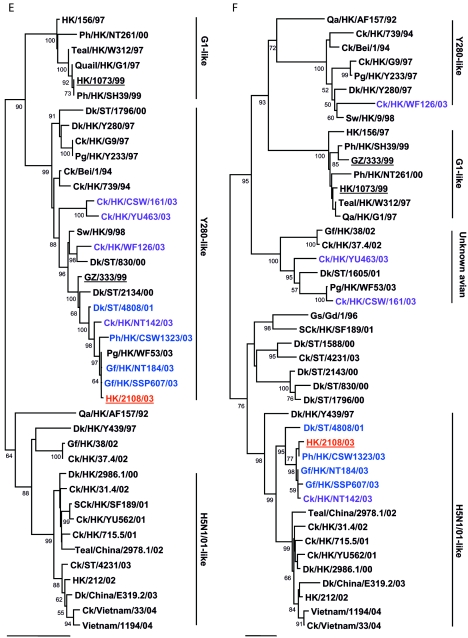
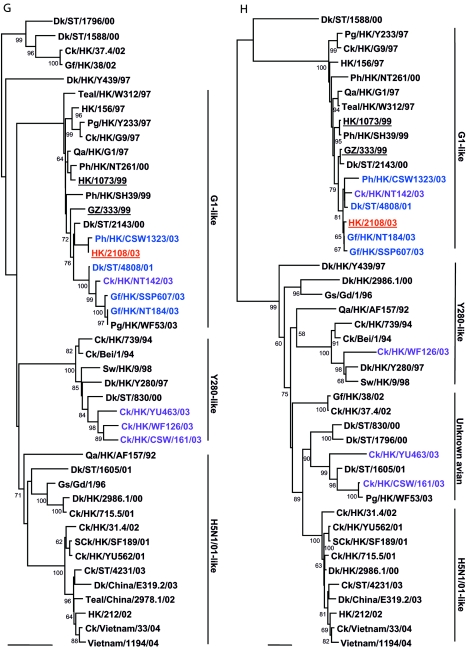
To determine the evolutionary relationship of human isolate HK/2108/03, other H9N2 viruses isolated from poultry markets and previous human isolates were genetically analyzed, and the viruses were categorized into genotypes, as described previously (1). Phylogenetic analysis revealed that human isolate HK/2108/03 and avian isolates WDk/ST/4808/01, Gf/HK/SSP607/03, and Ph/HK/CSW1323/03 belong to Y280-like genotype A. These viruses are closely related to another previous human isolate, isolate GZ/333/99, in seven of eight gene segments (Fig. 1A to H). The genetic homology of HK/2108/03 was highest to Gf/HK/SSP607/03 and Ph/HK/CSW1323/03 (greater than 99% in all eight gene segments; data not shown), both viruses of which were isolated from January to October 2003 from markets in Hong Kong. This result corresponds to that of the antigenic analysis included in our study.
FIG.1.
Phylogenetic relationships of the influenza A virus genes HA, nucleotide positions 55 to 1042 (A); NA, nucleotide positions 1 to 1372 (B); M (matrix), nucleotide positions 1 to 972 (C); NP (nucleoprotein), nucleotide positions 49 to 1009 (D); NS (nonstructural), nucleotide positions 23 to 823 (E); PA (polymerase acidic), nucleotide positions 1423 to 2144 (F); PB1 (polymerase basic 1), nucleotide positions 1 to 1218 (G); and PB2 (polymerase basic 2), nucleotide positions 1101 to 2220 (H). Trees were generated with MEGA2 by using a Tamura-Nei (gamma) neighbor-joining analysis. The HA phylogenetic tree is rooted to A/turkey/Wisconsin/66; the NA tree is rooted to A/Leningrad/134/57; and the PA and PB1 trees are rooted to A/Ann Arbor/6/60 and B/Lee/40, respectively. The M, NP, NS, and PB2 trees are rooted to A/equine/Prague/1/56. Scale bars, 0.05 (HA, NS, PB1) and 0.02 (NA, M, NP, PA, PB2) nucleotide changes per nucleotide. The numbers at the nodes indicate bootstrap values from 500 bootstrap replicates. All viruses sequenced in the present study are underlined, and the remaining sequences can be found in GenBank. Abbreviations: Bei, Beijing; Cu, chukar; Ck, chicken; Dk, duck; GD, Guangdong; Gf, Guinea fowl; Gs, goose; GZ, Guangzhou; HK, Hong Kong; Kor, Korea; Pg, pigeon; Ph, pheasant; Qa, quail; SCk, silky chicken; ST, Shantou; Sw, swine; Ty, turkey; WDk, wild duck.
Of the remaining isolates tested, Ck/HK/NT142/03 belongs to genotype B, while Ck/HK/WF126/03 and SCk/HK/WF285/03 are nonreassortant Y280-like viruses. Four viruses (SCk/HK/YU663/03, Ck/HK/CSW161/03, Gf/HK/NT101/03, and Ck/HK/YU577/03) belong to genotype D+, while Ck/HK/YU463/03 belongs to genotype D. No genotype E viruses were identified in this study.
Molecular analysis.
To try to identify possible determinants of interspecies transmission of H9N2 influenza virus from avian to human, the deduced amino acid sequences of viral proteins were analyzed. Sequence comparison showed that the human HK/2108/03 virus and all 22 viruses isolated from Hong Kong poultry markets had an identical connecting peptide R-S-S-R motif and amino acid residues 226-Leu and 228-Gly at the receptor binding site (H3 numbering used throughout); Gf/HK/NT184/03, however, has 226-Gln.
The amino acid residues at the antigenic sites on the globular head of the HA molecule were also analyzed. All genotype A viruses had an Asp-145-Asn mutation not seen in the other viruses investigated. In addition, of the genotype A viruses, HK/2108/03, Gf/HK/SSP607/03, and Ph/HK/CSW1323/03 had 226-Leu, while Gf/HK/NT184/03 and WDk/ST/4808/01 had 226-Gln. Genotype A viruses also lost a potential glycosylation site at the globular head of the HA molecule due to a Thr-212-Ala substitution.
DISCUSSION
The H9N2 subtype of influenza virus has repeatedly been isolated from human flu-like illness in China since 1998 (4, 7, 16, 21). However, so far there is little evidence for human-to-human transmission (23). The H9N2 lineages Y280-like and G1-like, which are established and dominant in terrestrial poultry in southern China, are predicted to have an affinity for the human receptor binding profile (14, 18) and have also been associated with human disease since the late 1990s. This suggests that these viruses have the ability to be introduced into humans. The current case suggests that the interspecies transmission of H9N2 influenza virus from poultry to humans continues to occur in this region.
Repeated interspecies transmission of H9N2 influenza viruses from poultry to humans raises concerns about a pandemic. Even though we identified closely related counterparts of HK/2108/03 from terrestrial poultry in Hong Kong markets from the same period, there was no known direct contact with poultry before the onset of infection with HK/2108/03. Thus, the source of the infection could have been from indirect contact with poultry or from another human. Previous studies during the late 1990s have already shown that the seroprevalence of H9 antibodies in southern China was at least 2% among the human population (5, 21). Furthermore, in a random screening study conducted in southern China, 2.6% of serum samples (n = 3,228) from voluntary blood donors tested positive for neutralizing antibody to Dk/HK/Y280/97 virus (authors' unpublished data). Since direct contact with poultry is common in southern China, this suggests that the interspecies transmission of H9N2 influenza virus into humans is not a rare event and it is not impossible that a low level of human-to-human transmission may occur. The question still remains as to why this influenza virus subtype has not become established or acquired an efficient means of human-to-human transmission.
Genotyping of HK/2108/03 revealed that it belongs to genotype A of H9N2 influenza viruses (1). Genetic and antigenic analyses showed that the counterpart of the human H9N2 isolate was present in live poultry markets of Hong Kong during the same period of time. In addition, another H9N2 influenza virus (GZ/333/99), which differs from genotype A only in the source of the PA gene, had caused human disease in 1999. This virus was isolated from a 22-month-old girl who developed fever, cough, and viral bronchitis (6, 7). In addition, a neutralization test of the serum from the mother had a titer of ≥640 against serum of GZ/333/99, indicating that the mother had also been infected by H9N2 influenza virus. Neither the 22-month-old baby nor her mother had a history of contact with animals (6). Whether this specific genotype has the ability to be transmitted to humans more easily than other genotypes currently circulating in poultry is still unknown.
Previous studies have shown an association between the addition of a glycosylation site on the globular head of the hemagglutinin molecule and a loss of virulence in H5 influenza virus (12) and decreased receptor binding and cell fusion activity in H2 influenza virus (22). Therefore, the loss of a potential glycosylation site in genotype A viruses may be a factor in the ability of these viruses to infect humans.
Surveillance of poultry for influenza detected the counterpart of this human H9N2 influenza virus in farmed wild ducks and domestic ducks from markets in southern China as early as 2000, demonstrating that this virus is not a novel reassortant and that it may have become a relatively stable genotype. The prevalence of H9N2 genotype A viruses in poultry in the broader region of southeast Asia is still to be determined.
H9N2 influenza virus usually causes relatively mild symptoms in humans that are clinically indistinguishable from the symptoms of common influenza caused by human H1N1 and H3N2 viruses. As H9N2 viruses are not highly pathogenic for poultry, the extent of infection in both poultry and humans is likely to remain underappreciated. Interaction or reassortment between the prevailing human and avian influenza viruses is considered the most probable scenario for the generation of new pandemic strains. Given the predicted affinity of H9N2 viruses for α-2,6-NeuAcGal receptors (8, 11, 19), they appear to have the potential to cross the species barrier to humans more efficiently than current H5N1 strains. The repeated introduction of H9N2 virus into humans provides increased opportunities for reassortment with human viruses and also increases the likelihood that this virus will acquire the ability to be transmitted from human to human. Two previous pandemic strains (H2N2 in 1957 and H3N2 in 1968) were also derived from viruses with low pathogenicities (10). Therefore, while the impact of H5N1 disease in poultry and in humans in Asia has been altogether more dramatic, it is important to have H9N2 viruses high on the list of candidate human pandemic strains and to carry out more studies on its ecology.
Acknowledgments
This work was supported by the Research Fund for the Control of Infectious Diseases of the Health, Welfare and Food Bureau of the Hong Kong SAR Government, the Li Ka Shing Foundation, the National Institutes of Health (NIAID contract AI95357), and Arthur Lui of Providence Foundation Limited.
REFERENCES
- 1.Choi, Y. K., H. Ozaki, R. J. Webby, R. G. Webster, J. S. Peiris, L. Poon, C. Butt, Y. H. C. Leung, and Y. Guan. 2004. Continuing evolution of H9N2 influenza viruses in southeastern China. J. Virol. 78:8609-8614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guan, Y., K. F. Shortridge, S. Krauss, and R. G. Webster. 1999. Molecular characterization of H9N2 influenza viruses: were they the donors of the “internal” genes of H5N1 viruses in Hong Kong? Proc. Natl. Acad. Sci. USA 96:9363-9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guan, Y., K. F. Shortridge, S. Krauss, P. S. Chin, K. C. Dyrting, T. M. Ellis, R. G. Webster, and M. Peiris. 2000. H9N2 influenza viruses possessing H5N1-like internal genomes continue to circulate in poultry in southeastern China. J. Virol. 74:9372-9380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guo, Y., J. Dong, M. Wang, Y. Zhang, J. Guo, and K. Wu. 2001. Characterization of hemagglutinin gene of influenza A virus subtype H9N2. Chinese Med. J. 114:76-79. [PubMed] [Google Scholar]
- 5.Guo, Y., J. Li, X. Cheng, M. Wang, Y. Zhou, C. Li, F. Chai, H. Liao, Y. Zhang, J. Guo, L. Huang, and D. Bei. 1999. Discovery of men infected by avian influenza A (H9N2) virus. Chinese J. Exp. Clin. Virol. 13:105-108. [PubMed] [Google Scholar]
- 6.Guo, Y., J. Xie, M. Wang., J. Dang., J. Guo., Y. Zhang, and K. Wu. 2000. A strain of influenza A H9N2 virus repeatedly isolated from human population in China. Chinese J. Exp. Clin. Virol. 14:209-212. [PubMed] [Google Scholar]
- 7.Guo, Y., J. Xie, K. Wu, J. Dong, M. Wang, Y. Zhang, J. Guo, J. Chen, Z. Chen, and Z. Li. 2002. Characterization of genome A/Guangzhou/333/99 (H9N2) virus. Chinese J. Exp. Clin. Virol. 16:142-145. [PubMed] [Google Scholar]
- 8.Ha, Y., D. J. Stevens, J. J. Skehel, and D. C. Wiley. 2001. X-ray structures of H5 avian and H9 swine influenza virus hemagglutinins bound to avian and human receptor analogs. Proc. Natl. Acad. Sci. USA 98:11181-11186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 10.Horimoto, T., and Y. Kawaoka. 2001. Pandemic threat posed by avian influenza A viruses. Clin. Microbiol. Rev. 14:129-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaverin, N. V., I. A. Rudneva, N. A. Ilyushina, A. S. Lipatov, S. Krauss, and R. G. Webster. 2004. Structural differences among hemagglutinins of influenza A virus subtypes are reflected in their antigenic architecture: analysis of H9 escape mutants. J. Virol. 78:240-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaverin, N. V., I. A. Rudneva, N. A. Ilyushina, N. L. Varich, A. S. Lipatov, Y. A. Smironv, E. A. Govorkova, A. K. Gitelman, D. K. Lvov, and R. G. Webster. 2002. Structure of antigenic sites at the haemagglutinin molecule of a H5 avian influenza virus and phenotypic variation of escape mutants. J. Gen. Virol. 83:2497-2505. [DOI] [PubMed] [Google Scholar]
- 13.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: Molecular Evolutionary Genetics Analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 14.Li, K. S., K. M. Xu, J. S. M. Peiris, L. L. M. Poon, K. Z. Yu, K. Y. Yuen, K. F. Shortridge, R. G. Webster, and Y. Guan. 2003. Characterization of H9 subtype influenza viruses from the ducks of southern China: a candidate for the next influenza pandemic in humans? J. Virol. 77:6988-6994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li, K. S., Y. Guan, J. Wang, G. J. D. Smith, K. M. Xu, L. Duan, A. P. Rahardjo, P. Puthavathana, C. Buranathai, T. D. Nguyen, A. T. S. Estoepangestie, A. Chaisingh, P. Auewarakui, H. T. Long, N. T. H. Hanh, R. J. Webby, L. L. M. Poon, H. Chen, K. F. Shortridge, K. Y. Yuen, R. G. Webster, and J. S. M. Peiris. 2004. Genesis of a highly pathogenic and potentially pandemic H5N1 influenza virus in eastern Asia. Nature 430:209-213. [DOI] [PubMed] [Google Scholar]
- 16.Lin, Y. P., M. Shaw, V. Gregory, K. Cameron, W. Lim, A. Klimov, K. Subbarao, Y. Guan, S. Krauss, K. Shortridge, R. Webster, N. Cox, and A. Hay. 2000. Avian-to-human transmission of H9N2 subtype influenza A viruses: relationship between H9N2 and H5N1 human isolates. Proc. Natl. Acad. Sci. USA 97:9654-9658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu, M., S. He, D. Walker, N. Zhou, D. R. Perez, B. Mo, F. Lai, X. Huang, R. G. Webster, and R. J. Webby. 2003. The influenza virus gene pool in a poultry market in south central China. Virology 305:267-275. [DOI] [PubMed] [Google Scholar]
- 18.Matrosovich, M. N., S. Krauss, and R. G. Webster. 2001. H9N2 influenza A viruses from poultry in Asia have human virus-like receptor specificity. Virology 281:156-162. [DOI] [PubMed] [Google Scholar]
- 19.Matrosovich, M. N., T. Y. Matrosovich, T. Gray, N. A. Roberts, and H. D. Klenk. 2004. Human and avian influenza viruses target different cell types in cultures of human airway epithelium. Proc. Natl. Acad. Sci. USA 101:4620-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peiris, J. S. M., Y. Guan, D. Markwell, P. Ghose, R. G. Webster, and K. F. Shortridge. 2001. Cocirculation of avian H9N2 and contemporary “human“ H3N2 influenza A viruses in pigs in southeastern China: potential for genetic reassortment? J. Virol. 75:9679-9686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peiris, M., K. Y. Yuen, C. W. Leung, K. H. Chan, P. L. S. Ip, R. W. M. Lai, W. K. Orr, and K. F. Shortridge. 1999. Human infection with influenza H9N2. Lancet 354:916-917. [DOI] [PubMed] [Google Scholar]
- 22.Tsuchiya, E., S. Sugawara, S. Hongo, Y. Matsuzaki, Y. Muraki, and K. Nakamura. 2002. Effect of addition of new oligosaccharide chains to the globular head of influenza A/H2N2 virus haemagglutinin on the intracellular transport and biological activities of the molecule. J. Gen. Virol. 83:1137-1146. [DOI] [PubMed] [Google Scholar]
- 23.Uyeki, T. M., Y. H. Chong, J. M. Katz, W. Lim, Y. Y. Ho, S. S. Wang, T. H. F. Tsang, W. W. Y. Au, S. C. Chan, T. Rowe, J. Hu-Primmer, J. C. Bell, W. W. Thompson, C. B. Bridges, N. J. Cox, K. H. Mak, and K. Fukuda. 2002. Lack of evidence of human-to-human transmission of avian influenza A (H9N2) viruses in Hong Kong, China 1999. Emerg. Infect. Dis. 8:154-159. [DOI] [PMC free article] [PubMed] [Google Scholar]