Abstract
Resistance to linezolid has been associated with a G2576U mutation in domain V of the 23S rRNA. We analyzed nine clinical isolates of linezolid-resistant enterococci and showed a clear association between the number of 23S rRNA genes containing this mutation and the level of linezolid resistance expressed.
Linezolid inhibits protein synthesis by interacting with the bacterial initiation complex (7). Resistance to linezolid has been selected in vitro and encountered in the clinical setting (3, 6, 9) and has been attributed to a G2576U (Escherichia coli numbering scheme) mutation in domain V of the 23S rRNA (6). The presence of multiple 23S rRNA genes in most species suggests that an increased percentage of genes with this mutation may be associated with increased levels of resistance expressed. We report an analysis of 23S rRNA genes from nine clinical Enterococcus faecium isolates and one Enterococcus faecalis isolate derived from patients during treatment with linezolid. Our results confirm previous reports regarding the importance of the G2576U mutation and suggest a direct correlation between the levels of resistance and the percentage of 23S rRNA genes possessing this mutation.
E. faecium LS-1 and LR-1 were clinical isolates cultured from the same patient before and after 49 days of linezolid therapy. E. faecalis LR-2 was isolated from a fecal sample as part of a screening program for linezolid-resistant enterococci in patients undergoing linezolid therapy. Seven additional E. faecium isolates representing linezolid treatment failures were kindly provided to us by John Quinn and Paul Schreckenberger of the University of Illinois at Chicago. For ease of reference, these isolates were designated LR-3 through LR-9 (original designations of these strains were M56114, M67118, X25750, T64825, T70210, 1225, and H11553, respectively). Five of these strains were described in a previous report (3).
Species determination for E. faecium LS-1 and LR-1 was performed by using an API Strep strip (bioMerieux, Inc., Hazelwood, Mo.). Species determination for E. faecium LR-3 through LR-9 was performed using the Vitek GPI card (bioMerieux, Inc.). E. faecalis LR-2 gave conflicting results with the API 20 Strep strip, and so definitive determination occurred by analysis of 16S rRNA sequence according to a previously published protocol (8). MICs were determined by broth macrodilution in brain heart infusion broth (5).
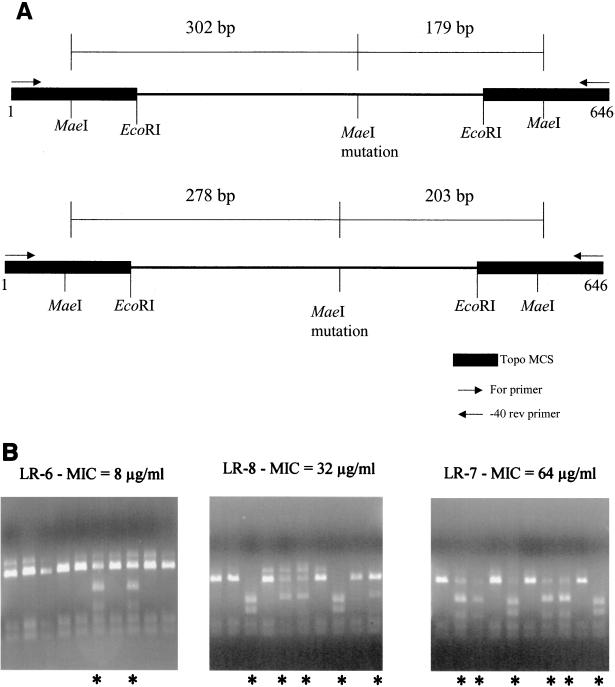
Enterococcal DNA was extracted, digested with restriction enzyme MaeI (Roche Applied Sciences, Indianapolis, Ind.), transferred, and hybridized with digoxigenin-labeled probes as previously described (1). Pulsed-field gel electrophoresis of SmaI-digested genomic DNA from LS-1 and LR-1 was performed as previously described (2). 23S rRNA amplification primers were commercially synthesized (Sigma Genosys, St. Louis, Mo.) based on the E. faecium 23S rRNA gene (primer A, 5′-GCAGAAGGGAGCTTGACTGCGAG-3′; primer B, 5′-ACCCAGCAATGCCCTTGGCAG-3′) and used to amplify a 389-bp internal segment flanking the 2576 site in both E. faecalis and E. faecium. Amplifications of 23S rRNA genes were performed with overnight broth cultures as templates and Tth polymerase (Perkin-Elmer Cetus, Branchburg, N.J.). Amplification conditions were 95°C for 10 min, followed by 30 cycles of 95°C (30 s), 60°C (30 s), and 68°C (1 min). Amplification products were ligated to pCR-XL-TOPO (Invitrogen, Carlsbad, Calif.) and transformed into electroporation-competent E. coli DH10B cells (Bethesda Research Laboratories, Gaithersburg, Md.). Cloned PCR products were sequenced for some experiments as previously described (1) by using an ALF automated sequencer (Amersham Pharmacia, Piscataway, N.J.). For most strains, cloned PCR products were amplified a second time with the forward and reverse primers of the vector and the amplification products were digested with MaeI. The presence of the 2576 mutation was inferred by the appearance of two additional digestion fragments, as outlined in Fig. 1A.
FIG. 1.
(A) Diagrammatic representation of the amplification and restriction strategy employed to assess the number of 23S rRNA genes. The thin line represents the PCR amplification product. The thick line represents the multiple cloning site of the pCR-XL-TOPO vector. The arrows represent the forward and reverse primers used to generate the PCR product containing the 23S insert and the multiple cloning site of the vector. MaeI sites within the insert (created by the presence of the G2576T mutation) and within the vector multiple cloning site are indicated. As the figure indicates, the size of the restriction fragments resulting from MaeI digestion of the PCR products will differ depending on the orientation of the insert. (B) MaeI digestion of cloned amplification products as described for panel A. Two additional bands are readily apparent in lanes with inserts containing the G2576T mutation (lanes marked by asterisks). Linezolid MICs for the strains are indicated above the panels and in Table 1.
E. faecium LS-1 was resistant to vancomycin and ampicillin but susceptible to linezolid (MIC = 2 μg/ml). The posttreatment isolate (E. faecium LR-1) was resistant to vancomycin, ampicillin, and linezolid (MIC = 32 μg/ml). Pulsed-field gel electrophoresis analysis revealed identical SmaI digestion patterns in the two strains (data not shown). 23S rRNA hybridization of restriction digestions of E. faecium LS-1 and LR-1 genomic DNA revealed six copies of the 23S rRNA gene with identical restriction patterns (data not shown). Of 22 PCR amplification products from LR-1 that were cloned and sequenced, eight (36%) contained the G2576T mutation (yielding the G2576U mutation in the 23S rRNA), suggesting that two of the six 23S rRNA genes were mutated. 23S rRNA hybridization of MaeI digests of LS-1 and LR-1 revealed digestion of two of six EcoRI bands in LR-1, consistent with the presence of the mutation in two of six genes (data not shown). The mutation was present in none of 20 PCR products derived from LS-1.
E. faecalis LR-2 was susceptible to both ampicillin and vancomycin but resistant to linezolid (MIC = 128 μg/ml). 23S rRNA hybridization of EcoRI digests of LR-2 genomic DNA revealed four copies of this gene (data not shown), consistent with the number identified in the E. faecalis V583 database (http://www.tigr.org/tigr-scripts/CMR2/GenomePage3.spl?database=gef). Twenty-two of 22 (100%) 23S rRNA clones derived from LR-2 amplification possessed the G2576T mutation, strongly suggesting that all four of the 23S rRNA genes in E. faecalis LR-2 were mutated.
The remaining seven isolates were analyzed using MaeI digestion of amplifications of the cloned 23S PCR products. In order to confirm the reliability of the restriction digestion for identifying the mutation, a Cy5-labeled oligonucleotide (5′-AGGGACGAAAGTCGGGCTTAGT-3′) corresponding to the sequence internal to the PCR product was used for sequencing 15 of the products resulting from amplification of PCR clones from strain LR-3. The correlation between the presence of the mutation and the appearance of the new restriction bands was 100%. We amplified inserts from 15 clones for both LR-3 (5 of 15 mutants) and LR-4 (14 of 15 mutants) and inserts from 10 clones for the remaining strains.
Results of the mutation analysis of the additional seven strains and the agar dilution MIC determinations are shown in Table 1, with examples of the restriction digestions shown in Fig. 1B. We observed an excellent correlation between the percentage of 23S rRNA genes with the G2576U mutation and the level of resistance to linezolid expressed by the clinical isolate.
TABLE 1.
Correlation of linezolid resistance levels with percentage of mutated 23S rRNA genesa
| Enterococcal strain | Linezolid MIC (μg/ml) | No. of mutated genes/total no. of 23S genes (%) |
|---|---|---|
| E. faecium LS-1 | 2 | 0/6 (0) |
| E. faecium LR-6 | 8 | 1/6 (17) |
| E. faecium LR-9 | 16 | 1/6 (17) |
| E. faecium LR-1 | 32 | 2/6 (33) |
| E. faecium LR-3 | 32 | 2/6 (33) |
| E. faecium LR-5 | 32 | 2/6 (33) |
| E. faecium LR-8 | 32 | 3/6 (50) |
| E. faecium LR-7 | 64 | 4/6 (67) |
| E. faecium LR-4 | 64 | 5/6 (83) |
| E. faecalis LR-2 | 128 | 4/4 (100) |
Detected by sequencing (or MaeI digestion) of PCR-amplified 23S rRNA genes.
Our data are consistent with previous work implicating the G2576U 23S rRNA mutation in resistance to linezolid, and the data suggest that the level of resistance expressed correlates with the percentage of 23S rRNA genes with the mutation (4, 6). Since all of the isolates analyzed in this study were derived from clinical specimens and since the entire 23S gene was not sequenced for all copies, it is possible that other mechanisms of resistance may coexist in these strains. However, substantial in vitro data suggest that this region of the 23S rRNA gene, and this site in particular, is important for linezolid resistance (4, 6). The direct correlation between the number of genes with the mutation and the level of resistance expressed suggests that the percentage of ribosomes in which this mutation is present is the primary determinant of resistance. The occurrence of several strains with multiple mutated 23S genes also suggests that mechanisms other than independent mutations of each 23S gene, such as homologous recombination between mutated and nonmutated copies, may help promote enterococcal evolution under persistent antimicrobial selective pressure.
Acknowledgments
These studies were supported by a Merit Review (L.B.R.) and a Career Development Award (C.J.D.) from the Research Service of the Department of Veterans Affairs.
We are grateful to Tanvir Chowdry for her help with the initial amplifications of the 23rRNA genes.
REFERENCES
- 1.Carias, L. L., S. D. Rudin, C. J. Donskey, and L. B. Rice. 1998. Genetic linkage and cotransfer of a novel vanB-containing transposon (Tn5382) and a low-affinity penicillin-binding protein 5 gene in a clinical vancomycin-resistant Enterococcus faecium isolate. J. Bacteriol. 180:4426-4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Donskey, C. J., J. R. Schreiber, M. R. Jacobs, R. Shekar, F. Smith, S. Gordon, R. A. Salata, C. Whalen, and L. B. Rice. 1999. A polyclonal outbreak of predominantly VanB vancomycin-resistant enterococci in northeast Ohio. Clin. Infect. Dis. 29:573-579. [DOI] [PubMed] [Google Scholar]
- 3.Gonzales, R. D., P. C. Schreckenberger, M. B. Graham, S. Kelkar, K. DenBesten, and J. P. Quinn. 2001. Infections due to vancomycin-resistant Enterococcus faecium resistant to linezolid. Lancet 357:1179. [DOI] [PubMed] [Google Scholar]
- 4.Kloss, P., L. Xiong, D. L. Shinabarger, and A. S. Mankin. 1999. Resistance mutations in 23S rRNA identify the site of action of the protein synthesis inhibitor linezolid in the ribosomal peptidyl transferase center. J. Mol. Biol. 294:93-101. [DOI] [PubMed] [Google Scholar]
- 5.National Committee for Clinical Laboratory Standards. 1997. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, p. M7-A4. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 6.Prystowsky, J., F. Siddiqui, J. Chosay, D. L. Shinabarger, J. Millichap, L. R. Peterson, and G. A. Noskin. 2001. Resistance to linezolid: characterization of mutations in rRNA and comparison of their occurrences in vancomycin-resistant enterococci. Antimicrob. Agents Chemother. 45:2154-2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shinabarger, D. L., K. R. Marotti, R. W. Murray, A. H. Lin, E. P. Melchior, S. M. Swaney, D. S. Dunyak, W. F. Demyan, and J. M. Buysse. 1997. Mechanism of action of oxazolidinones: effects of linezolid and eperezolid on translation reactions. Antimicrob. Agents Chemother. 41:2132-2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsiodras, S., H. S. Gold, E. P. Coakley, C. Wennersten, R. C. Moellering, Jr., and G. M. Eliopoulos. 2000. Diversity of domain V of 23S rRNA gene sequence in different Enterococcus species. J. Clin. Microbiol. 38:3991-3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsiodras, S., H. S. Gold, G. Sakoulas, G. M. Eliopoulos, C. Wennersten, L. Venkataraman, R. C. Moellering, Jr., and M. Ferraro. 2001. Linezolid resistance in a clinical isolate of Staphylococcus aureus. Lancet 358:207-208. [DOI] [PubMed] [Google Scholar]