Abstract
The mechanisms of intrinsic resistance of Mycoplasma hominis to 14- and 15-membered macrolides were investigated in comparison with those of M. pneumoniae, which is naturally susceptible to macrolides. Radiolabeled erythromycin was not accumulated by M. hominis PG21, but addition of an ABC transporter inhibitor increased the level of erythromycin uptake more than two times, suggesting the existence of an active efflux process. The affinity of [14C]erythromycin to ribosomes isolated from M. hominis was dramatically reduced relative to that to ribosomes isolated from M. pneumoniae. The nucleotide sequences of 23S rRNA of both ribosomal operons rrnA and rrnB and ribosomal proteins L4 and L22 of M. hominis were obtained. Compared to the sequence of M. pneumoniae, M. hominis harbored a G2057A transition in its 23S rRNA sequence, as did M. fermentans, another mycoplasma that is erythromycin resistant. An additional C2610U change was also found in the sequence of M. hominis. Moreover, two M. hominis clinical isolates with acquired resistance to 16-membered macrolides were examined for mutations in domain II and domain V of 23S rRNA and in ribosomal proteins L4 and L22. Compared to the sequence of reference strain PG21, one isolate harbored a A2059G transition and a C2611U transition in one of the two rrn operons, while the other one was mutated only at position 2059, also on the same operon. No mutation was found in the two ribosomal protein sequences. Overall, the present study is an exhaustive characterization of the intrinsic resistance of M. hominis to 14- and 15-membered macrolides and the first description of mycoplasma clinical isolates resistant to macrolide, lincosamide, and streptogramin antibiotics harboring a mutation at position 2611 in the 23S rRNA.
Human mycoplasmas are responsible for urogenital and respiratory tract infections. Macrolide, lincosamide, and streptogramin antibiotics (MLSs) are a class of antimicrobials commonly used for the treatment of these infections. The MICs of ketolides, a new class of antimicrobials derived from erythromycin, are low for these microorganisms (3). The MLSs and ketolide antibiotics (MLSKs) inhibit protein synthesis by binding to domain V and domain II of 23S rRNA (13, 21). Three main mechanisms of resistance have been reported: drug inactivation, active efflux, and modification of the target sites by methylation or mutation (46, 49). Resistance by ribosomal mutations in domain II and domain V of 23S rRNA has been described, and recently, mutations in ribosomal proteins L4 and L22 were also associated with resistance to MLSKs (7, 30, 33, 42, 43).
Mycoplasmas present different phenotypes of intrinsic resistance to macrolides. Mycoplasma pneumoniae, a respiratory mycoplasma, is susceptible to all MLSKs. In contrast, M. hominis, a genital species, is naturally resistant to 14- and 15-membered macrolides and ketolides but is susceptible to josamycin, a 16-membered macrolide, and lincosamides. Intrinsic resistance to 14-membered macrolides has been observed in other mycoplasma species, like M. fermentans, a human mycoplasma, and M. pulmonis, an animal mycoplasma. In M. hominis, this resistance has been associated with a guanine-to-adenine transition at position 2057 (G2057A; Escherichia coli numbering) in domain V of 23S rRNA (17), but other mechanisms of resistance remain to be explored.
For M. pneumoniae and M. hominis, strains with acquired resistance to macrolides have very rarely been described (16, 41). M. pneumoniae mutants isolated in vitro (27) or in vivo (31) have been found to harbor A2058G and A2059G transitions, and two in vitro-selected mutants of M. hominis resistant to 16-membered macrolides had mutations at position 2062 (16). Mycoplasmas possess a small number of rrn operons. While M. pneumoniae possesses only one copy of 23S rRNA, the M. hominis genome contains two rRNA operons (26), the complete sequences of which are not yet available. Consequently, the target-related mechanism of resistance through mutation is the most expected mechanism, and both heterozygous and homozygous mutants of M. hominis could be expected.
The purpose of this study was to obtain an understanding of the intrinsic resistance to 14- and 15-membered macrolides in M. hominis. Several approaches were undertaken including studies of the accumulation of radiolabeled erythromycin, measurement of drug-ribosome binding, and characterization of the molecular targets of macrolides, domains V and II of the 23S rRNA gene and ribosomal proteins L4 and L22. Thus, the two copies of the M. hominis 23S rRNA and the genes encoding the L4 and L22 ribosomal proteins were entirely sequenced. The sequences were compared to those of M. pneumoniae and M. pulmonis, for which the complete genome sequences were available, and to that of the M. fermentans 23S rRNA, which was obtained in this study. Finally, two M. hominis clinical isolates resistant to josamycin were examined for point mutations in domain II and domain V of 23S rRNA and in ribosomal proteins L4 and L22.
MATERIALS AND METHODS
Bacterial strains, antibiotics, and chemicals. (i) Strains.
Four reference strains of mycoplasmas, M. hominis PG21 (ATCC 23114), M. pneumoniae FH (ATCC 15531), M. fermentans PG18 (ATCC 19989), and M. pulmonis UAB CTIP (9), and one reference strain of E. coli (ATCC 25922) were used. Two clinical M. hominis isolates, isolates MHb1 and MHb2, resistant to josamycin and fluoroquinolones were described previously (6). Staphylococcus aureus ATCC 25923 and S. aureus RN4220 harboring plasmid pUL5054, which contains the msr(A) gene (35), were used as controls for accumulation assays. E. coli TG1 was used to amplify pGEM7ZF(+) recombinant plasmids X2, H1, and H20.
(ii) Antibiotics.
The antimicrobial agents used in the study were provided by the indicated manufacturers: erythromycin A, josamycin, spiramycin, pristinamycin, quinupristin, quinupristin-dalfopristin, and telithromycin, Aventis Pharma (Romainville, France); clarithromycin, Abbott (Rungis, France); azithromycin, Pfizer (Orsay, France); lincomycin and clindamycin, The Upjohn Co. (Guyancourt, France); midecamycin, Menarini (Rungis, France); and tylosin, Sigma (Saint Quentin Fallavier, France). [N-methyl-14C]erythromycin (47 mCi/mmol) was obtained from NEN Life Science Products (Boston, Mass.).
(iii) Chemicals.
Carbonyl cyanide m-chlorophenylhydrazone (CCCP) and sodium orthovanadate were purchased from Sigma.
MIC determinations.
Susceptibility testing was performed as described previously by an agar dilution method on Hayflick modified medium (2). M. pulmonis was grown in Hayflick modified medium containing 2% (vol/vol) PolyViteX (bioMérieux) before susceptibility testing.
PCR amplifications.
After alignment of seven mycoplasmal sequences of 23S rRNA with Clustal W software (Infobiogen), six primer pairs specific for conserved motifs were chosen to amplify six overlapping fragments ranging from 400 to 600 bp. PCRs were performed on M. hominis PG21 genomic DNA for 40 cycles in a Perkin-Elmer 480 thermal cycler as described previously (5). To amplify regions of interest from both the M. hominis and the M. fermentans 23S rRNAs, primer pair MH23S-3 and MH23S-19 was used for domain II and primer pair MH23S-11 and MH23S-25 was used for domain V (Table 1).
TABLE 1.
Oligonucleotides used in this study
| Primer target and designation | Primer sequence (5′→3′) | Position |
|---|---|---|
| 23S domain II | ||
| MH23S-3 | AGTACCGTGAGGGAAAGGTG | 457-476a |
| MH23S-19 | CTGAATCGTTACTCATACCG | 1258-1277a |
| 23S domain V | ||
| MH23S-11 | TAACTATAACGGTCCTAAGG | 1911-1930a |
| MH23S-25 | CCGCTTAGATGCTTTCAGCG | 2743-2762a |
| 23S 3′, MH23S-21 | GACTATGAGGTTGATAGGCTG | 2822-2842a |
| rrnA and rrnB | ||
| MH23S-7 | TCGTCCACCCAGGGTTAGTC | 1326-1345a |
| MH23S-A | TTGGCATCTTTTAATGCGCTC | 3117-3137b |
| MH23S-B | CTCTTGTCATATTAATTCCTCC | 3012-3033c |
| Ribosomal proteins | ||
| MHL4-U | AGATAATGAAGAACACGTTG | 499-518d |
| MHL4-R | AAAACCTTGTTTAGACTAGC | 929-948d |
| MHL22-U | CCTTCAATGGTCATGGTGCAG | 2717-2737d |
| MHL22-R | TTCCATAACGAATCCATTTGG | 3166-3186d |
Amplification of ribosomal protein L4 and L22 DNA fragments from M. hominis was performed with primer pair MH-L4U and MH-L4R and primer pair MH-L22U and MH-L22R, respectively (Table 1).
Cloning procedures for the two ribosomal operons and the two genes of ribosomal proteins L4 and L22.
Total DNA from strain PG21 was extracted as described previously (50) and digested with restriction endonucleases HindIII and XbaI. Representative libraries of HindIII- and XbaI-digested M. hominis total DNA were constructed with the pGEM7ZF(+) cloning vector (Promega). After transformation of E. coli by ligation mixtures, recombinant clones were selected by colony hybridization with the [α-32P]dATP-labeled PCR products including those of the 23S rRNA of domain V from M. hominis (nucleotides [nt] 1917 to 2769; GenBank accession no. AF443616) or the region coding for L4-L22-flanking ribosomal protein S19 of M. pulmonis (nt 55449 to 55724; GenBank accession no. AL445565). Manipulations of DNA, including electrophoresis, Southern blotting, in situ colony hybridization, propagation, and extraction of plasmids in E. coli, were carried out by standard procedures (37). For Southern and colony hybridization, DNA was radiolabeled with 50 μCi of [α-32P]dATP (3,000 Ci/mmol) by using the Random Primers DNA labeling system from Invitrogen.
Sequencing of the M. hominis 23S rRNA genes and the L4-L22 ribosomal protein-coding regions.
After purification with the Wizard PCR preps DNA purification system (Promega), both strands of the PCR products were directly sequenced by using an ABI PRISM dRhodamine terminator cycle sequencing ready reaction kit and an ABI PRISM 377 sequencer (Applied Biosystems), according to the instructions of the manufacturer. The complete sequences of both strands of clones X2, H1, and H20 were achieved by using the forward and reverse primers flanking the multiple-cloning-site polylinker of the pGEM7ZF(+) vector as well as internal primers. Specific primers located downstream of each ribosomal operon were determined from the DNA sequences of clones X2 and H1, and these were called MH23S-A for operon rrnA and MH23S-B for operon rrnB. Residues specific to each 23S rRNA copy, observed in domain III, were assigned by sequencing the PCR products obtained with internal primer MH23S-7, which is associated with primer MH23S-A or MH23S-B (Table 1).
PFGE.
Pulsed-field gel electrophoresis (PFGE) was performed as described previously by using restriction endonucleases BamHI, SalI, and XhoI (4). The locations of the rrnA and rrnB operons were determined by using as probes two [α-32P]dATP-labeled PCR products, which were obtained by PCR with primer MH23S-21, whose sequence is specific for the 3′ end of the 23S rRNA, and with either primer MH23S-A or primer MH23S-B.
Ribosome isolation and erythromycin binding assays.
Studies of the binding of radiolabeled erythromycin to mycoplasma and E. coli ribosomes were carried out as described previously, with slight modifications (10, 27). For each ribosome preparation, harvested cells were washed twice in phosphate-buffered saline, ground with alumina powder, and suspended in buffer A (10 mM Tris-HCl [pH 7.8], 10 mM MgCl2, 60 mM NH4Cl, 6 mM β-mercaptoethanol) containing DNase I at 2 μg/ml (Promega). Ribosomes were then isolated by three differential centrifugations (10) at 4°C. Ribosomes were stored in small aliquots at −80°C in buffer A. Binding assays were performed by incubating various concentrations of ribosomes (optical density at 260 nm, 1 to 4) with 50 pmol of [N-methyl-14C]erythromycin in 0.5 ml of binding buffer (10 mM Tris-HCl [pH 7.2], 4 mM MgCl2, 100 mM KCl, 10 mM NH4Cl) at 30°C for 30 min. The reactions were stopped by diluting the reaction mixture with 3 ml of cold washing buffer (50 mM Tris-HCl [pH 7.2], 25 mM MgCl2, 755 mM KCl). Ribosomes were collected on 0.45-μm-pore-size nitrocellulose filters (Millipore) and washed four times with 3.5 ml of cold washing buffer. The amount of [14C]erythromycin bound to the filter was determined by liquid scintillation counting.
Accumulation studies.
Uptake studies by a silicone oil method were first performed for the two S. aureus control strains, ATCC 25923 and RN4220/pUL5054, as described previously (35). For mycoplasmas, cells were grown at 37°C in Hayflick modified broth to the mid-log growth phase and harvested by centrifugation at 20,000 × g for 20 min at 4°C. The pellets were washed once, concentrated 20-fold in 50 mM phosphate-buffered saline, and preincubated for 5 min at 37°C. When an energizer (24 mM arginine for M. hominis or 27.8 mM glucose for M. pneumoniae) was used, it was added 5 min before the addition of the radiolabeled antibiotic. After addition of [14C]erythromycin at a final concentration of 0.9 μg/ml, duplicate 0.5-ml aliquots were removed at the times indicated in Fig. 1 and immediately centrifuged through 0.4 ml of silicone oil (density, 1.03; Fluka). The tubes were then frozen at −80°C and cut in the middle of the silicone layer, and residual oil was eliminated. The cell pellets were resuspended in 0.2 ml of glycine-HCl buffer (0.1 M; pH 3.0) and allowed to lyse overnight at room temperature. The radioactivities of the samples were determined by liquid scintillation counting. In some experiments, the efflux pumps inhibitors CCCP (0.1 mM) and sodium orthovanadate (2 mM) were added 10 and 20 min prior to the addition of radiolabeled erythromycin, respectively. When sodium orthovanadate was used, 50 mM sodium phosphate buffer was replaced by 50 mM potassium-HEPES buffer.
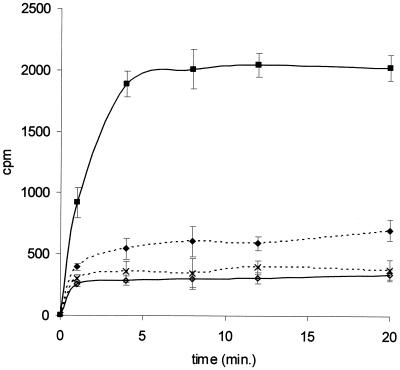
FIG. 1.
Uptake of [14C]erythromycin by M. hominis PG21 cells alone (⋄) and in the presence of sodium orthovanadate (♦) or CCCP (×) and by M. pneumoniae FH (▪). M. hominis cells were energized with 24 mM arginine. CCCP (0.1 mM) and sodium orthovanadate (2 mM) were added 10 and 20 min prior to the addition of the antibiotic, respectively. Results are presented as the means ± standard deviations (error bars) for three independent experiments.
Nucleotide sequence accession numbers.
The nucleotide sequences of the fragments encompassing the 23S rRNA of operons rrnA and rrnB of M. hominis have been assigned GenBank accession nos. AF443616 and AF443617, respectively, and the fragment encompassing the genes encoding the L4 and L22 ribosomal proteins has been assigned GenBank accession no. AY083306. The sequences of both domains II and V of the 23S rRNA gene of M. fermentans have been assigned GenBank accession nos. AY083307 and AY083308, respectively.
RESULTS
MLSK MICs for different mycoplasma species.
Table 2 summarizes the in vitro activities of the MLSKs against the M. pneumoniae, M. fermentans, M. pulmonis, and M. hominis reference strains and against M. hominis clinical isolates MHb1 and MHb2, which will be discussed below. As described previously (3), MLSKs are active against M. pneumoniae. M. hominis PG21 is resistant to 14- and 15-membered macrolides, to some of the 16-membered macrolides like spiramycin and tylosin, to quinupristin, a streptogramin B, and to ketolides. It remains sensitive to josamycin and midecamycin diacetate, two 16-membered macrolides, to clindamycin, and to the streptogramin A- and streptogramin B-associated antibiotics pristinamycin and quinupristin-dalfopristin. M. fermentans is also resistant to 14- and 15-membered macrolides, but the MICs for M. fermentans are 4 and 16 times lower than those for M. hominis, respectively. Among the 16-membered macrolides, in contrast to M. hominis, M. fermentans is susceptible to spiramycin and tylosin, for which MICs are 256- and 128-fold lower than those for M. hominis, respectively. M. pulmonis, an animal mycoplasma phylogenetically close to M. hominis and M. fermentans (25), harbors an MLSK susceptibility profile similar to that of M. fermentans except for the profile of susceptibility to 16-membered macrolides and quinupristin. Indeed, josamycin and midecamycin diacetate are significantly less active against M. pulmonis (MICs, 2 μg/ml) than against both M. fermentans and M. hominis (MIC range, 0.06 to 0.25 μg/ml). Spiramycin shows intermediate activity, with M. fermentans being susceptible and M. hominis being resistant (Table 2), while tylosin, a veterinary macrolide, is very active against M. pulmonis (MIC, 0.06 μg/ml). It should also be noticed that, in contrast to M. fermentans and M. hominis but like M. pneumoniae, M. pulmonis is susceptible to streptogramin B antibiotics (quinupristin MIC, 1 μg/ml). Moreover, among the lincosamides, as described for other bacteria, lincomycin is less potent than clindamycin against these mycoplasmas. Interestingly, M. fermentans and M. pulmonis are susceptible to telithromycin (MICs, 0.25 μg/ml), while M. hominis is resistant, with the telithromycin MIC being 128-fold higher (Table 2).
TABLE 2.
MICs of MLSKs for several mycoplasma species including M. hominis isolates resistant to josamycin
| Strain | MIC (μg/ml)a
|
Intrinsic changesb | Acquired mutations (hetero- zygosity rrnA:rrnB) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ERY | CLR | AZM | JOS | SPI | MID | TYL | LIN | CLI | QUI | Q-D | PRI | TEL | |||
| M. pneumoniae FH | 0.007 | 0.015 | 0.015 | 0.03 | 0.25 | 0.015 | 0.03 | 4 | 1 | 1 | 0.03 | 0.06 | 0.007 | None | None |
| M. fermentans PG18 | 128 | 32 | 2 | 0.25 | 1 | 0.06 | 0.25 | 1 | 0.12 | 64 | 0.25 | 0.25 | 0.25 | G2057A | None |
| M. pulmonis UAB CTIP | 128 | 32 | 8 | 2 | 8 | 2 | 0.06 | 1 | 0.12 | 1 | 0.12 | 0.25 | 0.25 | G2057A | None |
| M. hominis | |||||||||||||||
| PG21 | 512 | 128 | 32 | 0.25 | 256 | 0.25 | 32 | 4 | 0.12 | 64 | 0.25 | 0.5 | 32 | G2057A, C2610U | None |
| MHb1 | 512 | 256 | 512 | 512 | 512 | 128 | 512 | 256 | 64 | 32 | 0.25 | 0.5 | 256 | G2057A, C2610U | A2059G (1A:1G), C2611U (1C:1U) |
| MHb2 | 512 | 256 | 512 | 512 | 512 | 128 | 512 | 32 | 16 | 64 | 0.5 | 1 | 256 | G2057A, C2610U | A2059G (1A:1G) |
ERY, erythromycin; CLR, clarithromycin; AZM, azithromycin; JOS, josamycin; SPI, spiramycin; MID, midecamycin diacetate; TYL, tylosin; LIN, lincomycin; CLI, clindamycin; PRI, pristinamycin; QUI, quinupristin; Q-D, quinupristin-dalfopristin; TEL, telithromycin.
Intrinsic changes in 23S rRNA sequence proposed to be responsible for the macrolide resistance in comparison with the sequence of M. pneumoniae reference strain FH.
Accumulation studies.
Studies of the accumulation of radiolabeled erythromycin were first performed, as described previously for S. aureus (51), by filtering each sample removed on glass microfiber filters. However, as the filter pore size was not small enough to retain mycoplasmas, this method was not suitable. Thus, a silicone oil method (35) was used and validated with erythromycin-resistant S. aureus strain RN4220/pUL5054 containing the msr(A) gene, which confers a macrolide-streptogramin B-type efflux (35, 49). Figure 1 shows the ability of exponential-phase cells of M. hominis PG21 and M. pneumoniae FH to accumulate [14C]erythromycin during a 20-min incubation period. The levels of intracellular [14C]erythromycin increased in M. pneumoniae for up to 5 min before the achievement of a steady state, while the levels of [14C]erythromycin in M. hominis remained very low.
In order to reveal the activity of a putative efflux pump in M. hominis, cells were energized with arginine, the metabolic energy source for this mycoplasma, and the influences of inhibitors of efflux pumps were studied. The presence of the energizing agent did not significantly modify the uptake of the radiolabeled erythromycin in either M. hominis or M. pneumoniae cells (data not shown). As the majority of the bacterial efflux pumps are secondary transporters sensitive to decoupling agents such as CCCP, the protonophore at 0.1 mM was added to the culture 10 min before addition of [14C]erythromycin. The level of accumulation of the 14-membered macrolide was not significantly modified, with only a 10% increase at 20 min in M. hominis. In other assays, sodium orthovanadate, an inhibitor of ABC transporters, the second class of efflux pumps, was added 20 min before addition of labeled erythromycin. At 20 min, the level of uptake of erythromycin in M. hominis PG21 was twofold higher than that without 2 mM orthovanadate (Fig. 1). These data suggest the existence of an active process affecting erythromycin uptake by M. hominis, which is sensitive to some inhibitors of multidrug resistance efflux pumps like orthovanadate but not to CCCP.
Ribosome binding assays.
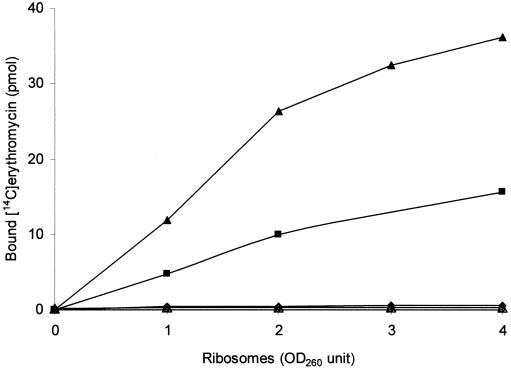
To compare the affinity of erythromycin for the ribosomes from M. hominis, M. fermentans, and M. pneumoniae, the binding of [14C]erythromycin to ribosomes isolated from reference strains PG21, PG18, and FH and from E. coli was measured. The results of these binding assays are shown in Fig. 2. Whatever the ribosome concentration used, the affinity of erythromycin for the ribosomes from both M. hominis and M. fermentans, which are resistant to 14-membered macrolides, was dramatically reduced relative to that for ribosomes from M. pneumoniae, which is naturally susceptible to erythromycin, and to that from ribosomes from E. coli, used as positive controls.
FIG. 2.
Binding of [14C]erythromycin to ribosomes isolated from E. coli (▴), M. pneumoniae FH (▪), M. fermentans PG18 (○), and M. hominis PG21 (⋄) and MHb1 (▵). Assays were performed by incubating various concentrations of ribosomes (optical density at 260 nm [OD260], 1 to 4) with 50 pmol of [14C]erythromycin, as described in Materials and Methods. The curves for strains PG18, PG21, and MHb1 are superposed.
Sequence analysis of the 23S rRNA from M. hominis.
With the aim of determining the complete sequence of M. hominis 23S rRNA, the corresponding gene sequences of all seven mycoplasmas available in databases were aligned. From the alignment, conserved sequence motifs were deduced and used to determine primer pairs suitable for use in PCRs. Six amplification products overlapping the whole 23S rRNA sequence were obtained and subsequently sequenced, leading to a M. hominis 23S rRNA sequence of 2,894 bp. Comparison of the complete M. hominis 23S rRNA sequence with those from the seven mycoplasmas available in databases with the BLAST program, M. genitalium (15), M. pneumoniae (28), M. gallisepticum (39), Ureaplasma urealyticum (19), M. pulmonis (9), M. flocculare (40), and M. hyopneumoniae (28), revealed percentages of identity ranging from 69.9% with the 23S rRNA sequence of M. genitalium to 84.6% with the 23S rRNA sequence of M. hyopneumoniae.
The secondary structure of the M. hominis 23S rRNA was established by comparison with that of the E. coli 23S rRNA (data not shown). The M. hominis 23S rRNA possesses all the characteristic helices described in the E. coli 23S rRNA, constituting the six catalytic domains, which are numbered according to the classical nomenclature. However, M. hominis possesses an extended domain I, with an additional helix in this domain.
Two primer pairs whose sequences were chosen from the complete sequence of the 23S rRNA of M. hominis, primer pair MH23S-3 and MH23S-19 and primer pair MH23S-11 and MH23S-25, were used to amplify domains II and V of M. fermentans PG18, respectively. After sequencing of the nucleotide sequences and comparison to the other mycoplasmal sequences available, the nucleotide sequences obtained from M. fermentans shared the best homology with the sequence of M. pulmonis, with 88% identity for domain II and 91% identity for domain V.
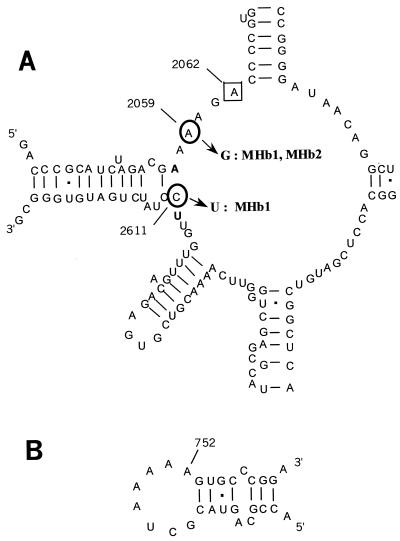
As the interaction sites for macrolides in many bacteria are mainly located in the peptidyltransferase region within domain V of the 23S rRNA (11, 21, 29, 38), the DNA sequences in this region of the intrinsically erythromycin-resistant species M. hominis and M. fermentans were compared to the DNA sequences of 23S rRNA from erythromycin-susceptible M. pneumoniae. M. hominis and M. fermentans both showed a G-to-A transition at position 2057 (E. coli numbering), while in M. pneumoniae, the guanine at position 2057 was conserved. Moreover, M. hominis harbored an additional C-to-U transition at position 2610 (Fig. 3). These two positions have previously been found to be mutated in macrolide-resistant bacteria (7, 46).
FIG. 3.
Secondary structure of the peptidyltransferase center in domain V of 23S rRNA (A) and hairpin 35 in domain II (B) (20) from M. hominis PG21. The nucleotides are numbered on the basis of the E. coli sequence. Mutations G2057A and C2610U, naturally present in the M. hominis 23S rRNA sequence, are indicated in boldface. The circled nucleotides indicate the positions of mutations encountered in clinical isolates MHb1 and MHb2 (positions 2059 and 2611). Framed position 2062 was associated with 16-membered macrolide resistance in M. hominis PG21 (16).
Characterization of the two fragments encompassing the 23S rRNA of ribosomal operons rrnA and rrnB and of the L4 and L22 ribosomal protein genes from M. hominis.
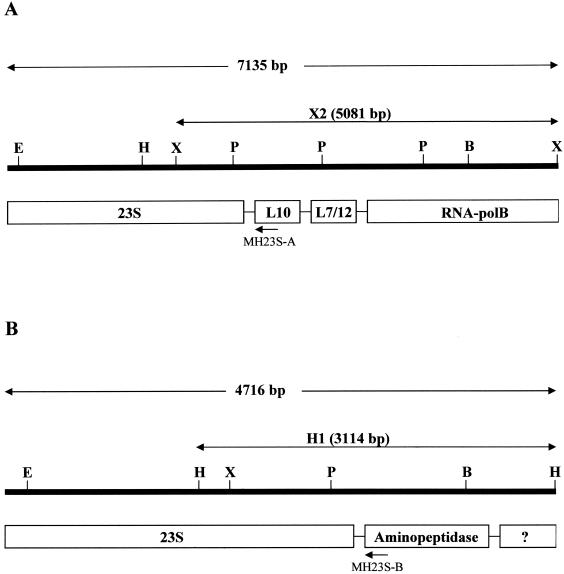
The presence of two ribosomal operons, rrnA and rrnB, within the M. hominis genome was previously suggested by Ladefoged and Christiansen (26). The analysis of the M. hominis 23S rRNA sequence confirmed this organization. Indeed, a mixture of bases C and T at nt 1548 (E. coli nt 1516) was observed on the sequencing traces covering domain III of the M. hominis 23S rRNA. Moreover, two distinct HindIII and XbaI fragments were revealed by Southern hybridization of the digested genomic DNA, with a labeled PCR product from M. hominis overlapping domain V. To determine the molecular organization in the vicinity of both 23S rRNA copies, two libraries were generated by using the HindIII and XbaI sites of the pGEM7ZF(+) vector, respectively. The screening of each library with the domain V-specific probe revealed two different recombinant plasmids, X2 and H1 (Fig. 4). Recombinant plasmid X2, issued from the XbaI library, carried a genomic fragment of 5,081 bp, while plasmid H1, issued from the HindIII library, contained an M. hominis fragment of 3,114 bp.
FIG. 4.
Restriction map and molecular organization of the two fragments encompassing the 23S rRNA of operon rrnA (A) and rrnB (B) from M. hominis PG21. E, EcoRI; H, HindIII; X, XbaI; P, PvuII; B, BamHI; ?, hypothetical protein. Operon-specific primers MH23S-A and MH23S-B are indicated by arrows.
Sequencing of the insert of plasmid X2 revealed at the 5′ end the presence of the last 840 nucleotides of the M. hominis 23S rRNA, followed by three putative open reading frames (ORFs) assumed to encode ribosomal proteins L10 and L7-L12 and the β subunit of the DNA-dependent RNA polymerase, based on similarities to the sequences of known genes from the GenBank database (Fig. 4A). Sequencing of the 3,114-bp fragment of plasmid H1 revealed a different molecular organization (Fig. 4B). At the 5′ end, the last 1,292 nt of the 23S rRNA are followed by two putative ORFs. Only the first ORF (nt 3024 to 4109) was functionally assigned as the gene of an aminopeptidase, namely, the endo-1,4-β-glucanase. These results are consistent with the presence of two copies of 23S rRNA genes in M. hominis. Two primers were then designed to amplify each of the two 23S rRNA genes; the sequence of primer MH23S-A is specific for the beginning of the L10 sequence of rrnA, and the sequence of primer MH23-B is specific for the beginning of the aminopeptidase-like sequence of rrnB (Fig. 4).
PFGE analysis and Southern blot hybridization with a probe containing the sequence for a part of the L10 gene and a probe containing the sequence for a part of the aminopeptidase-like gene confirmed that operons rrnA and rrnB are located within the BamHI, XhoI, and SalI genomic DNA fragments previously described by Ladefoged and Christiansen (26) (data not shown).
Sequencing of the PCR products obtained with 23S rRNA-specific internal primer MH23S-7, which is associated with primer MH23S-A or MH23S-B, clarified the double peaks of C and U residues observed in PCR products containing domain III. Thus, at position 1548, the rrnA sequence possessed a U residue, while rrnB harbored a C residue. Moreover, both alleles carried A2057 and U2610. It should be noted that the sequence analysis of M. fermentans also revealed a mixture of bases C and U at nt 598 and 991 (E. coli numbering) in domain II of 23S rRNA. This is consistent with the presence of two ribosomal operons previously described in M. fermentans (24).
The same cloning and sequencing strategy was conducted to obtain the nucleotide sequences of the genes encoding ribosomal proteins L4 and L22 from M. hominis. Ribosomal proteins are usually organized in a cluster in which the L4 and L22 genes flank the L23, L2, and S19 genes. Alignment of the sequences of the latter ribosomal proteins available from databases for mycoplasmas revealed that the S19 gene possessed the more conserved sequence. Screening of the HindIII library from M. hominis, using as a probe the amplified S19 gene from M. pulmonis, revealed the 5-kb recombinant clone H20. Sequencing of this fragment showed the presence of five complete ORFs which encoded ribosomal proteins L4, L23, L2, S19, and L22. The partial 5′ sequence of the L3 gene was also obtained upstream of the L4 gene, and the partial 3′ sequence of S3 was obtained downstream of the L22 gene. The 969-bp M. hominis L4 gene encoded a protein of 322 amino acids. Alignment with the corresponding proteins from mycoplasmas available in databases revealed identities ranging from 47.7% with the amino acid sequence of M. gallisepticum to 76.4% with the amino acid sequence of M. pulmonis. The 387-bp M. hominis L22 gene encoded a 128-amino-acid protein, which shared identities ranging between 60% with the amino acid sequence of U. urealyticum and 76.6% with the amino acid sequence of M. pulmonis.
Characterization of two clinical isolates of M. hominis resistant to josamycin.
M. hominis clinical isolates MHb1 and MHb2 presented an MLSB resistance phenotype and had similar profiles of resistance to macrolides, quinupristin, and telithromycin but distinct profiles of resistance to lincosamides (Table 2). For MHb1, the MICs of lincomycin and clindamycin were 256 and 64 μg/ml, respectively, while they were eight- and fourfold lower, respectively, for MHb2. Both strains remained susceptible to quinupristin-dalfopristin and pristinamycin (Table 2).
As MHb1 and MHb2 were isolated 6 months apart from two distinct lung aspirates from the same patient, we wondered whether these two isolates were issued from the same clone. PFGE analysis was performed with restriction endonucleases BamHI, SmaI, SalI, and XhoI to determine clonality. The migration patterns were significantly different according to the interpretation guidelines of Tenover et al. (44), while the total genome sizes of the two strains, deduced from the addition of the generated fragment lengths, were nearly identical (data not shown).
DNA sequence analysis of domain V of the 23S rRNA from MHb1 and MHb2 showed that both isolates had an A-to-G transition at position 2059. Moreover, MHb1 presented an additional C-to-U transition at position 2611 (Fig. 3). However, examination of the sequencing traces showed a mixture of bases at the altered residues. By using primers designed to amplify each 23S rRNA gene independently, the nature of the mutations in both isolates was confirmed to be heterozygous, with mutations located in one of the two alleles. Within the rrnB copy, MHb2 contained only the A2059G mutation, while MHb1 had both the A2059G and the C2611U mutations. For both isolates, rrnA was unchanged. As hairpin 35 in domain II of 23S rRNA is also a region where additional erythromycin interactions may occur (13, 21, 52), the DNA sequence of this region of both clinical isolates was analyzed. No difference from the sequence of this region of reference strain M. hominis PG21 was noted. Both isolates were also examined for any change in the ribosomal protein L4 and L22 sequences. In these two resistant strains, the sequences of both L4 and L22 were identical to those of M. hominis PG21. Moreover, as expected, in ribosome binding assays, [14C]erythromycin had no affinity for the ribosome from M. hominis clinical isolate MHb1 (Fig. 2).
DISCUSSION
To determine the role of 23S rRNA in macrolide resistance in M. hominis, we first had to determine its complete sequence, ascertain whether the two ribosomal operons were present, and differentiate the two ribosomal operons in M. hominis. The complete sequence of the M. hominis 23S rRNA obtained was 2,894 bp, which is average for the 23S rRNA lengths reported so far for mycoplasmas (19, 28, 39, 40).
Comparison of the M. hominis 23S rRNA sequence with those from other mycoplasmas available in databases led to the differentiation of two distinct groups. The sequences of the 23S rRNA from one group, including M. genitalium, M. pneumoniae, M. gallisepticum, and U. urealyticum, showed 69.9 to 74.8% identities with that of M. hominis. The sequences of the 23S rRNA from the second group, including M. pulmonis, M. hyopneumoniae, and M. flocculare, possessed higher levels of identity with that of M. hominis, ranging from 81.7 to 84.6%. This splitting into two groups is in agreement with the phylogenetic tree based on 16S rRNA sequences (25). Indeed, based on 16S rRNA analysis, M. hominis has been reported to be more closely related to M. pulmonis, M. fermentans, and M. hyopneumoniae than to M. genitalium, M. pneumoniae, and U. urealyticum. Moreover, this splitting was strengthened by the molecular organization of the ribosomal operons in these species. The typical bacterial organization of rRNA operons, 5′-16S rRNA-23S rRNA-5S rRNA-3′, was reported for M. pneumoniae and M. genitalium (one operon) and for U. urealyticum and M. gallisepticum (two operons). In contrast, M. pulmonis and M. hyopneumoniae possess one ribosomal operon including only the 16S and 23S rRNA genes, but not the 5S rRNA gene, as we observed for M. hominis. Indeed, for M. pulmonis (9), M. hyopneumoniae (40), and also M. fermentans (24), the 5S rRNA gene is known to be separated from the closely linked 16S and 23S rRNA genes. The location of the 5S rRNA genes(s) was not determined in M. hominis.
The interactions of macrolides have mainly been mapped by chemical footprinting experiments (11, 13, 21, 29) and recently by X-ray crystallography (38) and have been mapped to the peptidyltransferase region within domain V of 23S rRNA. Positions A2058 and A2059 (E. coli numbering) appeared to be essential for macrolide binding. Next to those nucleotides, position 2057 is occupied by a guanine in M. pneumoniae M129 (23), an erythromycin-susceptible mycoplasma; however, a G-to-A transition was found at this position in M. hominis PG21 and M. fermentans PG18, both of which are resistant to erythromycin. M. pulmonis (9), M. hyopneumoniae (28), and M. flocculare (40) harbored the same G2057A transition and were found to be intrinsically resistant to 14-membered macrolides but susceptible to 16-membered macrolides and lincosamides (45). Such a transition at position 2057 was also associated with erythromycin resistance but with susceptibility to 16-membered macrolides in E. coli and propionibacteria (14, 36). It should be noted that in E. coli, this mutation at position 2057 was also associated with chloramphenicol resistance (14), but this phenotype was not observed for M. hominis or M. pneumoniae, with both mycoplasmas harboring similar profiles of susceptibility to chloramphenicol (3). Nevertheless, the MICs of 14- and 15-membered macrolides, lincomycin, and telithromycin were higher for M. hominis than for M. fermentans and M. pulmonis. This difference could be associated with the additional C2610U transition. This mutation has recently been associated with macrolide resistance in Streptococcus pneumoniae (7). Moreover, Garza-Ramos et al. (18) described a direct interaction of ketolides with nt 2609 in domain V of 23S rRNA. The C2610U transition present in M. hominis 23S rRNA could modify the affinities of ketolides for the adjacent uracil at position 2609 and participate in the loss of potency of telithromycin against M. hominis.
Furthermore, as mutations in the 23S rRNA domain II and ribosomal proteins L4 and L22 were also associated with MLSK resistance in other bacteria like S. pneumoniae and E. coli (7, 30, 33, 42, 43, 52), the nucleotide sequences of these regions have been examined. Alignment of the sequences of the 23S rRNA genes of M. hominis and other mycoplasmas available in databases revealed that domain II of 23S rRNA is less conserved than the peptidyltransferase region of domain V. Thus, although M. hominis and M. fermentans harbored an adenine at position 752 whereas M. pneumoniae had a cytosine, it is difficult to correlate the intrinsic resistance to macrolides in these species to the nature of the nucleotide at position 752. Sequencing of the entire genes of proteins L4 and L22 did not reveal any of the amino acid alterations previously described to be associated with macrolide resistance in other bacteria.
In M. pneumoniae, susceptibility to erythromycin was associated with the intracellular accumulation of the radiolabeled macrolide and its binding to a preparation of ribosomes. On the contrary, in M. hominis, high-level resistance to erythromycin was associated with an absence of intracellular accumulation and ribosome binding of [14C]erythromycin. The most likely explanation for the higher level of accumulation in M. pneumoniae than in M. hominis is probably due to differences in the levels of drug binding to the intracellular target, as shown in Fig. 2. As macrolide transport into bacteria is a passive process (8), the lack of antibiotic binding to the molecular targets in M. hominis would lead to lower intracellular levels of drug. Thus, the G2057A and the C2610U transitions in M. hominis ribosomes were probably the main mechanism of intrinsic drug resistance. However, the participation of an efflux pump cannot be eliminated. In accumulation studies, addition of orthovanadate, an inhibitor of ATP hydrolysis, but not CCCP increased the rate of intracellular [14C]erythromycin approximately twofold, suggesting the possible presence of an endogenous ABC transporter-like pump. Moreover, this is consistent with the high number of ABC transporters found in mycoplasmas. Paulsen et al. (32) described 11 ABC transporters each in M. genitalium and M. pneumoniae, but only 1 major facilitator superfamily proton motive force-driven transporter. Furthermore, we recently described the physiological presence of a multidrug resistance-like efflux process in M. hominis that is implicated in resistance to ciprofloxacin and ethidium bromide and that probably belongs to the ABC transporter family (34).
The two josamycin-resistant clinical isolates of M. hominis, MHb1 and MHb2, were isolated from the same patient 6 months apart. One could speculate that they were both isolates of the same clonal strain, but their PFGE migration patterns were significantly different, indicating that this patient was infected with two different strains. MHb2 harbored high-level resistance to 16-membered macrolides and telithromycin and intermediate-level resistance to lincosamides, whereas MHb1 harbored high-level resistance to all these drugs. Using a strategy that allowed us to amplify each allele independently, MHb2 was found to contain an A2059G transition on the rrnB copy. This transition was also associated with macrolide resistance in mycobacteria (47), Helicobacter pylori (48), and M. pneumoniae (27). In the last mycoplasma species, which has only one 23S rRNA copy (46), Lucier et al. (27) found that this mutation mediates a resistance profile similar to that of MHb2. Mutants of propionibacteria with an A2059G transition showed the same resistance phenotype but were homozygous at all three loci (36). Recently, in vitro-selected strains and clinical isolates of S. pneumoniae with a macrolide-lincosamide resistance phenotype were described to be heterozygous for this A2059G mutation, with at least two of four alleles carrying the mutation (33, 43; S. Z. Doktor, D. Shortridge, P. Zhong, and R. K. Flamm, Abstr. 41st Intersci. Conf. Antimicrob. Agents Chemother., abstr. 1812, 2001). Thus, a macrolide-lincosamide resistance phenotype is conferred even when the A2059G mutation is heterozygous. In contrast to what is observed in pneumococci (7, 43), a high level of telithromycin resistance is observed in M. hominis concurrently with the A2059G transition. However, a wild-type strain of M. hominis already has a high level of resistance to telithromycin (MIC, 32 μg/ml).
The second M. hominis isolate, MHb1, contained two mutations, A2059G and C2611U. The C2611U mutation was described in Chlamydomonas reinhardtii (22) and was associated with erythromycin and lincosamide resistance. The same C2611U transition was recently described in a Streptococcus pyogenes clinical strain (B. Malbruny, K. Nagai, M. Coquemont, B. Bozdogan, R. Leclercq, and P. C. Appelbaum, Abstr. 41st Intersci. Conf. Antimicrob. Agents Chemother., abstr. 1814, 2001) and in four S. pneumoniae strains selected in vitro (7). In both cases, this mutation was associated with a significant increase in the lincosamide MIC. Interestingly, in M. hominis PG21, the G2057A substitution presumably results in a disruption of the rRNA structure with an opening of the stem preceding the single-stranded portion of the peptidyltransferase region. This structural modification was associated with erythromycin resistance (12). In clinical isolate MHb1, the C2611U transition appears to recreate a Watson-Crick base pair with the adenine at position 2057. However, even if this change could mediate an increase in susceptibility to MLSs, the presence of the A2059G mutation would likely mask it.
Thus, both clinical strains had mutations in domain V but not in domain II of 23S rRNA or in ribosomal protein L4 or L22. The mutations were found only within the rrnB allele, and this heterozygous condition is sufficient to confer macrolide resistance. In such a bacterium, with a small number of rrn operons, a resistance mechanism conferred by mutations was more expected than resistance mediated by an erm-encoded methyltransferase. In both clinical isolates, the absence of rRNA methylase was confirmed by PCR with universal primers for the amplification of an internal segment of every erm-related gene (1). However, these primers could detect only a subset of the numerous erm genes actually present. Thus, the presence of a methylase or a resistance mechanism by drug inactivation cannot be ruled out.
To summarize, the M. hominis 23S rRNA sequence is described here, and the presence of two distinct operons has been confirmed. The intrinsic erythromycin resistance was certainly linked to the G2057A transition, but the high-level resistance might be associated with the additional C2610U transition or with a putative efflux mechanism. Further studies will be required to demonstrate the eventual presence of primary transporters. Moreover, this is the first characterization of mycoplasma clinical isolates that are resistant to MLSKs and that harbor a mutation at position 2611 in the 23S rRNA.
Acknowledgments
We thank Roland Leclercq for providing the erythromycin-resistant S. aureus strain, Alain Blanchard for the M. pulmonis strain, and Florence Doucet-Populaire and Alessandra Occhialini for technical assistance with ribosome binding.
REFERENCES
- 1.Arthur, M., C. Molinas, C. Mabilat, and P. Courvalin. 1990. Detection of erythromycin resistance by the polymerase chain reaction using primers in conserved regions of erm rRNA methylase genes. Antimicrob. Agents Chemother. 34:2024-2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bébéar, C., and J. Robertson. 1996. Determination of minimal inhibitory concentration, p. 189-199. In J. G. Tully and S. Razin (ed.), Molecular and diagnostic procedures in mycoplasmology, vol. II. Academic Press, Inc., San Diego, Calif.
- 3.Bébéar, C. M., and C. Bébéar. 2002. Antimycoplasmal agents, p. 545-566. In S. Razin, and R. Herrmann (ed.), Molecular biology and pathogenicity of mycoplasmas. Kluwer Academic/Plenum Publishers, London, United Kingdom.
- 4.Bébéar, C. M., A. Charron, J. M. Bové, C. Bébéar, and J. Renaudin. 1998. Cloning and nucleotide sequences of the topoisomerase IV parC and parE genes of Mycoplasma hominis. Antimicrob. Agents Chemother. 42:2024-2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bébéar, C. M., O. Grau, A. Charron, H. Renaudin, D. Gruson, and C. Bébéar. 2000. Cloning and nucleotide sequence of the DNA gyrase (gyrA) gene from Mycoplasma hominis and characterization of quinolone-resistant mutants selected in vitro with trovafloxacin. Antimicrob. Agents Chemother. 44:2719-2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bébéar, C. M., J. Renaudin, A. Charron, H. Renaudin, B. de Barbeyrac, T. Schaeverbeke, and C. Bébéar. 1999. Mutations in the gyrA, parC, and parE genes associated with fluoroquinolone resistance in clinical isolates of Mycoplasma hominis. Antimicrob. Agents Chemother. 43:954-956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Canu, A., B. Malbruny, M. Coquemont, T. A. Davies, P. C. Appelbaum, and R. Leclercq. 2002. Diversity of ribosomal mutations conferring resistance to macrolides, clindamycin, streptogramin, and telithromycin in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 46:125-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Capobianco, J. O., and R. C. Goldman. 1990. Erythromycin and azithromycin transport into Haemophilus influenzae ATCC 19418 under conditions of depressed proton motive force (delta mu H). Antimicrob. Agents Chemother. 34:1787-1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chambaud, I., R. Heilig, S. Ferris, V. Barbe, D. Samson, F. Galisson, I. Moszer, K. Dybvig, H. Wroblewski, A. Viari, E. P. C. Rocha, and A. Blanchard. 2001. The complete genome sequence of the murine respiratory pathogen Mycoplasma pulmonis. Nucleic Acids Res. 29:2145-2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doucet-Populaire, F., C. Truffot-Pernot, J. Grosset, and V. Jarlier. 1995. Acquired resistance in Mycobacterium avium complex strains isolated from AIDS patients and beige mice during treatment with clarithromycin. J. Antimicrob. Chemother. 36:129-136. [DOI] [PubMed] [Google Scholar]
- 11.Douthwaite, S. 1992. Interaction of the antibiotics clindamycin and lincomycin with Escherichia coli 23S ribosomal RNA. Nucleic Acids Res. 20:4717-4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Douthwaite, S., and C. Aagaard. 1993. Erythromycin binding is reduced in ribosomes with conformational alterations in the 23S rRNA peptidyl transferase loop. J. Mol. Biol. 232:725-731. [DOI] [PubMed] [Google Scholar]
- 13.Douthwaite, S., L. H. Hansen, and P. Mauvais. 2000. Macrolide-ketolide inhibition of MLS-resistant ribosomes is improved by alternative drug interaction with domain II of 23S rRNA. Mol. Microbiol. 36:183-192. [DOI] [PubMed] [Google Scholar]
- 14.Ettayebi, M., S. M. Prasad, and E. A. Morgan. 1985. Chloramphenicol-erythromycin resistance mutations in a 23S rRNA gene of Escherichia coli. J. Bacteriol. 162:551-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fraser, C. M., J. D. Gocayne, O. White, M. D. Adams, R. A. Clayton, R. D. Fleischmann, C. J. Bult, A. R. Kerlavage, G. Sutton, J. M. Kelley, et al. 1995. The minimal gene complement of Mycoplasma genitalium. Science 270:397-403. [DOI] [PubMed] [Google Scholar]
- 16.Furneri, P. M., G. Rappazzo, M. P. Musumarra, P. Dipietro, L. S. Catania, and L. S. Roccasalva. 2001. Two new point mutations at A2062 associated with resistance to 16-membered macrolide antibiotics in mutant strains of Mycoplasma hominis. Antimicrob. Agents Chemother. 45:2958-2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Furneri, P. M., G. Rappazzo, M. P. Musumarra, G. Tempera, and L. S. Roccasalva. 2000. Genetic basis of natural resistance to erythromycin in Mycoplasma hominis. J. Antimicrob. Chemother. 45:547-548. [DOI] [PubMed] [Google Scholar]
- 18.Garza-Ramos, G., L. Q. Xiong, P. Zhong, and A. Mankin. 2001. Binding site of macrolide antibiotics on the ribosome: new resistance mutation identifies a specific interaction of ketolides with rRNA. J. Bacteriol. 183:6898-6907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glass, J. I., E. J. Lefkowitz, J. S. Glass, C. R. Heiner, E. Y. Chen, and G. H. Cassell. 2000. The complete sequence of the mucosal pathogen Ureaplasma urealyticum. Nature 407:757-762. [DOI] [PubMed] [Google Scholar]
- 20.Gutell, R. R. 1994. Collection of small subunit (16S- and 16S-like) ribosomal RNA structures. Nucleic Acids Res. 22:3502-3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hansen, L. H., P. Mauvais, and S. Douthwaite. 1999. The macrolide-ketolide antibiotic binding site is formed by structures in domains II and V of 23S ribosomal RNA. Mol. Microbiol. 31:623-631. [DOI] [PubMed] [Google Scholar]
- 22.Harris, E. H., B. D. Burkhart, N. W. Gillham, and J. E. Boynton. 1989. Antibiotic resistance mutations in the chloroplast 16S and 23S rRNA genes of Chlamydomonas reinhardtii: correlation of genetic and physical maps of the chloroplast genome. Genetics 123:281-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Himmelreich, R., H. Hilbert, H. Plagens, E. Pirkl, B. C. Li, and R. Herrmann. 1996. Complete sequence analysis of the genome of the bacterium Mycoplasma pneumoniae. Nucleic Acids Res. 24:4420-4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang, Y., J. A. Robertson, and G. W. Stemke. 1995. An unusual rRNA gene organization in Mycoplasma fermentans (incognitus strain). Can. J. Microbiol. 41:424-427. [DOI] [PubMed] [Google Scholar]
- 25.Johansson, K. E., M. Heldtander, and B. Petterson. 1998. Characterization of mycoplasmas by PCR and sequence analysis with universal 16S rDNA primers. Methods Mol. Biol. 104:145-165. [DOI] [PubMed] [Google Scholar]
- 26.Ladefoged, S. A., and G. Christiansen. 1992. Physical and genetic mapping of the genomes of five Mycoplasma hominis strains by pulsed-field gel electrophoresis. J. Bacteriol. 174:2199-2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lucier, T. S., K. Heitzman, S. K. Liu, and P. C. Hu. 1995. Transition mutations in the 23S rRNA of erythromycin-resistant isolates of Mycoplasma pneumoniae. Antimicrob. Agents Chemother. 39:2770-2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ludwig, W., G. Kirchof, N. Klugbauer, M. Weizenegger, D. Betzl, M. Ehrmann, C. Hertel, S. Jilg, R. Tatzel, H. Zitzelsberger, S. Liebl, M. Hochberger, J. Shah, D. Lane, and P. R. Wallnoef. 1992. Complete 23S ribosomal RNA sequences of gram-positive bacteria with a low DNA G+C content. Syst. Appl. Microbiol. 15:487-501. [Google Scholar]
- 29.Moazed, D., and H. F. Noller. 1987. Chloramphenicol, erythromycin, carbomycin and vernamycin B protect overlapping sites in the peptidyl transferase region of 23S ribosomal RNA. Biochimie 69:879-884. [DOI] [PubMed] [Google Scholar]
- 30.Nagai, K., P. C. Appelbaum, T. A. Davies, L. M. Kelly, D. B. Hoellman, A. T. Andrasevic, L. Drukalska, W. Hryniewicz, M. R. Jacobs, J. Kolman, J. Miciuleviciene, M. Pana, L. Setchanova, M. K. Thege, H. Hupkova, J. Trupl, and P. Urbaskova. 2002. Susceptibilities to telithromycin and six other agents and prevalence of macrolide resistance due to L4 ribosomal protein mutation among 992 pneumococci from 10 Central and Eastern European countries. Antimicrob. Agents Chemother. 46:371-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Okazaki, N., M. Narita, S. Yamada, K. Izumikawa, M. Umetsu, T. Kenri, Y. Sasaki, Y. Arakawa, and T. Sasaki. 2001. Characteristics of macrolide-resistant Mycoplasma pneumoniae strains isolated from patients and induced with erythromycin in vitro. Microbiol. Immunol. 45:617-620. [DOI] [PubMed] [Google Scholar]
- 32.Paulsen, I. T., L. Nguyen, M. K. Sliwinski, R. Rabus, and M. H. Saier, Jr. 2000. Microbial genome analyses: comparative transport capabilities in eighteen prokaryotes. J. Mol. Biol. 301:75-100. [DOI] [PubMed] [Google Scholar]
- 33.Pihlajamaki, M., J. Kataja, H. Seppala, J. Elliot, M. Leinonen, P. Huovinen, and J. Jalava. 2002. Ribosomal mutations in Streptococcus pneumoniae clinical isolates. Antimicrob. Agents Chemother. 46:654-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raherison, S., P. Gonzalez, H. Renaudin, A. Charron, C. Bébéar, and C. M. Bébéar. 2002. Evidence of active efflux in resistance to ciprofloxacin and to ethidium bromide by Mycoplasma hominis. Antimicrob. Agents Chemother. 46:672-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ross, J. I., E. A. Eady, J. H. Cove, W. J. Cunliffe, S. Baumberg, and J. C. Wootton. 1990. Inducible erythromycin resistance in staphylococci is encoded by a member of the ATP-binding transport super-gene family. Mol. Microbiol. 4:1207-1214. [DOI] [PubMed] [Google Scholar]
- 36.Ross, J. I., E. A. Eady, J. H. Cove, C. E. Jones, A. H. Ratyal, Y. W. Miller, S. Vyakrnam, and W. J. Cunliffe. 1997. Clinical resistance to erythromycin and clindamycin in cutaneous propionibacteria isolated from acne patients is associated with mutations in 23S rRNA. Antimicrob. Agents Chemother. 41:1162-1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook, J., F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Schlunzen, F., R. Zarivach, J. Harms, A. Bashan, A. Tocilj, R. Albrecht, A. Yonath, and F. Franceschi. 2001. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature 413:814-821. [DOI] [PubMed] [Google Scholar]
- 39.Skamrov, A. V., M. A. Gol'dman, E. S. Feoktistova, and R. Bibilashvili. 2000. Determination and analysis of the nucleotide sequence of a segment of a Mycoplasma gallisepticum strain A5969 chromosome, containing operons S10 and rrn23-5. Mol. Biol. (Moscow) 34:390-396. [PubMed] [Google Scholar]
- 40.Stemke, G. W., Y. Huang, F. Laigret, and J. M. Bové. 1994. Cloning the ribosomal RNA operons of Mycoplasma flocculare and comparison with those of Mycoplasma hyopneumoniae. Microbiology 140:857-860. [DOI] [PubMed] [Google Scholar]
- 41.Stopler, T., and D. Branski. 1986. Resistance of Mycoplasma pneumoniae to macrolides, lincomycin and streptogramin B. J. Antimicrob. Chemother. 18:359-364. [DOI] [PubMed] [Google Scholar]
- 42.Taitkamradt, A., T. Davies, P. C. Appelbaum, F. Depardieu, P. Courvalin, J. Petitpas, L. Wondrack, A. Walker, M. R. Jacobs, and J. Sutcliffe. 2000. Two new mechanisms of macrolide resistance in clinical strains of Streptococcus pneumoniae from Eastern Europe and North America. Antimicrob. Agents Chemother. 44:3395-3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taitkamradt, A., T. Davies, M. Cronan, M. R. Jacobs, P. C. Appelbaum, and J. Sutcliffe. 2000. Mutations in 23S rRNA and ribosomal protein L4 account for resistance in pneumococcal strains selected in vitro by macrolide passage. Antimicrob. Agents Chemother. 44:2118-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ter Laak, E. A., A. Pijpers, J. H. Noordergraaf, E. C. Schoevers, and J. H. Verheijden. 1991. Comparison of methods for in vitro testing of susceptibility of porcine Mycoplasma species to antimicrobial agents. Antimicrob. Agents Chemother. 35:228-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vester, B., and S. Douthwaite. 2001. Macrolide resistance conferred by base substitutions in 23S rRNA. Antimicrob. Agents Chemother. 45:1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wallace, R. J., Jr., A. Meier, B. A. Brown, Y. Zhang, P. Sander, G. O. Onyi, and E. C. Bottger. 1996. Genetic basis for clarithromycin resistance among isolates of Mycobacterium chelonae and Mycobacterium abscessus. Antimicrob. Agents Chemother. 40:1676-1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang, G., and D. E. Taylor. 1998. Site-specific mutations in the 23S rRNA gene of Helicobacter pylori confer two types of resistance to macrolide-lincosamide-streptogramin B antibiotics. Antimicrob. Agents Chemother. 42:1952-1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weisblum, B. 1995. Erythromycin resistance by ribosome modification. Antimicrob. Agents Chemother. 39:577-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Williamson, D. L., J. Renaudin, and J. M. Bové. 1991. Nucleotide sequence of the Spiroplasma citri fibril protein gene. J. Bacteriol. 173:4353-4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wondrack, L., M. Massa, B. V. Yang, and J. Sutcliffe. 1996. Clinical strain of Staphylococcus aureus inactivates and causes efflux of macrolides. Antimicrob. Agents Chemother. 40:992-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xiong, L. Q., S. Shah, P. Mauvais, and A. S. Mankin. 1999. A ketolide resistance mutation in domain II of 23S rRNA reveals the proximity of hairpin 35 to the peptidyl transferase centre. Mol. Microbiol. 31:633-639. [DOI] [PubMed] [Google Scholar]