Abstract
The human papillomaviruses (HPVs) are epitheliotropic viruses that require the environment of a differentiating squamous epithelium for their life cycle. HPV infection through abrasion of the skin or sexual intercourse causes benign warts and sometimes cancer. HPV DNA detected in the blood has been interpreted as having originated from metastasized cancer cells. The present study examined HPV DNA in banked, frozen peripheral blood mononuclear cells (PBMCs) from 57 U.S. human immunodeficiency virus (HIV)-infected pediatric patients collected between 1987 and 1996 and in fresh PBMCs from 19 healthy blood donors collected in 2002 to 2003. Eight patients and three blood donors were positive mostly for two subgroups of the HPV type 16 genome. The HPV genome detected in all 11 PBMC samples existed as an episomal form, albeit at a low DNA copy number. Among the eight patients, seven acquired HIV from transfusion (three associated with hemophilia) and one acquired HIV through vertical transmission; this patient also had received a transfusion before sampling. Our data suggest that PBMCs may be HPV carriers and might spread the virus through blood.
Sexual transmission of and infection with human papillomaviruses (HPVs) are widely recognized as a cause of anogenital warts and cervical cancer. The infection through abrasion of the skin or sexual intercourse is initiated when a viral particle gains entry into a basal epithelial cell. While all cells of a wart contain the viral genome, viral gene expression and multiplication occur exclusively in the nuclei of the infected cells and are tightly linked to the state of differentiation of the cells. In basal and parabasal cells, viral DNA replicates at a low level as an episome and only early genes are transcribed. Extensive viral DNA multiplication and transcription of all viral genes as well as capsid formation occur only in the most superficial layers of the epithelium (14). It has been widely accepted that HPVs are not disseminated to other sites by blood, i.e., there is no viremic phase in the course of HPV infection. However, successful transmission of bovine papillomavirus type 2 from peripheral blood (35) raises the possibility that HPVs might in some circumstances be spread via a hematogenous route. In addition, HPV DNA can be detected in the peripheral blood mononuclear cells (PBMCs) (29), sera (22), or plasma (9) of patients with cervical cancer or HPV-associated head and neck squamous cell carcinoma (5). It should therefore be considered whether PBMCs might serve as a carrier of HPV during the course of HPV infection.
Women with human immunodeficiency virus (HIV) infection have a high prevalence of cervical HPV infection and cervical cancer (10, 36). Although both HIV and HPV are sexually transmitted and this could partly account for the higher prevalence of HPV infection in HIV-positive patients, HIV-associated immunosuppression might contribute to reactivation of preexisting HPV infection and predispose patients to progression to high-grade squamous intraepithelial lesions (1, 25). Also, HIV infection of CD4+ cells might hypothetically reactivate HPV within the PBMCs if the HPV genome resides in the cells.
In this report, we have examined HPV DNA in PBMCs obtained from HIV-infected pediatric patients and healthy blood donors. Our data document that the HPV genome is associated with PBMCs and hence could potentially be spread through blood transfusion.
MATERIALS AND METHODS
PBMC acquisition and DNA extraction.
To determine the presence of HPV infection in PBMCs, a total of 76 banked, frozen PBMC samples obtained between 1987 and 1996 from 57 U.S. pediatric patients with vertical or transfusion-acquired HIV infection, with a median age of 13.2 years (Table 1), enrolled in National Cancer Institute (NCI) Institutional Review Board-approved protocols, were analyzed. All clinical blood specimens obtained from pediatric patients were obtained by nurses or phlebotomists wearing gloves. PBMCs were isolated from clinical specimens by the standard Ficoll-Hypaque gradient separation technique and cryopreserved in a vapor-phase liquid nitrogen storage freezer. A total of 24 PBMC samples from 19 healthy blood donors without clinical complaints at the time of donation were also collected from the NIH Clinical Center blood bank over a period of 6 months in 2002 to 2003 and were isolated by a centrifugal elutriation technique performed by the Cell Processing Section of the NIH Clinical Center blood bank. For HIV-positive samples, all PBMC samples, each at >2 × 106, were randomly coded and blinded, with random duplications as internal controls. DNA samples were extracted directly from PBMCs by brief centrifugation and homogenized in 1 ml of DNAzol (Molecular Research Center, Inc., Cincinnati, OH) according to the manufacturer's protocol. The isolated DNA was dissolved in ∼500 μl of 8 mM NaOH and adjusted to pH 7.0 with 1 M HEPES. Various precautions were taken to minimize sample-to-sample cross-contamination, including limiting HIV-PBMC sample processing and DNA extraction to a maximum of 10 samples per day.
TABLE 1.
Demographic characteristics and prevalence of HPV DNA in PBMCs of pediatric HIV patients and healthy blood donors
| Characteristic | No. (%) of patients or donorsa
|
Pb | ||
|---|---|---|---|---|
| Total | HPV-pos | HPV-neg | ||
| Pediatric HIV patients | 57 | 8c | 49 | |
| Mode of HIV acquisition | ||||
| Transfusion | 17 | 4 (23.5) | 13 (76.5) | 0.35* |
| Hemophilia | 21 | 3 (14.3) | 18 (85.7) | |
| Vertical | 19 | 1 (5.3) | 18 (94.7) | |
| CD4 count/mm3 | ||||
| Minimal (>500) | 4 | 0 | 4 | 0.78** |
| Moderate (200-500) | 10 | 2 (20) | 8 (80) | |
| Severe (<200) | 43 | 6 (14) | 37 (86) | |
| Gender | ||||
| Male | 40 | 5 (12.5) | 35 (87.5) | 0.68*** |
| Female | 17 | 3 (17.6) | 14 (82.4) | |
| Aged (yr) | ||||
| ≤13 | 27 | 3 (11.1) | 24 (88.9) | 0.38**** |
| >13 | 30 | 5 (16.7) | 25 (83.3) | |
| Healthy blood donors | 19 | 3 | 16 | |
| Gender | ||||
| Male | 16 | 2 (12.5) | 14 (87.5) | 0.42*** |
| Female | 3 | 1 (33.3) | 2 (66.7) | |
| Race | ||||
| White | 11 | 0 | 11 | 0.06*** |
| Black | 8 | 3 (37.5) | 5 (62.5) | |
| Aged (yr) | ||||
| 20-35 | 8 | 3 (37.5) | 5 (62.5) | 0.08**** |
| ≥36 | 11 | 0 | 11 (100) | |
HPV-pos and HPV-neg, HPV positive and negative, respectively.
Two-tailed P value for HPV positive versus HPV negative. *, by Mehta's version of Fisher's exact test for all three acquisition categories versus HPV positivity. P = 0.25 by Fisher's exact test for vertical versus transfusion/hemophilia acquired: odds ratio = 0.25 (95% exact confidence interval, 0.005 to 2.22). **, by exact Cochran-Armitage trend test. ***, by Fisher's exact test. ****, by Wilcoxon rank sum test.
P = 1.00 for 8/57 versus 3/19 HPV positive in pediatric HIV patients versus healthy blood donors.
Median ages, in years, with ranges in parentheses, were as follows: for total, HPV-positive, and HPV-negative pediatric HIV patients, 13.2 (2 to 29), 13.3 (10 to 18), and 13.0 (2 to 29), respectively; for total, HPV-positive, and HPV-negative healthy blood donors, 36 (23 to 71), 34 (23 to 35), and 40.5 (29 to 71), respectively.
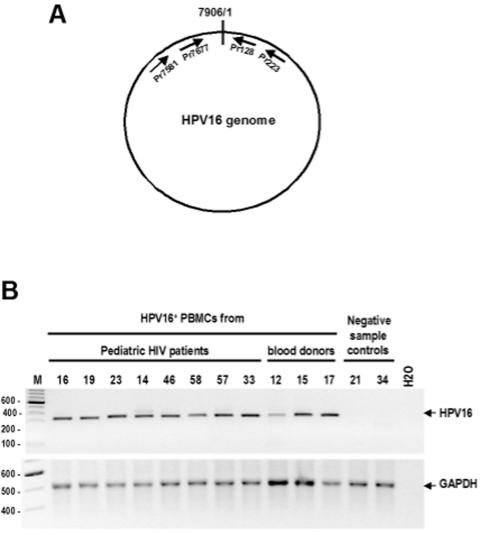
HPV DNA detection and sequencing. Detection of HPV L1 and HPV type 16 (HPV16) E2 and E6 genes either from randomly coded and blinded PBMC DNA samples or from blood donor PBMC DNA samples was performed by nested PCR as described previously (3). After the PCR products for L1 were sequenced and an HPV type was confirmed, two sets of HPV type-specific E6 and E2 primers for nested PCR were further applied for HPV type-specific detection and sequencing. By combining the detection for L1, E2, and E6 genes that cover the two ends and middle part of the virus genome, this strategy allowed us to analyze whether a full-length HPV genome existed in the PBMCs. Head-to-tail junctions of HPV genomes were further analyzed to determine the presence of an episomal HPV genome as detailed in Fig. 3. Two sets of HPV16-specific primers for nested PCR were used for the detection, including two forward primers, Pr7581 (5′-CACTGCTTGCCAACCATTCC-3′) and Pr7677 (5′-GCCAACGCCTTACATACCG-3′), and two backward primers, Pr128 (5′-GTCGCTCCTGTGGGTCCTG-3′) and Pr223 (5′-ACGTCGCAGTAACTGTTGC-3′).
FIG. 3.
Detection of head-to-tail junctions of episomal HPV16 genomes in HPV16-positive PBMCs from pediatric HIV patients and healthy blood donors. (A) Schematic diagram of circular, episomal HPV16 genome and relative positions of the PCR primers used in this study. Drawings are not to scale. (B) Head-to-tail junction products amplified from individual HPV16-positive PBMC DNA samples by the nested PCR in which the primers Pr7581 and Pr223 were used for the first PCR and the primers Pr7677 and Pr128 were used for the nested PCR. Shown on the top of the gel are patient or donor sample numbers. HPV16-negative PBMC DNA samples (21 and 34) in Fig. 1 were used as negative controls. See the description of GAPDH for DNA quality control in Fig. 1. Lane M, molecular size markers (sizes at left in base pairs).
Gel-purified PCR products with the expected sizes were used as DNA templates in cycle-sequencing reactions (BigDye Terminator cycle sequencing kit; Applied Biosystems, Foster City, CA) from two different directions. Sequencing reaction mixtures were purified using Centricep Spin columns (Princeton Separations, Adelphia, NJ) and were run in the Applied Biosystems model 377 sequencing apparatus. Sequence data compiled were analyzed using Sequencher sequence analysis software (Gene Codes Corp., Ann Arbor, MI).
Validation of PCRs.
Each DNA sample was screened for the presence of human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) DNA by PCR amplification with a primer set as previously described (3). This primer set amplifies a 496-bp product and provides an indication of good DNA quality for each sample. For HPV detection, two water controls were also included for both first-run and nested PCR. If either of the two water controls yielded a false positive in the nested PCR, the whole set of PCRs and nested PCRs were restarted. A sample was deemed to be HPV positive if it yielded PCR products at least twice in a total of three repeats with the expected sizes for both L1 and E6 that could be further confirmed by DNA sequencing. This high stringency allowed us to exclude any possibility of laboratory cross-contamination in the nested PCR. Sample unblinding was then performed after completion of the detection and sequencing.
RESULTS
Presence of HPV genome in PBMCs of pediatric HIV patients.
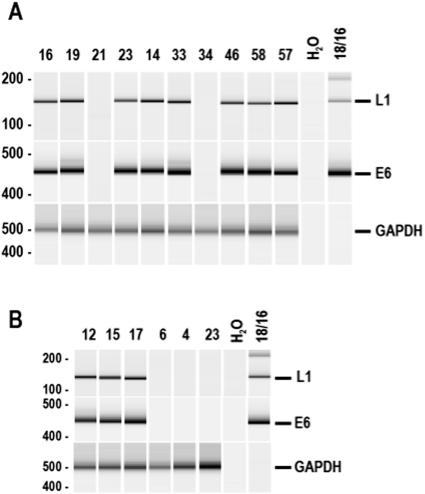
Of the 76 samples from 57 pediatric HIV patients, 10 samples (two duplicates from the same draw date) from eight patients were positive for HPV16 DNA (Fig. 1). Of the eight patients (14%) with HPV detected, seven had transfusion-acquired HIV (three associated with hemophilia) and one had vertical infection with a history of transfusion before sampling (Table 1). Although there was no significant difference in this small study in the rate of HPV DNA detection according to the patients' mode of HIV acquisition (P = 0.35) or in the overall ages of patients with or without HPV (P = 0.38), the data suggest that the PBMC HPV DNA in these patients might be acquired through blood products. Among those eight patients with PBMC HPV DNA, three were 13 years of age, two 11, one 14, one 17, and one 18 years of age when the blood samples were drawn. According to clinical history, all of the HPV-positive pediatric patients were sexually naive at that time.
FIG. 1.
Electrophoreticprofiles of HPV L1 and HPV16 E6 DNA products amplified from banked PBMCs of pediatric HIV patients (A) and from PBMCs of healthy blood donors (B). HPV L1 and HPV16 E6 genes were detected, respectively, from purified total PBMC DNA by nested PCR with primer sets as described previously (3). PCR amplification was performed as described previously (3) with a primer set for the human GAPDH gene serving as a DNA quality control for each sample. Gel images are representatives of amplified products analyzed with an Agilent 2100 Bioanalyzer. Shown on the top of each panel are patient sample numbers, water controls, and HPV18 DNA (for L1) as well as HPV16 DNA (for E6) controls. Numbers at left of panels are molecular sizes in base pairs.
Analysis of 19 duplicate samples from 13 patients with more than one sample tested showed that five patients had duplicate samples with the same results, with two duplicates positive and three duplicates negative for HPV DNA, indicating reliable HPV DNA detection. Other patients with samples from different time points, including samples from two patients positive for HPV DNA, were negative for HPV DNA at another draw date at least 5 months distant, implying a fluctuation of HPV DNA levels in HIV patients' PBMCs. All eight patients with PBMC HPV DNA had moderate (two patients) or severe (six patients) immune suppression (Table 1).
Detection of HPV genome in PBMCs of healthy blood donors.
To further explore the possibility that blood transfusion could be a source of acquired PBMC HPV DNA, we obtained 24 PBMC samples from 19 healthy blood donors over a period of 6 months and examined them for HPV DNA using the same HPV DNA detection strategy as described above. Three donors (15.8%) were also positive for HPV16 DNA in their PBMCs (Table 1; Fig. 1). Interestingly, two donors with multiple samples at different time points were documented to have HPV16 DNA only once, again suggesting that detection of HPV16 DNA in PBMCs may be transient.
PBMCs carry an episomal HPV genome.
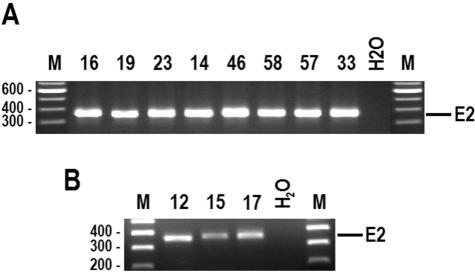
To determine the physical status of HPV DNA detected in the PBMCs, the HPV16 E2 gene from all 11 HPV16 DNA-positive PBMCs was examined. In general, an intact E2 gene is disrupted upon HPV integration, thus distinguishing the episomal form of HPV DNA from the integrated form by detection of an intact E2 gene. Although it is indirect, amplification of the E2 region indicates the presence of episomal HPV DNA in the PBMCs; otherwise, it is assumed that the DNA has integrated (18, 39). Using this approach, we demonstrated that all 11 HPV16 DNA-positive PBMC specimens had the intact HPV16 E2 gene (Fig. 2), suggesting that the detected HPV genome in the PBMCs is episomal.
FIG. 2.
Detection of intact HPV16 E2 gene in all of 11 HPV16-positive PBMCs. Total PBMC DNA was detected by nested PCR with HPV16 E2 primer sets as described previously (3). (A) Intact E2 in pediatric HIV patients with HPV-positive PBMCs. (B) Intact E2 in healthy blood donors with HPV-positive PBMCs. Shown on the top of each panel are patient (A) or donor (B) sample numbers and water controls. Lanes M, molecular size markers (sizes at left in base pairs).
To further confirm the presence of episomal HPV16 DNA in the PBMCs, we detected the head-tail junctions of episomal HPV16 genomes in assuming that an episomal genome should be circular. We found that all of the 11 HPV16-positive PBMC DNA samples from pediatric HIV patients and healthy blood donors rendered an amplicon with correct sizes by nested PCR with two sets of primers, the forward primers covering the end of the HPV16 genome and the backward primers being positioned at the beginning of the genome (Fig. 3). All of the 11 amplicons were sequenced and showed a correct head-tail junction (…TAATACTAA-7906/1-ACTACAA.……), demonstrating the existence of the circular (episomal) HPV16 genomes in the PBMCs.
Two subgroups of HPV genomes in PBMCs.
Sequence analysis of all HPV16 E6 and E2 amplicons from PBMCs indicated that they were European variants and amplified mainly from two subgroups of the HPV16 genome that have not been reported in genital or cervical variants and are different from our laboratory strains (HPV16R, CaSki and SiHa) (Table 2), convincingly indicating that the detected HPV16 DNA in PBMCs was not a result of cross-contamination in our laboratory. One subgroup (five isolates) has an A-to-T change at the nucleotide (nt) 362 position in conjunction with a C-to-A change at the nt 3684 position, subsequently resulting in a missense mutation in the E6 (T87S) and E2 (T310K) proteins, respectively. The other subgroup (four isolates) has prototype HPV16R sequences in the corresponding positions, but three of them have a T-to-G change at the nt 350 position, leading to an amino acid change (L83V) in the E6 protein. Two other isolates from pediatric HIV patients could not be grouped: one (patient 19) has the same nucleotide sequences at those positions as seen in CaSki HPV16, but the nucleotide sequence at the nt 350 position could not be determined as a T or G in multiple sequencing reactions, and the other (patient 33) had all three nucleotide sequences identical to HPV16R at the corresponding positions except the one at nt 362, at which an A-to-T change leads to an amino acid change in the E6 (T87S).
TABLE 2.
Sequence variations in HPV16 E6 and E2 DNA obtained from PBMCsa
| Source of HPV16 DNA | Nucleotide variation in HPV16 genome at nt:b
|
|||
|---|---|---|---|---|
| 350 | 362 | 442 | 3684 | |
| PBMCs of pediatric HIV patient: | ||||
| 14 | G (V) | A (T) | A (E) | C (T) |
| 23 | N | A (T) | A (E) | C (T) |
| 58 | G (V) | A (T) | A (E) | C (T) |
| 19 | N | A (T) | A (E) | A (K) |
| 16 | G (V) | T (S) | A (E) | A (K) |
| 46 | N | T (S) | A (E) | A (K) |
| 57 | T (L) | T (S) | A (E) | A (K) |
| 33 | T (L) | T (S) | A (E) | C (T) |
| PBMCs of heathy blood donor: | ||||
| 12 | T (L) | T (S) | A (E) | A (K) |
| 17 | T (L) | T (S) | A (E) | A (K) |
| 15 | G (V) | A (T) | A (E) | C (T) |
| CaSki cellsb | G (V) | A (T) | A (E) | A (K) |
| SiHa cellsb | G (V) | A (T) | C (D) | A (K) |
| HPV16R | T (L) | A (T) | A (E) | C (T) |
Positions available in the E6 (nt 350, 362, and 442) and E2 (nt 3684) PCR products. N, ambiguous G or T. Letter in parentheses, a resultant amino acid residue in HPV16 E6 or E2 protein.
Both CaSki and SiHa cells are HPV16-positive cervical cancer cell lines.
DISCUSSION
There are two groups of HPVs based on clinical infection: cutaneous HPVs and mucosal HPVs. These include approximately 200 types of HPVs that have less than 90% similarity with each other at the nucleotide level (2, 26). Cutaneous HPV infection, commonly via abrasion of skin, often causes benign skin warts but, in some rare situations, has been associated with skin cancer (30) and is usually related to HPV5 and HPV8 infection (24). Anogenital HPV infection is the most common type of mucosal HPV infection acquired through sexual intercourse. The oncogenic potential of high-risk HPVs, such as HPV16, -18, and -31, has been well documented in the development of anogenital cancer (27), particularly cancer of the cervix (27, 33) and anus (11). Recent studies suggest that HPV infection may play a role in the development of oral cancer (19, 32, 41), head and neck cancer (12, 31), esophageal cancer (34, 38), lung cancer (8, 16), and colorectal cancer (3, 7). In addition, other reports document the presence of HPV DNA in prostatic tissue (46), sperm cells (20, 21), and breast cancer tissue (43, 45). The latter observations raise questions as to how HPV could localize to these organ tissues given the lack of direct infection and the historical presumption that HPV viremia and hematogenous dissemination do not occur.
Perhaps there is no better interpretation than the finding of HPV DNA in PBMCs to address how HPV could spread and infect epithelial cells in other organs. Previously, several laboratories have demonstrated that HPV DNA could exist in PBMCs of patients with genital HPV infection (29), in the peripheral blood of patients with cervical cancer (15, 17, 28, 40), and in the sera or plasma of patients with cervical cancer (9, 22) or head and neck squamous cell carcinoma (5). However, HPV DNA detected in the peripheral blood has historically been presumed to have originated from metastasized cancer cells in the blood or from virus-containing cell debris being shed into the blood from local HPV infection. Our study demonstrates that the HPV16 genome exists in PBMCs of pediatric HIV patients who acquired HIV infection via transfusion and vertical transmission (one patient with a history of transfusion before sampling) and who were, according to clinical history, sexually naive. Further study demonstrated that the HPV16 genome is also present in PBMCs of “healthy” blood donors, suggesting a potential for transmission via the bloodstream. To our knowledge, these HPV DNA-positive donors had no clinical complaints or history of genital HPV infection when their blood samples were drawn. However, the possible existence of asymptomatic HPV infection in these donors at the time of blood sample donation cannot be excluded. More importantly, the presence of HPV DNA in PBMCs in this study, albeit at a low DNA copy number, is very unlikely to be a result of cross-contamination since extremely stringent criteria had been used to establish HPV DNA positivity (see Materials and Methods) and subsequent HPV sequencing confirmed the detection of unique HPV variants distinct from laboratory strains.
Although the HPV genome replicates as an episome in benign and most preinvasive lesions, it is integrated into the cellular DNA in most cancers. Prior to integration, the episomal, circular viral genome undergoes linearization by a break, which most frequently occurs in the E2 region (18, 39). Thus, the viral E2 gene is often disrupted during HPV DNA integration. In this report, we show that the HPV16 genome in all PBMCs positive for HPV DNA has an intact E2 and thus exists as an episomal form. This conclusion was further supported by the presence of circular HPV16 genome forms with a head-to-tail linkage in all HPV16-positive PBMC DNAs. Most interestingly, the episomal HPV16 genome in the PBMCs described in this report can be grouped into two subgroups based on nucleotide sequence variations in the E6 and E2 regions. Although mutations in nt 350 and nt 442 in the E6 region and nt 3684 in the E2 region of HPV16 have been documented (23, 37, 42), one subgroup of HPV16 genome characterized from PBMCs in our study contains an additional novel mutation at nt 362 (A to T) which has not been reported before. HPV16 intratypic heterogeneity has been an important focus of phylogenetic studies, and the distribution of HPV16 variants has been geographically grouped into five distinct phylogenetic branches: European, Asian, Asian-American, African-1, and African-2 (6, 13). Recently studies suggest that HPV intratypic sequence variation might be a risk factor for the development of high-grade cervical intraepithelial neoplasia (44) and different forms of cervical cancer (4). Specifically, women with HLA-B*44, HLA-B*51, or HLA-B*57 who were infected with the HPV16 E6 variant L83V had an approximately four- to fivefold-increased risk for cervical cancer (47). Thus, it will be interesting to know whether the two subgroups of HPV16 variants identified from PBMCs in our study are biologically different from other common variants in pathogenicity and immunogenicity.
The results from this study have important implications regarding HPV transmission and pathogenesis. However, we were unable to detect HPV transcripts from HPV DNA-positive PBMCs or to define which cell subpopulation (monocytes or lymphocytes) preferentially harbors HPV genomes, indicating that PBMCs likely function as nonpermissive carriers. Although detection of HPV DNA in PBMCs is not synonymous with the presence of virions in these cells, its association with PBMCs in this study cannot be attributed to malignant lesions as has been previously hypothesized (9, 22, 28, 40). Since PBMCs migrate to sites of tissue inflammation and also take up microorganisms from tissues or the bloodstream, we speculate that PBMCs execute this function for HPV infection, as they do for many other viral infections. Consequently, PBMCs might serve as a source of HPV in the infection of epithelial cells and contribute to their nonsexual spread. However, additional work is needed to confirm this as a possible mode of HPV transmission. Further studies of specimens from linked donor-recipient repositories will be essential to establish a direct linkage.
Acknowledgments
This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. S.B. was supported by an NCI intramural grant 8340201 (to Z.-M.Z.).
We thank Douglas Lowy and Robert Yarchoan at NCI for their critical reading of the manuscript and Lori Wiener at NCI for histories of sexual activity of pediatric patients with PBMC HPV DNA.
All specimen samples were obtained from patients enrolled in NCI Institutional Review Board-approved clinical protocols, with Philip A. Pizzo and Ian Magrath as the principal investigators.
REFERENCES
- 1.Ahdieh, L., R. S. Klein, R. Burk, S. Cu-Uvin, P. Schuman, A. Duerr, M. Safaeian, J. Astemborski, R. Daniel, and K. Shah. 2001. Prevalence, incidence, and type-specific persistence of human papillomavirus in human immunodeficiency virus (HIV)-positive and HIV-negative women. J. Infect. Dis. 184:682-690. [DOI] [PubMed] [Google Scholar]
- 2.Bernard, H. U., S. Y. Chan, M. M. Manos, C. K. Ong, L. L. Villa, H. Delius, C. L. Peyton, H. M. Bauer, and C. M. Wheeler. 1994. Identification and assessment of known and novel human papillomaviruses by polymerase chain reaction amplification, restriction fragment length polymorphisms, nucleotide sequence, and phylogenetic algorithms. J. Infect. Dis. 170:1077-1085. [DOI] [PubMed] [Google Scholar]
- 3.Bodaghi, S., K. Yamanegi, S. Y. Xiao, M. Da Costa, J. M. Palefsky, and Z. M. Zheng. 2005. Colorectal papillomavirus infection in patients with colorectal cancer. Clin. Cancer Res. 11:2862-2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burk, R. D., M. Terai, P. E. Gravitt, L. A. Brinton, R. J. Kurman, W. A. Barnes, M. D. Greenberg, O. C. Hadjimichael, L. Fu, L. McGowan, R. Mortel, P. E. Schwartz, and A. Hildesheim. 2003. Distribution of human papillomavirus types 16 and 18 variants in squamous cell carcinomas and adenocarcinomas of the cervix. Cancer Res. 63:7215-7220. [PubMed] [Google Scholar]
- 5.Capone, R. B., S. I. Pai, W. M. Koch, M. L. Gillison, H. N. Danish, W. H. Westra, R. Daniel, K. V. Shah, and D. Sidransky. 2000. Detection and quantitation of human papillomavirus (HPV) DNA in the sera of patients with HPV-associated head and neck squamous cell carcinoma. Clin. Cancer Res. 6:4171-4175. [PubMed] [Google Scholar]
- 6.Chan, S. Y., L. Ho, C. K. Ong, V. Chow, B. Drescher, M. Durst, J. ter Meulen, L. Villa, J. Luande, H. N. Mgaya, and H.-U. Bernard. 1992. Molecular variants of human papillomavirus type 16 from four continents suggest ancient pandemic spread of the virus and its coevolution with humankind. J. Virol. 66:2057-2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng, J. Y., L. F. Sheu, C. L. Meng, W. H. Lee, and J. C. Lin. 1995. Detection of human papillomavirus DNA in colorectal carcinomas by polymerase chain reaction. Gut 37:87-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng, Y. W., H. L. Chiou, G. T. Sheu, L. L. Hsieh, J. T. Chen, C. Y. Chen, J. M. Su, and H. Lee. 2001. The association of human papillomavirus 16/18 infection with lung cancer among nonsmoking Taiwanese women. Cancer Res. 61:2799-2803. [PubMed] [Google Scholar]
- 9.Dong, S. M., S. I. Pai, S. H. Rha, A. Hildesheim, R. J. Kurman, P. E. Schwartz, R. Mortel, L. McGowan, M. D. Greenberg, W. A. Barnes, and D. Sidransky. 2002. Detection and quantitation of human papillomavirus DNA in the plasma of patients with cervical carcinoma. Cancer Epidemiol. Biomarkers Prev. 11:3-6. [PubMed] [Google Scholar]
- 10.Ellerbrock, T. V., M. A. Chiasson, T. J. Bush, X. W. Sun, D. Sawo, K. Brudney, and T. C. Wright, Jr. 2000. Incidence of cervical squamous intraepithelial lesions in HIV-infected women. JAMA 283:1031-1037. [DOI] [PubMed] [Google Scholar]
- 11.Frisch, M., B. Glimelius, A. J. van den Brule, J. Wohlfahrt, C. J. Meijer, J. M. Walboomers, S. Goldman, C. Svensson, H. O. Adami, and M. Melbye. 1997. Sexually transmitted infection as a cause of anal cancer. N. Engl. J. Med. 337:1350-1358. [DOI] [PubMed] [Google Scholar]
- 12.Gillison, M. L., W. M. Koch, R. B. Capone, M. Spafford, W. H. Westra, L. Wu, M. L. Zahurak, R. W. Daniel, M. Viglione, D. E. Symer, K. V. Shah, and D. Sidransky. 2000. Evidence for a causal association between human papillomavirus and a subset of head and neck cancers. J. Natl. Cancer Inst. 92:709-720. [DOI] [PubMed] [Google Scholar]
- 13.Ho, L., S. Y. Chan, R. D. Burk, B. C. Das, K. Fujinaga, J. P. Icenogle, T. Kahn, N. Kiviat, W. Lancaster, P. Mavromara-Nazos, V. Labropoulou, S. Mitrani-Rosenbaum, B. Norrild, M. R. Pillai, J. Stoerker, K. Syrjaenen, S. Syrjaenen, S.-K. Tay, L. L. Villa, C. M. Wheeler, A.-L. Williamson, and H.-U. Bernard. 1993. The genetic drift of human papillomavirus type 16 is a means of reconstructing prehistoric viral spread and the movement of ancient human populations. J. Virol. 67:6413-6423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Howley, P. M., and D. R. Lowy. 2001. Papillomaviruses and their replication, p. 2197-2229. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 15.Kay, P., B. Allan, L. Denny, M. Hoffman, and A. L. Williamson. 2005. Detection of HPV 16 and HPV 18 DNA in the blood of patients with cervical cancer. J. Med. Virol. 75:435-439. [DOI] [PubMed] [Google Scholar]
- 16.Kaya, H., E. Kotiloglu, S. Inanli, G. Ekicioglu, S. U. Bozkurt, A. Tutkun, and S. Kullu. 2001. Prevalence of human papillomavirus (HPV) DNA in larynx and lung carcinomas. Pathologica 93:531-534. [PubMed] [Google Scholar]
- 17.Kedzia, H., A. Gozdzicka-Jozefiak, M. Wolna, and E. Tomczak. 1992. Distribution of human papillomavirus 16 in the blood of women with uterine cervix carcinoma. Eur. J. Gynaecol. Oncol. 13:522-526. [PubMed] [Google Scholar]
- 18.Klaes, R., S. M. Woerner, R. Ridder, N. Wentzensen, M. Duerst, A. Schneider, B. Lotz, P. Melsheimer, and D. M. von Knebel. 1999. Detection of high-risk cervical intraepithelial neoplasia and cervical cancer by amplification of transcripts derived from integrated papillomavirus oncogenes. Cancer Res. 59:6132-6136. [PubMed] [Google Scholar]
- 19.Kojima, A., H. Maeda, Y. Sugita, S. Tanaka, and Y. Kameyama. 2002. Human papillomavirus type 38 infection in oral squamous cell carcinomas. Oral Oncol. 38:591-596. [DOI] [PubMed] [Google Scholar]
- 20.Lai, Y. M., J. F. Lee, H. Y. Huang, Y. K. Soong, F. P. Yang, and C. C. Pao. 1997. The effect of human papillomavirus infection on sperm cell motility. Fertil. Steril. 67:1152-1155. [DOI] [PubMed] [Google Scholar]
- 21.Lai, Y. M., F. P. Yang, and C. C. Pao. 1996. Human papillomavirus deoxyribonucleic acid and ribonucleic acid in seminal plasma and sperm cells. Fertil. Steril. 65:1026-1030. [PubMed] [Google Scholar]
- 22.Liu, V. W., P. Tsang, A. Yip, T. Y. Ng, L. C. Wong, and H. Y. Ngan. 2001. Low incidence of HPV DNA in sera of pretreatment cervical cancer patients. Gynecol. Oncol. 82:269-272. [DOI] [PubMed] [Google Scholar]
- 23.Meissner, J. D. 1999. Nucleotide sequences and further characterization of human papillomavirus DNA present in the CaSki, SiHa and HeLa cervical carcinoma cell lines. J. Gen. Virol. 80:1725-1733. [DOI] [PubMed] [Google Scholar]
- 24.Meyer, T., R. Arndt, I. Nindl, C. Ulrich, E. Christophers, and E. Stockfleth. 2003. Association of human papillomavirus infections with cutaneous tumors in immunosuppressed patients. Transplant. Int. 16:146-153. [DOI] [PubMed] [Google Scholar]
- 25.Moscicki, A. B., J. H. Ellenberg, S. Farhat, and J. Xu. 2004. Persistence of human papillomavirus infection in HIV-infected and -uninfected adolescent girls: risk factors and differences, by phylogenetic type. J. Infect. Dis. 190:37-45. [DOI] [PubMed] [Google Scholar]
- 26.Munger, K., A. Baldwin, K. M. Edwards, H. Hayakawa, C. L. Nguyen, M. Owens, M. Grace, and K. Huh. 2004. Mechanisms of human papillomavirus-induced oncogenesis. J. Virol. 78:11451-11460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Munoz, N., F. X. Bosch, S. de Sanjose, R. Herrero, X. Castellsague, K. V. Shah, P. J. Snijders, and C. J. Meijer. 2003. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N. Engl. J. Med. 348:518-527. [DOI] [PubMed] [Google Scholar]
- 28.Pao, C. C., J. J. Hor, F. P. Yang, C. Y. Lin, and C. J. Tseng. 1997. Detection of human papillomavirus mRNA and cervical cancer cells in peripheral blood of cervical cancer patients with metastasis. J. Clin. Oncol. 15:1008-1012. [DOI] [PubMed] [Google Scholar]
- 29.Pao, C. C., S. S. Lin, C. Y. Lin, J. S. Maa, C. H. Lai, and T. T. Hsieh. 1991. Identification of human papillomavirus DNA sequences in peripheral blood mononuclear cells. Am. J. Clin. Pathol. 95:540-546. [DOI] [PubMed] [Google Scholar]
- 30.Pfister, H. 2003. Chapter 8: human papillomavirus and skin cancer. J. Natl. Cancer Inst. Monogr. 2003(31):52-56. [DOI] [PubMed] [Google Scholar]
- 31.Ringstrom, E., E. Peters, M. Hasegawa, M. Posner, M. Liu, and K. T. Kelsey. 2002. Human papillomavirus type 16 and squamous cell carcinoma of the head and neck. Clin. Cancer Res. 8:3187-3192. [PubMed] [Google Scholar]
- 32.Ritchie, J. M., E. M. Smith, K. F. Summersgill, H. T. Hoffman, D. Wang, J. P. Klussmann, L. P. Turek, and T. H. Haugen. 2003. Human papillomavirus infection as a prognostic factor in carcinomas of the oral cavity and oropharynx. Int. J. Cancer 104:336-344. [DOI] [PubMed] [Google Scholar]
- 33.Schiffman, M., and S. K. Kjaer. 2003. Chapter 2: natural history of anogenital human papillomavirus infection and neoplasia. J. Natl. Cancer Inst. Monogr. 2003(31):14-19. [DOI] [PubMed] [Google Scholar]
- 34.Shen, Z. Y., S. P. Hu, L. C. Lu, C. Z. Tang, Z. S. Kuang, S. P. Zhong, and Y. Zeng. 2002. Detection of human papillomavirus in esophageal carcinoma. J. Med. Virol. 68:412-416. [DOI] [PubMed] [Google Scholar]
- 35.Stocco dos Santos, R. C., C. J. Lindsey, O. P. Ferraz, J. R. Pinto, R. S. Mirandola, F. J. Benesi, E. H. Birgel, C. A. Pereira, and W. Becak. 1998. Bovine papillomavirus transmission and chromosomal aberrations: an experimental model. J. Gen. Virol. 79:2127-2135. [DOI] [PubMed] [Google Scholar]
- 36.Sun, X. W., L. Kuhn, T. V. Ellerbrock, M. A. Chiasson, T. J. Bush, and T. C. Wright, Jr. 1997. Human papillomavirus infection in women infected with the human immunodeficiency virus. N. Engl. J. Med. 337:1343-1349. [DOI] [PubMed] [Google Scholar]
- 37.Swan, D. C., M. Rajeevan, G. Tortolero-Luna, M. Follen, R. A. Tucker, and E. R. Unger. 2005. Human papillomavirus type 16 E2 and E6/E7 variants. Gynecol. Oncol. 96:695-700. [DOI] [PubMed] [Google Scholar]
- 38.Syrjanen, K. J. 2002. HPV infections and oesophageal cancer. J. Clin. Pathol. 55:721-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tonon, S. A., M. A. Picconi, P. D. Bos, J. B. Zinovich, J. Galuppo, L. V. Alonio, and A. R. Teyssie. 2001. Physical status of the E2 human papilloma virus 16 viral gene in cervical preneoplastic and neoplastic lesions. J. Clin. Virol. 21:129-134. [DOI] [PubMed] [Google Scholar]
- 40.Tseng, C. J., C. C. Pao, J. D. Lin, Y. K. Soong, J. H. Hong, and S. Hsueh. 1999. Detection of human papillomavirus types 16 and 18 mRNA in peripheral blood of advanced cervical cancer patients and its association with prognosis. J. Clin. Oncol. 17:1391-1396. [DOI] [PubMed] [Google Scholar]
- 41.Uobe, K., K. Masuno, Y. R. Fang, L. J. Li, Y. M. Wen, Y. Ueda, and A. Tanaka. 2001. Detection of HPV in Japanese and Chinese oral carcinomas by in situ PCR. Oral Oncol. 37:146-152. [DOI] [PubMed] [Google Scholar]
- 42.Veress, G., K. Szarka, X. P. Dong, L. Gergely, and H. Pfister. 1999. Functional significance of sequence variation in the E2 gene and the long control region of human papillomavirus type 16. J. Gen. Virol. 80:1035-1043. [DOI] [PubMed] [Google Scholar]
- 43.Widschwendter, A., T. Brunhuber, A. Wiedemair, E. Mueller-Holzner, and C. Marth. 2004. Detection of human papillomavirus DNA in breast cancer of patients with cervical cancer history. J. Clin. Virol. 31:292-297. [DOI] [PubMed] [Google Scholar]
- 44.Xi, L. F., L. A. Koutsky, D. A. Galloway, J. Kuypers, J. P. Hughes, C. M. Wheeler, K. K. Holmes, and N. B. Kiviat. 1997. Genomic variation of human papillomavirus type 16 and risk for high grade cervical intraepithelial neoplasia. J. Natl. Cancer Inst. 89:796-802. [DOI] [PubMed] [Google Scholar]
- 45.Yu, Y., T. Morimoto, M. Sasa, K. Okazaki, Y. Harada, T. Fujiwara, Y. Irie, E. Takahashi, A. Tanigami, and K. Izumi. 2000. Human papillomavirus type 33 DNA in breast cancer in Chinese. Breast Cancer 7:33-36. [DOI] [PubMed] [Google Scholar]
- 46.Zambrano, A., M. Kalantari, A. Simoneau, J. L. Jensen, and L. P. Villarreal. 2002. Detection of human polyomaviruses and papillomaviruses in prostatic tissue reveals the prostate as a habitat for multiple viral infections. Prostate 53:263-276. [DOI] [PubMed] [Google Scholar]
- 47.Zehbe, I., J. Mytilineos, I. Wikstrom, R. Henriksen, L. Edler, and M. Tommasino. 2003. Association between human papillomavirus 16 E6 variants and human leukocyte antigen class I polymorphism in cervical cancer of Swedish women. Hum. Immunol. 64:538-542. [DOI] [PubMed] [Google Scholar]