Abstract
More than 700 bacterial species or phylotypes, of which over 50% have not been cultivated, have been detected in the oral cavity. Our purposes were (i) to utilize culture-independent molecular techniques to extend our knowledge on the breadth of bacterial diversity in the healthy human oral cavity, including not-yet-cultivated bacteria species, and (ii) to determine the site and subject specificity of bacterial colonization. Nine sites from five clinically healthy subjects were analyzed. Sites included tongue dorsum, lateral sides of tongue, buccal epithelium, hard palate, soft palate, supragingival plaque of tooth surfaces, subgingival plaque, maxillary anterior vestibule, and tonsils. 16S rRNA genes from sample DNA were amplified, cloned, and transformed into Escherichia coli. Sequences of 16S rRNA genes were used to determine species identity or closest relatives. In 2,589 clones, 141 predominant species were detected, of which over 60% have not been cultivated. Thirteen new phylotypes were identified. Species common to all sites belonged to the genera Gemella, Granulicatella, Streptococcus, and Veillonella. While some species were subject specific and detected in most sites, other species were site specific. Most sites possessed 20 to 30 different predominant species, and the number of predominant species from all nine sites per individual ranged from 34 to 72. Species typically associated with periodontitis and caries were not detected. There is a distinctive predominant bacterial flora of the healthy oral cavity that is highly diverse and site and subject specific. It is important to fully define the human microflora of the healthy oral cavity before we can understand the role of bacteria in oral disease.
The oral cavity is comprised of many surfaces, each coated with a plethora of bacteria, the proverbial bacterial biofilm. Some of these bacteria have been implicated in oral diseases such as caries and periodontitis, which are among the most common bacterial infections in humans. For example, it has been estimated that at least 35% of dentate U.S. adults aged 30 to 90 years have periodontitis (1). In addition, specific oral bacterial species have been implicated in several systemic diseases, such as bacterial endocarditis (4), aspiration pneumonia (26), osteomyelitis in children (8), preterm low birth weight (6, 20), and cardiovascular disease (2, 34). Surprisingly, little is known about the microflora of the healthy oral cavity.
By using culture-independent molecular methods, we previously detected over 500 species or phylotypes in subgingival plaque of healthy subjects and subjects with periodontal diseases (21), necrotizing ulcerative periodontitis in human immunodeficiency virus-positive subjects (23), dental plaque in children with rampant caries (3), noma (22), and on the tongue dorsum of subjects with and without halitosis (15). Other investigators have used similar techniques to determine the bacterial diversity of saliva (25), subgingival plaque of a subject with gingivitis (16), and dentoalveolar abscesses (10, 29). Over half of the species detected have not yet been cultivated. Data from these studies have implicated specific species or phylotypes in a variety of diseases and oral infections, but still only limited information is available on species associated with health. Recently, Mager et al. (19) demonstrated significant differences in the bacterial profiles of 40 oral cultivable species on soft and hard tissues in healthy subjects. They also found that the profiles of the soft tissues were more similar to each other than those of supragingival and subgingival plaques.
Our purposes were as follows: (i) to utilize culture-independent molecular techniques to extend our knowledge on the breadth of bacterial diversity in the healthy human oral cavity, including not-yet-cultivated phylotypes, and (ii) to determine the site and subject specificity of bacterial colonization.
MATERIALS AND METHODS
Subjects.
Five subjects, representing both genders, ranging in age from 23 to 55 and with no clinical signs of oral mucosal disease were included in the study. Subjects did not suffer from severe halitosis. The periodontia were healthy in that all periodontal pockets were less than 3 mm deep with no redness or inflammation of the gums. Subjects did not have active white spot lesions or caries on the teeth. Our results were consistent with these clinical observations in that species typically found in caries subjects were not detected (3). The subjects had not used antibiotics for the last 6 months.
Sample collection.
Samples from the following nine sites were analyzed for each subject: dorsum of the tongue, lateral sides of the tongue, buccal fold, hard palate, soft palate, labial gingiva and tonsils of soft tissue surfaces, and supragingival and subgingival plaques from tooth surfaces. Microbiological samples of supragingival and subgingival plaque samples were taken with a sterile Gracey curette. Other samples were collected with sterile swab brushes.
Sample lysis.
Plaque samples were directly suspended in 50 μl of 50 mM Tris buffer (pH 7.6), 1 mM EDTA, pH 8, and 0.5% Tween 20. Proteinase K (200 μg/ml; Roche Applied Science, Indianapolis, IN) was added to the mixture. The samples were then heated at 55°C for 2 h. Proteinase K was inactivated by heating at 95°C for 5 min. Detection of species is dependent upon obtaining DNA that can be amplified. Thus, a difficult-to-lyse bacterium may not be detected. However, by using our lysis technique, we were able to detect many hard-to-lyse species, such as species of Actinomyces and Streptococcus.
Amplification of 16S rRNA genes by PCR and purification of PCR products.
The 16S rRNA genes were amplified under standardized conditions using a universal primer set (forward primer, 5′-GAG AGT TTG ATY MTG GCT CAG-3′; reverse primer, 5′-GAA GGA GGT GWT CCA RCC GCA-3′) (21). Primers were synthesized commercially (Operon Technologies, Alameda, CA). The PCR primers do not necessarily include all bacterial species. Nevertheless, a wide range of phylogenetic types were obtained in this study and our previous studies by using this universal primer set (3, 15, 21-23). PCR was performed in thin-walled tubes with a GeneAmp PCR system 9700 (ABI, Foster City, CA). One microliter of the lysed sample was added to a reaction mixture (final volume, 50 μl) containing 20 pmol of each primer, 40 nmol of deoxynucleoside triphosphates, and 1 U of Platinum Taq polymerase (Invitrogen, San Diego, CA). In a hot-start protocol, the samples were preheated at 95°C for 4 min, followed by amplification under the following conditions: denaturation at 95°C for 45 s, annealing at 60°C for 45 s, and elongation at 72° for 1.5 min, with an additional 15 s for each cycle. A total of 30 cycles were performed; this was followed by a final elongation step at 72°C for 15 min. The results of the PCR amplification were examined by electrophoresis in a 1% agarose gel. DNA was stained with ethidium bromide and visualized under short-wavelength UV light.
Cloning procedures.
Cloning of PCR-amplified DNA was performed with the TOPO TA cloning kit (Invitrogen) according to the instructions of the manufacturer. Transformation was done with competent Escherichia coli TOP10 cells provided by the manufacturer. The transformed cells were then plated onto Luria-Bertani agar plates supplemented with kanamycin (50 μg/ml), and the plates were incubated overnight at 37°C. Each colony was placed into 40 μl of 10 mM Tris. Correct sizes of the inserts were determined in a PCR with an M13 (−20) forward primer and an M13 reverse primer (Invitrogen). Prior to sequencing of the fragments, the PCR-amplified 16S rRNA fragments were purified and concentrated with Microcon 100 (Amicon, Bedford, MA), followed by use of the QIAquick PCR purification kit (QIAGEN, Valencia, CA).
16S rRNA gene sequencing.
Purified PCR-amplified 16S rRNA inserts were sequenced using an ABI Prism cycle sequencing kit (BigDye terminator cycle sequencing kit with AmpliTaq DNA polymerase FS, GeneAmp PCR system 9700; ABI). The primers used for sequencing have been described previously (21). Quarter dye chemistry was used with 80 μM primers and 1.5 μl of PCR product in a final volume of 20 μl. Cycle sequencing was performed with a GeneAmp PCR system 9700 (ABI) with 25 cycles of denaturation at 96°C for 10 s, annealing at 55° for 5 s, and extension at 60°C for 4 min. The sequencing reactions were run on an ABI 3100 DNA sequencer (ABI).
16S rRNA gene sequencing and data analysis of unrecognized inserts.
A total of 2,589 clones with an insert of the correct size of approximately 1,500 bases were analyzed. The number of clones per subject that were sequenced ranged from 42 to 69, with an average of 57.5 and a standard deviation of 7.1. A sequence of approximately 500 bases was obtained first to determine identity or approximate phylogenetic position. Full sequences of about 1,500 bases were obtained by using five to six additional sequencing primers (15) for those species deemed novel. For identification of closest relatives, the sequences of the unrecognized inserts were compared to the 16S rRNA sequences of over 10,000 microorganisms in our database and over 100,000 sequences in the Ribosomal Database Project (7) and the GenBank databases. Our cutoff for species differentiation was 2%, or approximately 30 bases for a full sequence. The similarity matrices were corrected for multiple base changes at single positions by the method of Jukes and Cantor (13). Similarity matrices were constructed from the aligned sequences by using only those sequence positions for which data were available for 90% of the strains tested. Phylogenetic trees were constructed by the neighbor-joining method of Saitou and Nei (24). TREECON, a software package for the Microsoft Windows environment, was used for the construction and drawing of evolutionary trees (28). We are aware of the potential creation of 16S rRNA chimera molecules assembled during the PCR (18). The percentage of chimeric inserts in 16S rRNA gene libraries ranged from 1 to 15%. Chimeric sequences were identified by using the Chimera Check program in the Ribosomal Database Project, by treeing analysis, or by base signature analysis. Species identification of chimeras was obtained, but the sequences were not examined for phylogenetic analysis.
Nucleotide sequence accession numbers.
The complete 16S rRNA gene sequences of clones representing novel phylotypes defined in this study, sequences of known species not previously reported, and published sequences are available for electronic retrieval from the EMBL, GenBank, and DDBJ nucleotide sequence databases under the accession numbers shown in Fig. 1 to 9.
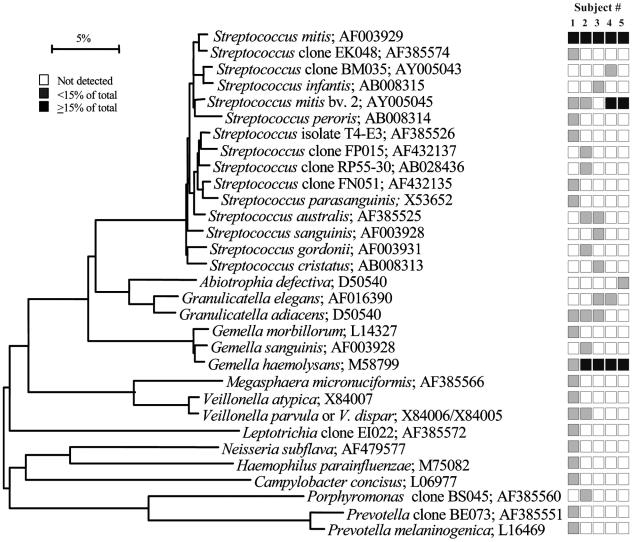
FIG. 1.
Bacterial profiles of the buccal epithelium of healthy subjects. Distribution and levels of bacterial species/phylotypes among five subjects are shown by the columns of boxes to the right of the tree as either not detected (clear box), <15% of the total number of clones assayed (shaded box), or ≥15% of the total number of clones assayed (darkened box). The 15% was chosen arbitrarily. GenBank accession numbers are provided. Marker bar represents a 5% difference in nucleotide sequences.
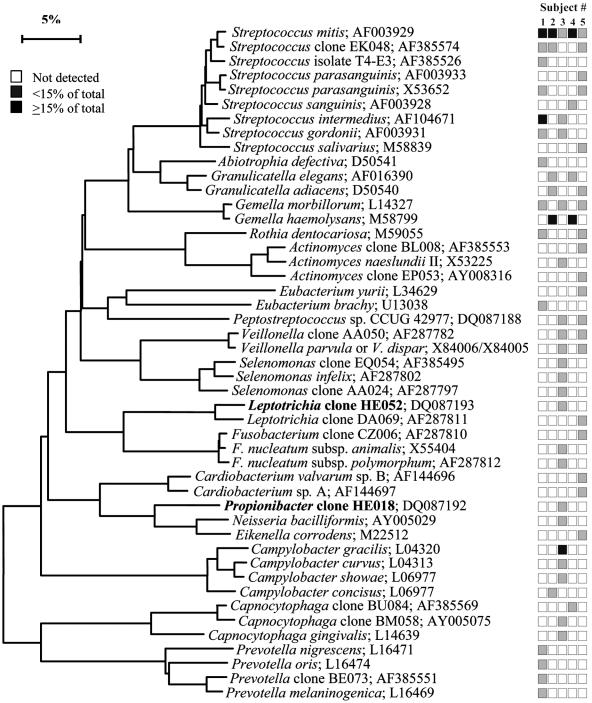
FIG. 9.
Bacterial profiles of the subgingival plaque of healthy subjects. Distribution and levels of bacterial species/phylotypes among five subjects are as described for Fig. 1. Novel phylotypes identified in this study are indicated in bold. GenBank accession numbers are provided. Marker bar represents a 5% difference in nucleotide sequences.
RESULTS AND DISCUSSION
It has long been known that oral bacteria preferentially colonize different surfaces in the oral cavity as a result of specific adhesins on the bacterial surface binding to complementary specific receptors on a given oral surface (11, 12). Indeed, the profiles of 40 cultivable bacterial species differed markedly on different oral soft tissue surfaces, saliva, and supragingival and subgingival plaques from healthy subjects (19). The purpose of this study was to define the predominant bacterial flora of the healthy oral cavity by identifying and comparing the cultivable and the not-yet-cultivated bacterial species on different soft tissues and supra- and subgingival plaques.
Based on the analysis of 2,589 16S rRNA clones, the bacterial diversity of the microflora from nine different sites of five clinically healthy subjects was striking—a total of 141 different bacterial taxa representing six different bacterial phyla were detected. The six phyla included the Firmicutes (previously referred to as the low-G+C gram positives, such as species of Streptococcus, Gemella, Eubacterium, Selenomonas, Veillonella, and related ones), the Actinobacteria (previously referred to as the high-G+C gram positives, such as species of Actinomyces, Atopobium, Rothia, and related ones), the Proteobacteria (e.g., species of Neisseria, Eikenella, Campylobacter, and related ones), the Bacteroidetes (e.g., species of Porphyromonas, Prevotella, Capnocytophaga, and related ones), the Fusobacteria (e.g., species of Fusobacterium and Leptotrichia), and the TM7 phylum, for which there are no cultivable representatives. As determined in our previous studies using culture-independent molecular techniques (15, 21, 22), over 60% of the bacterial flora was represented by not-yet-cultivated phylotypes. Thirteen new phylotypes (see Fig. 2 through 9) were identified for the first time in this study. In a comparable ongoing study of caries in permanent teeth, using the same techniques, we detected 224 species in the analysis of 1,275 16S rRNA clones with about 60% as not-yet-cultivable phylotypes (J. A. Aas, S. R. Dardis, A. L. Griffen, L. N. Stokes, A. M. Lee, I. Olsen, F. E. Dewhirst, E. J. Leys, and B. J. Paster. J. Dent. Res. 84:abstract 2805, 2005). Consequently, it is important to identify both the cultivable and not-yet-cultivated bacterial flora in a given environment before we can ascribe association to health or disease status. There is no evidence to suggest that the not-yet-cultivated segment is any less important than the cultivable segment.
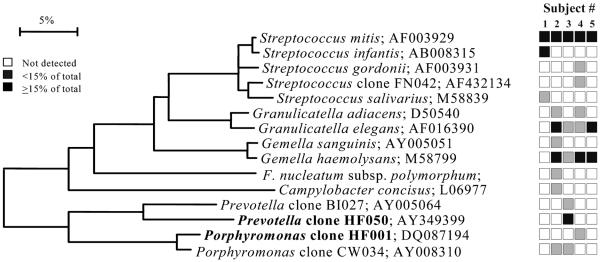
FIG. 2.
Bacterial profiles of the maxillary anterior vestibule of healthy subjects. Distribution and levels of bacterial species/phylotypes among five subjects are as described for Fig. 1. Novel phylotypes identified in this study are indicated in bold. GenBank accession numbers are provided. Marker bar represents a 5% difference in nucleotide sequences.
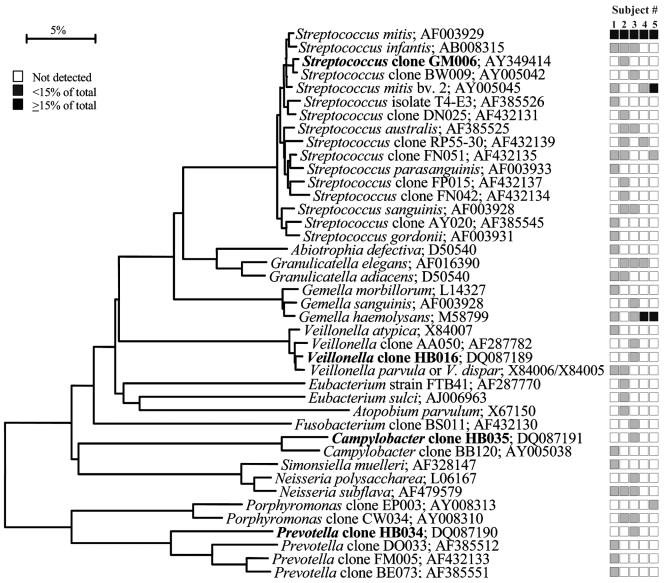
Bacterial profiles for each site tested for each subject are shown in nine phylogenetic trees (see Fig. 1 through 9). In these dendrograms, the distribution of bacterial species or phylotypes in each subject can be observed at a glance. For example, it is clear that Streptococcus mitis, S. mitis bv. 2, and Gemella hemolysans are the predominant species of the buccal epithelium (Fig. 1). Each of these species was detected in most or all of the subjects and represented ≥15% of the total number of assayed clones (Fig. 1). Similarly, the predominant bacterial flora for the other eight oral sites was examined. In the maxillary anterior vestibule, S. mitis, Granulicatella spp., and Gemella spp. predominated (Fig. 2). On the tongue dorsum, several species of Streptococcus, such as S. mitis, Streptococcus australis, Streptococcus parasanguinis, Streptococcus salivarius, Streptococcus sp. clone FP015, and Streptococcus sp. clone FN051, Granulicatella adiacens, and Veillonella spp. were the predominant species (Fig. 3). On the lateral tongue surface (Fig. 4), S. mitis, S. mitis bv. 2, Streptococcus sp. clone DP009, Streptococcus sp. clone FN051, S. australis, G. adiacens, G. hemolysans, and Veillonella spp. predominated. It is interesting that there were considerable differences in the bacterial profiles of the tongue dorsum and the lateral tongue surface. For example, S. mitis bv. 2 was well represented at the lateral side of the tongue but not detected on the tongue dorsum. Conversely, S.parasanguinis strain 85-81 was present on the tongue dorsum, but was absent on the lateral side. This was not surprising, because these surfaces are known to be different in ultrastructure and function. For instance, the lateral side of the tongue has a smooth nonkeratinized surface, in contrast to the dorsum of the tongue, which is a keratinized, highly papillated surface with a large surface area and underlying serous glands. These anatomic differences likely influence the ecology of these habitats and create microbial environmental differences.
FIG. 3.
Bacterial profiles of the tongue dorsum of healthy subjects. Distribution and levels of bacterial species/phylotypes among five subjects are as described for Fig. 1. Novel phylotypes identified in this study are indicated in bold. GenBank accession numbers are provided. Marker bar represents a 5% difference in nucleotide sequences.
FIG. 4.
Bacterial profiles of the lateral tongue surface of healthy subjects. Distribution and levels of bacterial species/phylotypes among five subjects are as described for Fig. 1. Novel phylotypes identified in this study are indicated in bold. GenBank accession numbers are provided. Marker bar represents a 5% difference in nucleotide sequences.
On the hard palate, the predominant bacterial species included S. mitis, S. mitis biovar 2, Streptococcus sp. clone FN051, Streptococcus infantis, Granulicatella elegans, G. hemolysans, and Neisseria subflava (Fig. 5). On the soft palate, S. mitis, other cultivable and not-yet-cultivable species of Streptococcus, G. adiacens and G. hemolysans were predominant (Fig. 6). The tonsil bacterial flora (Fig. 7) was rather diverse—some subjects had Prevotella and Porphyromonas spp. (subjects 1 and 5), and others had Firmicutes species (subjects 2, 3, and 4). On the tooth surface, several species of Streptococcus, including Streptococcus sp. clone EK048, S. sanguinis, and S. gordonii, and Rothia dentocariosa, G. hemolysans, G. adiacens, Actinomyces sp. clone BL008, and Abiotrophia defectiva were often detected (Fig. 8). Finally, in subgingival plaque, several species of Streptococcus and Gemella were often found (Fig. 9).
FIG. 5.
Bacterial profiles of the hard palate of healthy subjects. Distribution and levels of bacterial species/phylotypes among five subjects are as described for Fig. 1. Novel phylotypes identified in this study are indicated in bold. GenBank accession numbers are provided. Marker bar represents a 5% difference in nucleotide sequences.
FIG. 6.
Bacterial profiles of the soft palate of healthy subjects. Distribution and levels of bacterial species/phylotypes among five subjects are as described for Fig. 1. Novel phylotypes identified in this study are indicated in bold. GenBank accession numbers are provided. Marker bar represents a 5% difference in nucleotide sequences.
FIG. 7.
Bacterial profiles of the tonsils of healthy subjects. Distribution and levels of bacterial species/phylotypes among five subjects are as described for Fig. 1. Novel phylotypes identified in this study are indicated in bold. GenBank accession numbers are provided. Marker bar represents a 5% difference in nucleotide sequences.
FIG. 8.
Bacterial profiles of the tooth surfaces of healthy subjects. Distribution and levels of bacterial species/phylotypes among five subjects are as described for Fig. 1. Novel phylotypes identified in this study are indicated in bold. GenBank accession numbers are provided. Marker bar represents a 5% difference in nucleotide sequences.
The number of cultivable and not-yet-cultivable species detected in each subject for each site is also shown in Fig. 1 through 9. For example, in Fig. 1, only four species (S. mitis, S.mitis bv. 2, A. defectiva, and G. hemolysans) were detected on the buccal epithelium in subject 5. For convenience, the number of bacterial species detected in each oral site for each subject is summarized in Table 1. Surprisingly, the highest number of different species was found on the tonsils in subject 1, with 28 predominant species; collectively, 57 species were detected on the tonsils (Table 1; Fig. 7). In contrast, the maxillary anterior vestibule had the lowest diversity of the bacterial flora compared with the other sites, with as few as three to nine species detected among the subjects (Table 1; Fig. 2). A common question asked is how many bacterial species are present in the oral cavity of a single individual. The last column of Table 1 indicates the number of predominant species in each healthy individual from all nine different sites; thus, the numbers ranged from a low of 34 in subject 4 to a high of 72 in subject 3.
TABLE 1.
Number of predominant bacterial species per site and subject
| Subject no. | Total no. of species/site
|
Total no. of species/subject | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buccal | Maxillary vestibule | Tongue dorsum | Tongue lateral | Hard palate | Soft palate | Tonsils | Tooth surface | Subgingival | ||
| 1 | 20 | 5 | 23 | 14 | 21 | 16 | 28 | 27 | 14 | 66 |
| 2 | 12 | 6 | 13 | 18 | 18 | 20 | 14 | 12 | 6 | 45 |
| 3 | 11 | 9 | 10 | 9 | 15 | 14 | 22 | 21 | 22 | 72 |
| 4 | 5 | 7 | 10 | 8 | 4 | 6 | 11 | 12 | 4 | 34 |
| 5 | 4 | 3 | 17 | 20 | 6 | 13 | 18 | 16 | 21 | 64 |
| Total | 32 | 15 | 40 | 34 | 42 | 38 | 59 | 52 | 47 | |
Overall, cultivable and not-yet-cultivable species of Gemella, Granulicatella, Streptococcus, and Veillonella were commonly detected in most sites. S. mitis was the most commonly found species in essentially all sites and subjects (Fig. 1 through 9). In one subject, 79% of the clones identified were S. mitis (data not shown in figures). Note that in this study, S. mitis and Streptococcus pneumoniae have been grouped together. Some members of the mitis group share more than 99% 16S rRNA sequence similarity, although DNA-DNA similarity values for the entire chromosomes are estimated to be less than 60% (14). Both S. mitis and Streptococcus oralis have been associated with bacterial endocarditis, especially in patients with prosthetic valves (9). In addition, they are now recognized as frequent causes of infection in immunocompromised patients, particularly immediately after tissue transplants and in neutropenic cancer patients (30). G. adiacens, often considered an opportunistic pathogen, was also detected at all sites in the healthy oral cavity. G. adiacens isolates have been reported from bacteremia/septicemia in patients with infective endocarditis/atheroma (33) and in primary bacteremia in patients with neutropenic fever (33). A high mortality rate for endocarditis by G. adiacens has been reported (5).
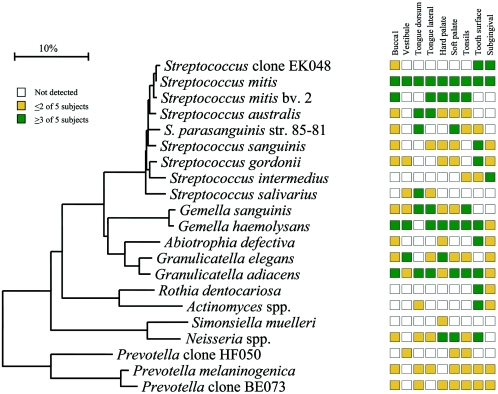
Figure 10 represents the overall summary of this study—namely, that there are emerging bacterial profiles that help define the healthy oral cavity. As already discussed, several species, such as S. mitis and G. adiacens, were detected in most or all oral sites, whereas several species were quite site specific. For example, R. dentocariosa, Actinomyces spp., S. sanguinis, S.gordonii, and A. defectiva appeared to preferentially colonize the teeth, while S. salivarius was found mostly on the tongue dorsum. Some species appeared to have a predilection for soft tissue, e.g., S. sanguinis and S. australis did not colonize the teeth or subgingival crevice. S. intermedius preferentially colonized the subgingival plaque in most of the subjects but was not detected in most other sites. On the other hand, Neisseria spp. were not found in subgingival plaque but were present in most other sites. Simonsiella muelleri colonized only the hard palate. Indeed, S. muelleri was initially isolated from the human hard palate (32), although it has been isolated from a neonate with dental cyst and early eruption of teeth (31). Several Prevotella species were detected in most sites, but only in one or two subjects. For example, P. melaninogenica and Prevotella sp. clone BE073 were abundant in seven out of nine sites of one subject and were detected sporadically in other subjects (Fig. 10). Prevotella sp. clone HF050 was found in the maxillary anterior vestibule of one subject, dominating the bacterial flora as 44% of the clones (Fig. 2 and 10). This clone was also found in lower proportions on the soft palate and tonsils of another subject (Fig. 6, 7, and 10).
FIG. 10.
Site specificity of predominant bacterial species in the oral cavity. In general, bacterial species or phylotypes were selected on the basis of their detection in multiple subjects for a given site. Distributions of bacterial species in oral sites among subjects are indicated by the columns of boxes to the right of the tree as follows: not detected in any subject (clear box), <15% of the total number of clones assayed (yellow box), ≥15% of the total number of clones assayed (green box). The 15% cutoff for low and high abundance was chosen arbitrarily. Marker bar represents a 10% difference in nucleotide sequences.
In conclusion, there is a distinctive bacterial flora in the healthy oral cavity which is different from that of oral disease. For example, many species specifically associated with periodontal disease, such as Porphyromonas gingivalis, Tannerella forsythia, and Treponema denticola, were not detected in any sites tested. In addition, the bacterial flora commonly thought to be involved in dental caries and deep dentin cavities, represented by Streptococcus mutans, Lactobacillus spp., Bifidobacterium spp., and Atopobium spp., were not detected in supra- and subgingival plaques from clinically healthy teeth. A more detailed description of bacterial species associated with oral disease is discussed elsewhere (17). As noted in this study on healthy subjects, some species are site specific at one or multiple sites, while other species are subject specific. As much as 60% of the species detected are not presently cultivable. Overall, there are still more species to be discovered, although the number of new species is beginning to reach saturation.
As we have previously asserted (21), to rigorously assess the association of specific species or phylotypes with oral health or disease, it is necessary to analyze larger numbers of clinical samples for the levels of essentially all oral bacteria in well-controlled clinical studies. The bacterial complexes involved in periodontal disease as defined by Socransky et al. (27) were based on the microbial analyses of 185 subjects representing about 13,000 plaque samples using DNA probes to 40 bacterial cultivable species in checkerboard hybridization assays. We are currently developing DNA probes for approximately 500 known bacterial species and novel phylotypes for use in similar studies. These DNA probes are being developed for use in the checkerboard hybridization assay (3, 22, 23) and DNA microarray formats (S. K. Boches, A. M. Lee, B. J. Paster, and F. E. Dewhirst, J. Dent. Res. 83:abstract 2263, 2004). Our intent was to first establish the full bacterial diversity of the oral cavity and then to determine variation and reproducibility using the microarrays. Consequently, it will be relatively easy to compare the bacterial composition of a statistically significant number of samples to more precisely identify those species that are associated with health and with oral disease from different anatomic sites. It is necessary to first define the bacterial flora of the healthy oral cavity before we can determine the role of oral bacteria in disease.
Acknowledgments
This study was supported by NIH grant DE-11443 from the National Institute of Dental and Craniofacial Research and grants from the Dental Faculty, University of Oslo, Norway.
REFERENCES
- 1.Albandar, J. M., J. A. Brunelle, and A. Kingman. 1999. Destructive periodontal disease in adults 30 years of age and older in the United States, 1988-1994. J. Periodontol. 70:13-29. [DOI] [PubMed] [Google Scholar]
- 2.Beck, J., R. Garcia, G. Heiss, P. S. Vokonas, and S. Offenbacher. 1996. Periodontal disease and cardiovascular disease. J. Periodontol. 67:1123-1137. [DOI] [PubMed] [Google Scholar]
- 3.Becker, M. R., B. J. Paster, E. J. Leys, M. L. Moeschberger, S. G. Kenyon, J. L. Galvin, S. K. Boches, F. E. Dewhirst, and A. L. Griffen. 2002. Molecular analysis of bacterial species associated with childhood caries. J. Clin. Microbiol. 40:1001-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berbari, E. F., F. R. Cockerill III, and J. M. Steckelberg. 1997. Infective endocarditis due to unusual or fastidious microorganisms. Mayo Clin. Proc. 72:532-542. [DOI] [PubMed] [Google Scholar]
- 5.Bouvet, A., and J. F. Acar. 1984. New bacteriological aspects of infective endocarditis. Eur. Heart J. 5(Suppl. C):45-48. [DOI] [PubMed] [Google Scholar]
- 6.Buduneli, N., H. Baylas, E. Buduneli, O. Turkoglu, T. Kose, and G. Dahlen. 2005. Periodontal infections and pre-term low birth weight: a case-control study. J. Clin. Periodontol. 32:174-181. [DOI] [PubMed] [Google Scholar]
- 7.Cole, J. R., B. Chai, R. J. Farris, Q. Wang, S. A. Kulam, D. M. McGarrell, G. M. Garrity, and J. M. Tiedje. 2005. The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 33:D294-D296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dodman, T., J. Robson, and D. Pincus. 2000. Kingella kingae infections in children. J. Paediatr. Child. Health 36:87-90. [DOI] [PubMed] [Google Scholar]
- 9.Douglas, C. W., J. Heath, K. K. Hampton, and F. E. Preston. 1993. Identity of viridans streptococci isolated from cases of infective endocarditis. J. Med. Microbiol. 39:179-182. [DOI] [PubMed] [Google Scholar]
- 10.Dymock, D., A. J. Weightman, C. Scully, and W. G. Wade. 1996. Molecular analysis of microflora associated with dentoalveolar abscesses. J. Clin. Microbiol. 34:537-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gibbons, R. J. 1989. Bacterial adhesion to oral tissues: a model for infectious diseases. J. Dent. Res. 68:750-760. [DOI] [PubMed] [Google Scholar]
- 12.Gibbons, R. J., D. M. Spinell, and Z. Skobe. 1976. Selective adherence as a determinant of the host tropisms of certain indigenous and pathogenic bacteria. Infect. Immun. 13:238-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jukes, T. H., and C. R. Cantor. 1969. Evolution of protein molecules, vol. 3, p. 21-132. In H. N. Munro (ed.), Mammalian protein metabolism. Academic Press, Inc., New York, N.Y. [Google Scholar]
- 14.Kawamura, Y., X. G. Hou, F. Sultana, H. Miura, and T. Ezaki. 1995. Determination of 16S rRNA sequences of Streptococcus mitis and Streptococcus gordonii and phylogenetic relationships among members of the genus Streptococcus. Int. J. Syst. Bacteriol. 45:406-408. [DOI] [PubMed] [Google Scholar]
- 15.Kazor, C. E., P. M. Mitchell, A. M. Lee, L. N. Stokes, W. J. Loesche, F. E. Dewhirst, and B. J. Paster. 2003. Diversity of bacterial populations on the tongue dorsa of patients with halitosis and healthy patients. J. Clin. Microbiol. 41:558-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kroes, I., P. W. Lepp, and D. A. Relman. 1999. Bacterial diversity within the human subgingival crevice. Proc. Natl. Acad. Sci. USA 96:14547-14552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumar, P. S., A. L. Griffen, J. A. Barton, B. J. Paster, M. L. Moeschberger, and E. J. Leys. 2003. New bacterial species associated with chronic periodontitis. J. Dent. Res. 82:338-344. [DOI] [PubMed] [Google Scholar]
- 18.Liesack, W., H. Weyland, and E. Stackebrandt. 1991. Potential risk of gene amplification by PCR as determined by 16S rDNA analysis of a mixed-culture of strict barophilic bacteria. Microb. Ecol. 21:191-198. [DOI] [PubMed] [Google Scholar]
- 19.Mager, D. L., L. A. Ximenez-Fyvie, A. D. Haffajee, and S. S. Socransky. 2003. Distribution of selected bacterial species on intraoral surfaces. J. Clin. Periodontol. 30:644-654. [DOI] [PubMed] [Google Scholar]
- 20.Offenbacher, S., H. L. Jared, P. G. O'Reilly, S. R. Wells, G. E. Salvi, H. P. Lawrence, S. S. Socransky, and J. D. Beck. 1998. Potential pathogenic mechanisms of periodontitis associated pregnancy complications. Ann. Periodontol. 3:233-250. [DOI] [PubMed] [Google Scholar]
- 21.Paster, B. J., S. K. Boches, J. L. Galvin, R. E. Ericson, C. N. Lau, V. A. Levanos, A. Sahasrabudhe, and F. E. Dewhirst. 2001. Bacterial diversity in human subgingival plaque. J. Bacteriol. 183:3770-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paster, B. J., W. A. Falkler, Jr., C. O. Enwonwu, E. O. Idigbe, K. O. Savage, V. A. Levanos, M. A. Tamer, R. L. Ericson, C. N. Lau, and F. E. Dewhirst. 2002. Prevalent bacterial species and novel phylotypes in advanced noma lesions. J. Clin. Microbiol. 40:2187-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paster, B. J., M. K. Russell, T. Alpagot, A. M. Lee, S. K. Boches, J. L. Galvin, and F. E. Dewhirst. 2002. Bacterial diversity in necrotizing ulcerative periodontitis in HIV-positive subjects. Ann. Periodontol. 7:8-16. [DOI] [PubMed] [Google Scholar]
- 24.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 25.Sakamoto, M., M. Umeda, I. Ishikawa, and Y. Benno. 2000. Comparison of the oral bacterial flora in saliva from a healthy subject and two periodontitis patients by sequence analysis of 16S rDNA libraries. Microbiol. Immunol. 44:643-652. [DOI] [PubMed] [Google Scholar]
- 26.Scannapieco, F. A. 1999. Role of oral bacteria in respiratory infection. J. Periodontol. 70:793-802. [DOI] [PubMed] [Google Scholar]
- 27.Socransky, S. S., A. D. Haffajee, M. A. Cugini, C. Smith, and R. L. Kent, Jr. 1998. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 25:134-144. [DOI] [PubMed] [Google Scholar]
- 28.Van de Peer, Y., and R. De Wachter. 1994. TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput. Appl. Biosci. 10:569-570. [DOI] [PubMed] [Google Scholar]
- 29.Wade, W. G., D. A. Spratt, D. Dymock, and A. J. Weightman. 1997. Molecular detection of novel anaerobic species in dentoalveolar abscesses. Clin. Infect. Dis. 25(Suppl. 2):S235-S236. [DOI] [PubMed] [Google Scholar]
- 30.Whatmore, A. M., A. Efstratiou, A. P. Pickerill, K. Broughton, G. Woodard, D. Sturgeon, R. George, and C. G. Dowson. 2000. Genetic relationships between clinical isolates of Streptococcus pneumoniae, Streptococcus oralis, and Streptococcus mitis: characterization of “atypical” pneumococci and organisms allied to S. mitis harboring S. pneumoniae virulence factor-encoding genes. Infect. Immun. 68:1374-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Whitehouse, R. L., H. Jackson, M. C. Jackson, and M. M. Ramji. 1987. Isolation of Simonsiella sp. from a neonate. J. Clin. Microbiol. 25:522-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whitehouse, R. L., H. Merrill, M. C. Jackson, and H. Jackson. 1990. A stable variant of Simonsiella muelleri with unusual colonial and cellular morphology. J. Bacteriol. 172:1673-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Woo, P. C., A. M. Fung, S. K. Lau, B. Y. Chan, S. K. Chiu, J. L. Teng, T. L. Que, R. W. Yung, and K. Y. Yuen. 2003. Granulicatella adiacens and Abiotrophia defectiva bacteraemia characterized by 16S rRNA gene sequencing. J. Med. Microbiol. 52:137-140. [DOI] [PubMed] [Google Scholar]
- 34.Wu, T., M. Trevisan, R. J. Genco, J. P. Dorn, K. L. Falkner, and C. T. Sempos. 2000. Periodontal disease and risk of cerebrovascular disease: the first national health and nutrition examination survey and its follow-up study. Arch. Intern. Med. 160:2749-2755. [DOI] [PubMed] [Google Scholar]