Abstract
The aim of this study was to use molecular identification methods, such as 16S RNA gene sequence and reverse-capture checkerboard hybridization, for identification of the bacteria associated with dental caries and with dental health in a subset of 204 twins aged 1.5 to 7 years old. A total of 448 plaque samples (118 collected from caries-free subjects and 330 from caries-active subjects) were used for analysis. We compared the bacteria found in biofilms of children exhibiting severe dental caries, with different degrees of lesion severity, with those found in biofilms of caries-free children. A panel of 82 bacterial species was selected, and a PCR-based reverse-capture checkerboard method was used for detection. A simple univariate test was used to determine the overabundance and underabundance of bacterial species in the diseased and in the healthy groups. Features identified with this univariate test were used to construct a probabilistic disease prediction model. Furthermore, a method for the analysis of global patterns of gene expression was performed to permit simultaneous analysis of the abundance of significant species by allowing cross-bacterial comparisons of abundance profiles between caries-active and caries-free subjects. Our results suggested that global patterns of microbial abundance in this population are very distinctive. The top bacterial species found to be overabundant in the caries-active group were Actinomyces sp. strain B19SC, Streptococcus mutans, and Lactobacillus spp., which exhibited an inverse relationship to beneficial bacterial species, such as Streptococcus parasanguinis, Abiotrophia defectiva, Streptococcus mitis, Streptococcus oralis, and Streptococcus sanguinis.
The mechanisms of dental caries manifestation are complex and are triggered at various levels, i.e., genetic, behavioral, environmental, and microbial. Understanding the role of specific bacterial species and subspecies is important for creating a complete model of caries etiology. Dental plaque is a microbial biofilm community consisting of hundreds of distinct organisms that are ubiquitous in the oral cavity and that colonize the tooth surfaces. Cariogenic microorganisms initially colonize the dental biofilm early in life and can subsequently emerge, under favorable environmental conditions, to cause disease (12). Conversely, studies have shown that if pits and fissures in occlusal surfaces are initially colonized by a noncariogenic bacterial flora, these microorganisms may confer protection to the host by physically occupying the niche and blocking the colonization of cariogenic organisms, such as Streptococcus mutans, thereby preventing the onset and development of dental decay (4, 5).
In previous studies, conventional culturing methods have been used to show that well-known species, such as Streptococcus mutans and Lactobacillus spp., are associated with dental caries (13). These species have been reported as potential contributors to caries onset and development. More recently, advanced molecular methods of bacterial identification, such as PCR techniques and 16S rRNA gene sequencing analysis, have become available and have revealed that the bacterial involvement in the development of dental caries is more complex than previously believed (2).
The aim of this study was to use molecular identification methods, such as 16S rRNA gene sequence and reverse-capture checkerboard hybridization, for identification of the bacteria associated with dental caries and health in infants and children. We compared the bacteria found in biofilms of caries-active children with different degrees of lesion severity with those found in biofilms of caries-free children. In addition, we compared biofilms of healthy surfaces of caries-active subjects with biofilms of healthy surfaces of caries-free subjects. A simple univariate test was used to determine the overabundance and underabundance of bacterial species in the diseased and in the healthy groups. Features identified with this univariate test were used to construct a probabilistic disease prediction model. With proper machine learning-based evaluation, we found that this model was successful in utilizing biofilm bacterial risk indicators to predict disease and health. Our modeling approach splits a data set into two groups of samples, a training and a test set. Because we evaluated the model by performing learning (classifier construction) in the training set and evaluated in the test set, the performance of the prediction model could be statistically validated and, thus, could be generalized.
MATERIALS AND METHODS
Subject population and sample.
The study population consisted of a twin cohort from low socioeconomic urban families who resided in the city of Montes Claros, State of Minas Gerais, Brazil. City water supplies have fluoride levels of <0.2 ppm, and parents reported 91% of the children having never visited a dentist. Parents of the twins signed consent forms, and the study protocol was approved by human subjects' institutional review boards at the University of Pittsburgh, Harvard University, Forsyth Institute, and UNIMONTES.
Study design and sampling.
Two examiners conducted dental caries examinations according to National Institute of Dental and Craniofacial Research criteria (3) modified to distinguish caries lesions with a chalky whitish/yellowish opaque appearance, without clinically detectable loss of substance (white spot lesions), from cavitated carious lesions. Interproximal surface caries were assessed using digital imaging fiber-optic transillumination (DIFOTI, Irvington, NY). About 20% of the total cohort was caries free, 25% had low to moderate levels of dental caries rates, and 55% had high to rampant caries rates.
Plaque samples were collected in the morning, and the children were asked not to brush their teeth or eat the night before the exam. Supragingival dental plaque samples were collected from the entire cohort over a period of 4 weeks. The caries-free twins had pooled plaque samples collected from three healthy surfaces, including anterior and posterior teeth. Caries-active subjects had plaque samples collected separately from four types of surfaces: surface of intact enamel (site 1), surface of white spot lesions (site 2), surface of initial enamel lesions (site 3), and excavated plaque from deep dentinal lesions (site 4) (Fig. 1). All caries-active subjects provided three to four sites of plaque collected separately from different teeth according to the severity of disease. For intact enamel and white spot lesions, plaque was collected by swiping the tooth surface with a Stimudent (Johnson & Johnson, New York, NY), whereas plaque from cavitated lesions was collected by means of a small Gracey curette (1-2; Hu-Friedy, Chicago, IL).
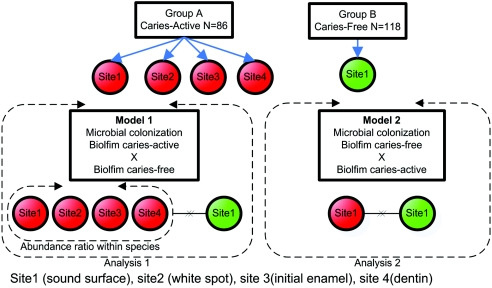
FIG. 1.
Study design. Model 1 compared site 1 (biofilm of surfaces of intact enamel) in the caries-free group with all sites combined in the caries-active group: site 1 (biofilm of surfaces of intact enamel), site 2 (white spot lesions), site 3 (dentinal lesions), and site 4 (deep dentinal lesions). Model 2 compared site 1 (biofilm present on surfaces of intact enamel) from the caries-free group to only site 1 (biofilm present on surfaces of intact enamel) from the caries-active group.
A total of 204 subjects, with an average age of 3.83 ± 2.55 years, were selected to comprise the caries-active and caries-free groups, and they were matched by gender. The two groups consisted of caries-free twins (n = 118; surface-based caries prevalence rate, 0) and a group of highly caries-active twins with no existing restorations (n = 86; mean surface-based caries prevalence rate, 17.23% ± 10.70%). A total of 448 plaque samples (118 collected from caries-free subjects and 330 from caries-active subjects) were used for analysis.
Isolation of bacterial DNA.
Samples were placed inside a sterile microcentrifuge tube already containing 50 μl of TE buffer (50 mM Tris-HCl, pH 7.6, 1 mM EDTA) and immediately processed for DNA extraction. Five microliters of 5% Tween 20 and 1 μl of 10-mg/ml proteinase K (100 mg; V3021; Promega) were added to each sample. Samples were incubated at 55°C for 2 h. The proteinase K was inactivated by heating the samples at 95°C for 10 min. Samples were frozen at −20°C and transported in dry ice from Brazil to the United States. Samples were frozen at −80°C for further analysis.
Cloning, sequencing, and clonal analysis.
Initially, four random samples from different types of lesions (intact healthy surfaces, white spot lesions, surfaces of initial enamel lesions, and surfaces of deep dentinal lesions) were selected for clonal analysis for initial characterization of microbial profiles in this cohort and to further determine the target species to be used for checkerboard analysis. Cloning procedures, which included 16S rRNA sequencing and data analysis, were performed with the TOPO TA cloning kit (Invitrogen, San Diego, CA) as previously described (10). A total of 200 clones with the insert of the correct size of approximately 1,500 bases were analyzed (100 per sample) and compared to known 16S rRNA gene sequences in the Ribosomal Database Project (7).
Amplification of bacterial DNA.
The 16S rRNA genes were amplified under standard conditions using a universal forward primer and a universal reverse primer (10). PCR was performed in thin-walled tubes with a Perkin-Elmer 9700 thermocycler. One microliter of the DNA template was added to 22.5 μl of a PCR SuperMix (Invitrogen, Carlsbad, CA). In a hot start protocol, samples were preheated at 94°C for 4 min, followed by amplification under the following conditions: denaturation at 94°C for 45 s, annealing at 60°C 45 s, and elongation for 1.5 min with an additional 5 s for each cycle. A total of 30 cycles were performed and were then followed by a final elongation step at 72°C for 10 min. The results of PCR amplification were examined by electrophoresis in 1% agarose gel. DNA was stained with ethidium bromide and visualized under short-wavelength UV light. For checkerboard hybridization, the resulting PCR product from each sample was used to run three sets of PCR amplification using a forward primer labeled at the 5′ end with digoxigenin and a universally conserved reverse primer. This approach was used based on the fact that for each sample we needed to run three different checkerboards (we could only fit 30 probes on each checkerboard). Using the same PCR product for the subsequent labeled PCR reduced the variation in the samples.
Checkerboard hybridization.
The reverse-capture checkerboard hybridization assay was used, as previously described (11), to detect levels (abundance) of 82 oral bacterial species or groups (see the Appendix, below). Briefly described, reverse-capture DNA probes (complementary oligonucleotide DNAs of known sequence) are used to target polynucleotides of unknown sequence (16S rRNA) bacterial genes in the biological sample solution. Probes were placed on a nylon membrane in separate horizontal lanes using a Mini Slot apparatus. 16S rRNA genes from plaque samples were PCR amplified using a specific labeled primer. Hybridizations were performed in vertical channels in a Miniblotter apparatus with labeled amplicons (target 16S rRNA genes) for up to 45 samples. A total of 1,350 hybridizations were performed simultaneously using a single membrane. Standard chemifluorescence detection was performed using the Storm system (Amersham, Piscataway, NJ). For each spot on the membranes, signal levels were extracted from their background by applying spot edge detection methodology. This method locates the average intensity around the spot's outline and then applies this as the background for the spot. The background was therefore calculated independently for each spot, and signal levels (normalized to mean counts) were calculated independently for each spot (ImageQuant software; Amersham, Piscataway, NJ). Low-quality spots were also filtered for quality control, and background noise was eliminated from the analysis. Two lanes in each run had the universal probes to serve as standards, and signal levels were converted to mean counts by comparison with standards on the membrane. Signal levels were then adjusted for abundance by comparing them to the universal control probes. This approach allowed for computing the abundance of the target species individually by adjusting the DNA concentration in each sample.
Computation of microbial abundance patterns.
To determine if differences in bacterial levels were present between caries-free subjects and caries-active subjects, the Wilcoxon rank sum test for nonparametric distribution was first used for mean comparisons across all 82 bacterial species. However, classification approaches employed in biological samples should be capable of accurately representing not only differences across groups but also patterns that reflect true relationships among samples and disease profiles. For that reason, the Gene Expression Data Analysis software (9) was used for the classification of global patterns of microbial colonization and abundance levels in the two groups, and data were evaluated as follows: (i) the data were cube root transformed as a means of stabilizing the variance of the data across the full range of bacterial levels combined from different checkerboards, (ii) a simple univariate test was used to determine the overabundance and underabundance of bacterial species in the diseased and in the healthy groups, (iii) significant bacterial species in the two groups were identified using the Baïve Bayes classifier, and (iv) computation validation (random resampling validation) was performed using the significant species as features. In this model, we used a classification framework named PACE (permutation-achieved classification error) (6), which allowed us to compare the performance statistic of the classifier on true data samples and to perform validation procedures in the same data with randomly reassigned class labels. Our modeling approach split the data set into two groups of samples, a training and a test set, by using a 70/30 ratio. Learning (classifier construction) was conducted on the training set and evaluated on the test set. Features (differentially expressed bacterial species) were selected using the specific test and threshold, and class prediction was performed on the test data set (Baïve Bayes posterior probability). Sensitivity and specificity of the model were measured using the area under the receiver/operator curve (ROC). A statistically significant result increases the chance of a discriminative signal in the data. The objective of performing classification analyses was to build a predictive model to determine whether, and how, the differentially abundant species could be used to determine if differences were present in the biofilm of the caries-active group relative to the caries-free group.
Two approaches were used for analysis (Fig. 1). Model 1 compared site 1 (biofilm of surfaces of intact enamel) in the caries-free group with all sites combined in the caries-active group: site 1 (biofilm of surfaces of intact enamel), site 2 (white spot lesions), site 3 (dentinal lesions), and site 4 (deep dentinal lesions) (1). In this model, a ratio was created to adjust bacterial levels within species by the total number of sites collected for each caries-active subject. Model 2 compared site 1 (biofilm present on surfaces of intact enamel) from the caries-free group to only site 1 (biofilm present on surfaces of intact enamel) from the caries-active group.
RESULTS
In order to exemplify the diversity of the microbial flora of the population under study, four randomly selected samples from caries-active subjects were cloned and sequenced. A minimum of 50 clones were sequenced from each sample. A total of 200 sequences were analyzed. A panel of 82 known bacterial species or phylotypes was selected, and a PCR-based reverse-capture checkerboard method was used for detection of bacterial levels (1, 11). The evaluation result from analysis through additional statistical validation tests (PACE) was able to determine the differential abundance of significant species in the caries-active group compared to the caries-free group. By using this model, a number of informative species were determined for each analytical model.
Model 1.
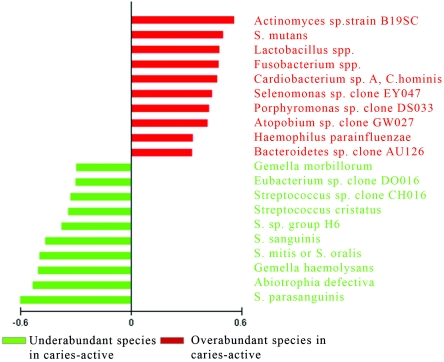
Model 1 compared site 1 (biofilm of surfaces of intact enamel) in the caries-free group with all sites combined in the caries-active group, i.e., site 1 (biofilm of surfaces of intact enamel), site 2 (white spot lesions), site 3 (dentinal lesions), and site 4 (deep dentinal lesions) (1). The achieved classification error was statistically significant under ROC (specificity, 0.86; sensitivity, 0.84) with a permutation test and PACE analysis at the 99% level. A total of 10 species were found to be significantly overabundant in the caries-active subjects, and 10 species were found to be significantly underabundant in this group compared to caries-free subjects (Fig. 2).
FIG. 2.
Overabundance and underabundance of bacterial species in caries-active subjects. Using model 1, the combined sites (intact enamel [1], white spot lesions [2], enamel lesions [3], and dentin lesions [4]) in caries-active subjects were compared to the biofilm of caries-free subjects. Species that are overabundant in the caries-active group relative to the caries-free group are shown in red; species that are underabundant in the caries-active group relative to the caries-free group are shown in green.
Model 2.
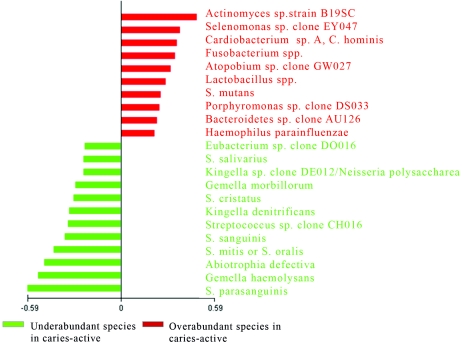
In model 2, only surfaces of intact enamel (site 1) in caries-active subjects were compared to surfaces of intact enamel (site 1) in caries-free subjects (Fig. 3). The achieved classification error was statistically significant under ROC (specificity, 0.81; sensitivity, 0.75), with a permutation test and PACE analysis at the 99% level. A total of 10 species were found to be significantly overabundant in the caries-active subjects, and 12 species were found to be significantly underabundant in this group compared to caries-free subjects. The top significant bacterial species (Fig. 2 and 3) in the caries-active group, compared to the caries-free group, in both models resulted in a noteworthy resemblance in which overabundance of disease-related species, such as Actinomyces sp., Lactobacillus sp., and S. mutans, in combination with an underabundance of beneficial, health-related species, such as Streptococcus parasanguinis, Streptococcus sanguis, and Streptococcus mitis/oralis, was observed for both models. We next determined species that were significantly elevated or decreased in caries-active subjects relative to caries-free subjects by employing the traditional Wilcoxon rank sum test. These analyses were performed when species were detected in >10 subjects by the checkerboard hybridization procedure. A number of species were deemed statistically significantly different in caries-active subjects relative to caries-free subjects (Table 1). Moreover, we compared the results in Table 1 to the PACE analysis that was employed for both analytical models (Fig. 2 and 3). Notably, it was found that many species deemed significant by the Wilcoxon test would not have been included as significant risk indicators of microbial overabundance and underabundance under PACE analysis in caries-active subjects relative to caries-free subjects.
FIG. 3.
Overabundance and underabundance of bacterial species in caries-active subjects. Using model 2, the biofilm of caries-active subjects was compared to the biofilm of caries-free subjects. Species that are overabundant in the caries-active group relative to the caries-free group are shown in red; species that are underabundant in the caries-active group relative to the caries-free group are shown in green.
TABLE 1.
Detection of bacterial species in subjectsa
| Bacterial species | Model 1: caries active (sites 1-4) compared to caries free (site 1) | Model 2: caries active (site 1) compared to caries free (site 1) | ||||||
|---|---|---|---|---|---|---|---|---|
| PACEb | Active | Free | P value | PACEb | Active | Free | P value | |
| Abiotrophia defectiva | UA | 60 | 107 | <0.0001 | UA | 50 | 107 | <0.0001 |
| Actinomyces georgiae | 28 | 23 | NS | 19 | 23 | NS | ||
| Actinomyces gerencseriae | 34 | 34 | NS | 29 | 34 | NS | ||
| Actinomyces naes/undii serotype II | 34 | 19 | 0.0007 | 22 | 19 | NS | ||
| Actinomyces sp. strain ATCC 49338 | 38 | 40 | NS | 32 | 40 | NS | ||
| Actinomyces sp. strain B19SC | OA | 86 | 75 | <0.0001 | OA | 86 | 75 | <0.0001 |
| Actinomyces sp. oral clone EP005 | 24 | 12 | 0.0032 | 15 | 12 | NS | ||
| Atopobium sp. clone GW027 | OA | 27 | 2 | <0.0001 | OA | 18 | 2 | <0.0001 |
| Bacteriodes sp. clone AU126 | OA | 64 | 42 | <0.0001 | OA | 43 | 42 | 0.0065 |
| Bacteriodes forsythus-like clone BU063 | 12 | 10 | NS | 8 | 10 | NS | ||
| Campylobacter concisus | 79 | 66 | 0.0003 | 56 | 66 | NS | ||
| Campylobacter showae | 69 | 56 | 0.0119 | 53 | 56 | NS | ||
| Capnocytophaga gingivalis | 12 | 9 | NS | 6 | 9 | NS | ||
| Capnocytophaga granulosa | 86 | 111 | NS | 36 | 111 | NS | ||
| Capnocytophaga sputigena | 29 | 18 | 0.0177 | 18 | 18 | NS | ||
| Cardiobacterum sp. A, Cardiobacterum hominis | OA | 25 | 1 | <0.0001 | OA | 15 | 1 | <0.0001 |
| Coryne bacterium matruchotii | 33 | 29 | NS | 27 | 29 | NS | ||
| Eubacterium sabboreum GT038 | 34 | 31 | NS | 15 | 31 | NS | ||
| Eubacterium sp. clone DO016 | UA | 68 | 99 | <0.0001 | UA | 63 | 99 | 0.0007 |
| Eubacterium sp. clone EI074 | 29 | 22 | NS | 18 | 22 | NS | ||
| Eubacterium sp. strain C27KA | 15 | 8 | 0.0302 | 9 | 8 | NS | ||
| Fusobacterium spp. | OA | 19 | 1 | <0.0001 | OA | 10 | 1 | 0.0002 |
| Fusobacterium animalis | 22 | 15 | NS | 13 | 15 | NS | ||
| Fusobacterium nucleatum subsp. polymorphum | 23 | 34 | NS | 16 | 34 | NS | ||
| Gemella haemolysans | UA | 35 | 94 | <0.0001 | UA | 28 | 94 | <0.0001 |
| Gemella morbillorum | UA | 73 | 106 | <0.0001 | UA | 62 | 106 | <0.0001 |
| Gemella sp. strain 933-88 | 14 | 9 | NS | 8 | 9 | NS | ||
| Haemophilus parainfluenzae | OA | 60 | 39 | <0.0001 | OA | 40 | 39 | 0.0137 |
| Kingella denitrificans | 27 | 57 | 0.003 | UA | 13 | 57 | <0.0001 | |
| Kingella oralis | 36 | 46 | NS | 22 | 46 | 0.0284 | ||
| Kingella sp. clone DE012/Neisseria polysaccharea | 56 | 76 | 0.0077 | UA | 40 | 76 | 0.0004 | |
| Lactobacillus spp. | OA | 28 | 1 | <0.0001 | OA | 14 | 1 | <0.0001 |
| Lactobacillus gasseri | 17 | 9 | 0.0123 | 6 | 9 | NS | ||
| Lautropia mirabolis | 33 | 21 | 0.0047 | 19 | 21 | NS | ||
| Leptotrichia spp. | 29 | 24 | NS | 19 | 24 | NS | ||
| Leptotrichia sp. strain A39FD | 44 | 25 | 0.0015 | 29 | 25 | NS | ||
| Neisseria mucosa/flavescens | 22 | 24 | NS | 10 | 24 | NS | ||
| Porphyromonas sp. clone DS033 | OA | 38 | 6 | <0.0001 | OA | 19 | 6 | 0.003 |
| Prevotella melaninogenica | 19 | 14 | NS | 10 | 14 | NS | ||
| Prevotella nigrescens | 9 | 10 | NS | 6 | 10 | NS | ||
| S. cristatus | UA | 70 | 112 | <0.0001 | UA | 61 | 112 | 0.0002 |
| S. mitis | 42 | 44 | NS | 35 | 44 | NS | ||
| S. mitis biovar 2 | 59 | 92 | 0.0079 | 55 | 92 | 0.0171 | ||
| S. mitis/oralis | UA | 77 | 117 | <0.0001 | UA | 70 | 117 | <0.0001 |
| S. mutans | OA | 56 | 23 | <0.0001 | OA | 36 | 23 | 0.0006 |
| S. parasanguinis | UA | 53 | 110 | <0.0001 | UA | 45 | 110 | <0.0001 |
| S. salivarius | 54 | 85 | 0.0234 | UA | 39 | 85 | 0.0004 | |
| S. sanguinis | UA | 83 | 118 | <0.0001 | UA | 79 | 118 | <0.0001 |
| Streptococcus sp. clone CH016 | UA | 74 | 112 | <0.0001 | UA | 66 | 112 | <0.0001 |
| Streptococcus sp. group H6 | 47 | 31 | NS | 36 | 31 | NS | ||
| Selenomonas infelix | 19 | 37 | NS | 14 | 37 | 0.0115 | ||
| Selenomonas noxia | 47 | 29 | 0.0059 | 30 | 29 | NS | ||
| Selenomonas sp. caries clone DS071 | 37 | 22 | 0.0051 | 28 | 22 | NS | ||
| Selenomonas sp. clone AA024 | 17 | 9 | 0.0212 | 12 | 9 | NS | ||
| Selenomonas sp. clone EY047 | OA | 66 | 41 | <0.0001 | OA | 56 | 41 | <0.0001 |
| Selenomonas sp. clone GT010 | 15 | 5 | 0.0031 | 12 | 5 | NS | ||
| Selenomonas sp. clone GT052 | 17 | 15 | NS | 11 | 15 | 0.0329 | ||
| Selenomonas sp. oral clone CS024 | 16 | 9 | 0.0382 | 15 | 9 | NS | ||
| Selenomonas sputigena | 23 | 12 | 0.0055 | 16 | 12 | NS | ||
| Veillonella dispar or V. parvula | 79 | 107 | 0.0037 | 68 | 107 | NS | ||
| Veillonella atypica | 34 | 31 | NS | 21 | 31 | NS | ||
| Veillonella sp. oral clone BU083 | 67 | 82 | NS | 54 | 82 | NS | ||
Species described were detected by the checkerboard hybridization procedure in >10 subjects. P values reflect significant species when levels were compared by the Wilcoxon rank sum test. NS, not significant.
OA, significant species overabundant in caries-active subjects; UA, significant species underabundant in caries-active subjects.
DISCUSSION
This study used a model for quantification of the abundance of different bacterial species colonizing the tooth surfaces of caries free-subjects and caries-active subjects. The methodology commonly used for gene expression analysis (9) was adapted to the checkerboard technology (11) to permit simultaneous analysis of the abundance of significant species by allowing cross-bacterial comparisons of abundance profiles between caries-active and caries-free subjects. This study represents an application of a statistical machine learning principle to predictive model construction. Unlike other types of analysis, our estimates of classifier performance are unbiased (or, at most, low bias) and are therefore generalizable, providing a promising source of subject-specific information manifesting potential impact on the early detection and classification of disease profiles. This is due to the use of training and test sets, instead of using all the data to merely identify species that exhibit statistical differences. The use of all the data to produce a predictive model would likely lead to model overfit (bias).
The results of our study suggest that an abundance of the cariogenic microflora, i.e., Actinomyces sp., S. mutans, and Lactobacillus sp., in concert with an underabundance of health-associated species (S. parasanguinis, Abiotrophia defectiva, Gemella hemolysans, S. mitis/oralis, and S. sanguinis) represented a profile of the caries-active group (Fig. 2 and 3). By using this model, it was possible to identify microbial profiles that distinguished caries-active subjects from caries-free subjects.
In this study we employed two approaches of analysis. The first model looked at bacterial colonization present in the biofilm of caries-free subjects compared to the different biofilms of sites, with different lesion severity, in the caries-active subjects (Fig. 1). In the second model the microflora present in the biofilms of caries-free subjects was compared with only the microflora present in biofilms of surfaces of intact enamel in the caries-active subjects (Fig. 1). The same patterns of bacterial overabundance and underabundance were found to be significant in both models (Fig. 2 and 3). Notably, by employing model 2 one could conceivably use bacterial biofilms of healthy surfaces of caries-active subjects to profile subjects who may be at increased risk for developing caries. Thus, this model predicted that biofilm bacteria abundance on enamel intact surfaces may provide useful information for early diagnosis and detection of dental caries.
The results of this study confirmed those of previous reports (1, 8) by indicating that the microflora associated with caries is dominated by gram-positive bacteria, particularly the genera Actinomyces, Lactobacillus, and Streptococcus, and more predominantly by Streptococcus mutans (1). Earlier culture-based studies (13) implicated mutans streptococci in caries initiation, with the lactobacilli as important contributory bacteria. These results corroborate our findings in that S. mutans was found to be overabundant in about 90% of caries-active subjects and underabundant in the caries-free subjects (Fig. 2 and 3). Similarly, Lactobacillus spp. were found with high frequency in caries-active subjects but found only in one subject in the caries-free group (Table 1). Most studies (13) have found that although Actinomyces spp. are commonly detected in the human mouth, their role in carious lesions is variable and inconclusive. However, in our study Actinomyces spp. were found in abundance in both caries-active and caries-free subjects but with statistically significant differences between the groups (Table 1). One particular species (Actinomyces sp. strain B19SC) was found in very high levels and was significantly overabundant in caries-active subjects relative to caries-free subjects (both models) (Fig. 2 and 3).
Caries-active subjects in this population had a distinctive microflora represented by an underabundance of beneficial health-associated species (Fig. 2 and 3). Conversely, caries-free subjects exhibited an overabundance of known beneficial species (data not shown), i.e., S. parasanguinis, A. defectiva, G. hemolysans, S. mitis/oralis, Streptococcus cristatus, and S. sanguinis. Streptococcus sanguinis has long been associated with health (13). To understand what constitutes health, before disease ensues, requires ascertaining which health-associated species play an essential role, since beneficial species may protect the host against harmful pathogens, thereby preventing the onset and development of dental caries.
Lastly, results from our study suggest that research is needed to identify ways to translate these findings into clinically useful diagnostic models. We speculate that the most effective treatment strategies and preventive measures may well depend on elucidation of specific microbial profiles in health and disease. Biofilm bacteria can be very resistant to antibiotic treatment, and the mechanisms by which the biofilm-grown bacteria attain this resistance are still unknown. Moreover, biofilm bacteria infections are rarely resolved by the host's immune system, and they are resistant to the host's defense mechanisms (2). Knowing which bacteria are beneficial can help in the development of new treatment strategies for caries, whereby one manipulates these communities so as to prevent disease and promote health as an adjunct to standard prevention treatments.
Acknowledgments
This work was supported by NIH grants DE14528 and DE15351.
APPENDIX
The bacterial species and groups used in the checkerboard analysis are shown in Table A1.
TABLE A1.
Bacterial species and groups used in checkerboard analysis
| Bacterial species or group |
|---|
| Streptococcus sp. clone CH016 |
| Streptococcus sp. group H6 |
| Actinomyces sp. strain B27SC |
| Actinomyces sp. oral clone EP005 |
| Actinomyces sp. strain ATCC 49338 |
| Actinomyces sp. strain B19SC |
| Abiotrophia defectiva |
| Actinomyces georgiae |
| Actinomyces gerencseriae |
| Actinomyces israelii |
| Actinomyces naeslundii serotype II |
| Actinomyces odontolyticus |
| Atopobium sp. clone GW027 |
| Bacteroides sp. clone AU126 |
| Bacteroides forsythus-like clone BU063 |
| Bifidobacterium all |
| Bifidobacterium sp. oral clone CX010 |
| Campylobacter concisus |
| Campylobacter showae |
| Capnocytophaga gingivalis |
| Capnocytophaga ochracea W089 |
| Capnocytophaga sp. clone DS022 |
| Capnocytophaga sputigena |
| Cardiobacterium sp. A, Cardiobacterium hominis |
| Corynebacterium matruchotii |
| Eubacterium sabboreum GT038 |
| Eubacterium sp. clone DO016 |
| Eubacterium sp. clone EI074 |
| Eubacterium sp. strain C27KA |
| Eubacterium yurii |
| Fusobacterium all |
| Fusobacterium animalis |
| Fusobacterium nucleatum subsp. polymorphum |
| Gemella haemolysans |
| Gemella morbillorum |
| Gemella sp. strain 933-88 |
| Granulicatella adiacens |
| Haemophilus parainfluenzae |
| Kingella denitrificans |
| Kingella oralis |
| Kingella sp. clone DE012/Neisseria polysaccharea |
| Lactobacillus all |
| Lactobacillus fermentum |
| Lactobacillus gasseri |
| Lautropia mirabilis |
| Leptotrichia sp. strain GT018 |
| Leptotrichia all |
| Leptotrichia sp. clone DE081 |
| Leptotrichia sp. clone DT031 |
| Leptotrichia sp. strain A39FD |
| Neisseria mucosa or N. flavescens |
| Peptostreptococcus sp. clone CK035 |
| Porphyromonas sp. clone DS033 |
| Prevotella loescheii |
| Prevotella melaninogenica |
| Prevotella nigrescens |
| Prevotella oris |
| Rothia dentocariosa |
| Streptococcus anginosus |
| Streptococcus cristatus |
| Streptococcus gordonii |
| Streptococcus intermedius |
| Streptococcus mitis |
| Streptococcus mitis biovar 2 |
| Streptococcus mitis or S. oralis |
| Streptococcus mutans |
| Streptococcus parasanguinis |
| Streptococcus salivarius |
| Streptococcus sanguinis |
| Streptococcus sp. strain 7A or Streptococcus sp. strain H6 |
| Selenomonas infelix |
| Selenomonas noxia |
| Selenomonas sp. oral clone CS024 |
| Selenomonas sp. caries clone DS071 |
| Selenomonas sp. clone AA024 |
| Selenomonas sp. clone EY047 |
| Selenomonas sp. clone GT010 |
| Selenomonas sp. clone GT052 |
| Selenomonas sputigena |
| Veillonella dispar or V. parvula |
| Veillonella atypica |
| Veillonella sp. oral clone BU083 |
REFERENCES
- 1.Becker, M. R., B. J. Paster, E. J. Leys, M. L. Moeschberger, S. G. Kenyon, J. L. Galvin, S. K. Boches, F. E. Dewhirst, and A. L. Griffen. 2002. Molecular analysis of bacterial species associated with childhood caries. J. Clin. Microbiol. 40:1001-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davey, M. E., and G. A. Otoole. 2000. Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 64:847-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kastle, L. M., R. H. Selwitz, R. J. Oldakowski, J. A. Brunelle, D. M. Winn, and L. J. Brown. 1996. Coronal caries in the primary and permanent dentition of children and adolescents 1-17 years of age: United States, 1988-1991. J. Dent. Res. 75:631-641. [DOI] [PubMed] [Google Scholar]
- 4.Loesche, W. J. 1986. Role of Streptococcus mutans in human dental decay. Microbiol. Rev. 50:353-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Loesche, W. J., S. Eklund, R. Earnest, and B. Burt. 1984. Longitudinal investigation of bacteriology of human fissure decay: epidemiological studies in molars shortly after eruption. Infect. Immun. 46:765-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lyons-Weiler, J., R. Pelikan, H. J. Zeh III, D. C. Whitcomb, D. E. Malehorn, William L. Bigbee, and M. Hauskrecht. 2005. Assessing the statistical significance of the achieved classification error of classifiers constructed using serum peptide profiles, and a prescription for random sampling repeated studies for massive high-throughput genomic and proteomic studies. Cancer Informatics 2005:5377. [PMC free article] [PubMed]
- 7.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker, Jr., P. R. Saxman, J. M. Stredwick, G. M. Garrity, B. Li, G. J. Olsen, S. Pramanik, T. M. Schmidt, and J. M. Tiedje. 2000. The RDP (Ribosomal Database Project) continues. Nucleic Acids Res. 28:109-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Munson, M. A., A. Banerjee, T. F. Watson, and W. G. Wade. 2004. Molecular analysis of the microflora associated with dental caries. J. Clin. Microbiol. 42:3023-3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patel, S., and J. Lyons-Weiler. 2004. caGEDA: a web application for the integrated analysis of global gene expression patterns in cancer. Appl. Bioinformatics 3:49-62. [DOI] [PubMed] [Google Scholar]
- 10.Paster, B. J., S. K. Boches, J. L. Galvin, R. E. Ericson, C. E. Lau, V. A. Levanos, A. Sahasrabudhe, and F. E. Dewhirst. 2001. Bacterial diversity in human subgingival plaque. J. Bacteriol. 183:3770-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paster, B. J., I. M. Bartoszyk, and F. E. Dewhirst. 1998. Identification of oral streptococci using PCR-based, reverse-capture, checkerboard hybridization. Methods Cell Sci. 20:223-231. [Google Scholar]
- 12.Scheie, A. A., and F. C. Petersen. 2004. The biofilm concept: consequences for future prophylaxis of oral diseases? Crit. Rev. Oral Biol. Med. 15:4-12. [DOI] [PubMed] [Google Scholar]
- 13.Tanzer, J. M., J. Livingston, and A. M. Thompson. 2001. The microbiology of primary dental caries in humans. J. Dent. Educ. 65:1028-1037. [PubMed] [Google Scholar]