Abstract
We previously reported that the group III histidine kinase Dic1p in the maize pathogen Cochliobolus heterostrophus is involved in resistance to dicarboximide and phenylpyrrole fungicides and in osmotic adaptation. In addition, exposure to the phenylpyrrole fungicide fludioxonil led to improper activation of Hog1-type mitogen-activated protein kinases (MAPKs) in some phytopathogenic fungi, including C. heterostrophus. Here we report, for the first time, the relationship between the group III histidine kinase and Hog1-related MAPK: group III histidine kinase is a positive regulator of Hog1-related MAPK in filamentous fungi. The phosphorylation pattern of C. heterostrophus BmHog1p (Hog1-type MAPK) was analyzed in wild-type and dic1-deficient strains by Western blotting. In the wild-type strain, phosphorylated BmHog1p was detected after exposure to both iprodione and fludioxonil at a concentration of 1 μg/ml. In the dic1-deficient strains, phosphorylated BmHog1p was not detected after exposure to 10 μg/ml of the fungicides. In response to osmotic stress (0.4 M KCl), a trace of phosphorylated BmHog1p was found in the dic1-deficient strains, whereas the band representing active BmHog1p was clearly detected in the wild-type strain. Similar results were obtained for Neurospora crassa Os-2p MAPK phosphorylation in the mutant of the group III histidine kinase gene os-1. These results indicate that group III histidine kinase positively regulates the activation of Hog1-type MAPKs in filamentous fungi. Notably, the Hog1-type MAPKs were activated at high fungicide (100 μg/ml) and osmotic stress (0.8 M KCl) levels in the histidine kinase mutants of both fungi, suggesting that another signaling pathway activates Hog1-type MAPKs in these conditions.
All living cells must appropriately sense and respond to various environmental stimuli. In eukaryotic cells, the mitogen-activated protein (MAP) kinase cascades are vital to the intracellular transmission of a variety of extracellular signals and to the regulation of growth and differentiation processes (7, 36). The MAP kinase cascades generally consist of three protein kinases that act in series: MAP kinase (MAPK), MAPK kinase (MAPKK), and MAPKK kinase (MAPKKK). In mammalian cells, a variety of receptor types, such as G-protein-coupled receptors, receptor tyrosine kinases, and integral membrane proteins, are known to perceive the extracellular signals and then promote the activation of MAPK cascades (36). The activated MAPK phosphorylates substrates, which include transcriptional activators, resulting in altered patterns of gene expression and protein activity.
Cellular signaling pathways that are mediated by MAPK cascades have also been characterized in many fungi (9, 21, 22, 23, 33, 42, 44, 47). In Neurospora crassa, for example, three MAPK modules have been identified by genome sequence analysis. os-2 mutants of N. crassa, which contain a null mutation in the MAPK gene that is homologous to Saccharomyces cerevisiae HOG1, were found to be unable to adapt to high osmolarity (47). Also, a Mak-2p MAPK of N. crassa that is related to the S. cerevisiae Fus3p/Kss1p was found to be required for hyphal fusion events (33). As another example, Colletotrichum lagenarium, which causes cucumber anthracnose, was discovered to have a Maf1p MAPK that is related to S. cerevisiae Mpk1p (Slt2p) and a Cmk1p MAPK that is related to S. cerevisiae Fus3p/Kss1p, which regulate diverse aspects of fungal pathogenesis (22, 42). Despite extensive studies of the functions of individual MAPKs in filamentous fungi, many unsolved problems remain concerning the signaling mechanisms of MAPK cascades. The upstream activators acting on the cascade and the mechanisms of dephosphorylation of MAPK modules are still unclear.
In contrast to eukaryotes, prokaryotic organisms commonly employ a His-Asp phosphorelay signaling strategy known as the two-component regulatory system (3, 34, 41). The prototypical system is composed of a histidine protein kinase and a response regulator protein. Extracellular signals are sensed by a histidine kinase that transfers a phosphoryl group to a response regulator, which results in the activation of a downstream effector that elicits the specific response. Although some eukaryotes, especially fungi, also use two-component signaling systems, the two proteins (a histidine kinase and a response regulator) are typically fused (1, 6, 32).
The results of fungal genome sequencing projects (http://www.broad.mit.edu) and recent studies (5, 16) have revealed that filamentous ascomycetes, such as the model filamentous fungi N. crassa and Aspergillus nidulans, and phytopathogenic fungi, such as Botryotinia fuckeliana (anamorph: Botrytis cinerea), Cochliobolus heterostrophus (anamorph: Bipolaris maydis), Gibberella moniliformis, and Magnaporthe grisea, possess several genes for hybrid histidine kinases, which are typically classified into eleven groups. Among them, six groups (III, V, VI, VIII, IX, and X) contain histidine kinases that are highly conserved in those fungal species, and other groups are more divergent (5). To date, however, there have been few reports regarding the functional analysis of the histidine kinase genes in filamentous fungi except for the group III histidine kinase gene.
The group III histidine kinases, e.g., N. crassa Os-1p (also known as Nik1p), B. fuckeliana Daf1p (also known as BcOs1p), and C. heterostrophus Dic1p (also known as ChNik1p), are characterized by a unique N-terminal region consisting of a HAMP domain repeat (5). The mutation of these histidine kinase genes resulted in resistance to the fungicides phenylpyrrole and dicarboxyimide and also in increased osmosensitivity (8, 30, 45, 46). In C. heterostrophus, null mutants for dic1 and mutants with a deletion or point mutation in the HAMP domain repeat were highly sensitive to osmotic stress and highly resistant to the previously mentioned fungicides (46). Similar effects resulting from mutations were observed in N. crassa os-1 and B. fuckeliana daf1 (8, 30). A single amino acid change within the kinase domain or the regulator domain of C. heterostrophus dic1 altered the sensitivity to osmotic stress and conferred a moderate resistance to the fungicides. Thus, the group III histidine kinase is considered to be a putative osmosensor (38). However, no membrane-spanning domain is predicted in the group III histidine kinase, so the protein is likely cytoplasmic. The precise function and mechanism of the group III histidine kinase-mediated response to extracellular osmotic stress are still obscure.
In S. cerevisiae, osmotic stress is processed by the osmoregulatory system HOG (high-osmolarity glycerol) pathway MAPK cascade that is controlled by a His-Asp phosphorelay signaling system employing the group VI histidine kinase Sln1p (19, 20). Sln1 is the sole histidine kinase occurring in S. cerevisiae and is essential. The group VI histidine kinases are also conserved and are found in several fungal species (5). However, the mutant of the Sln1 orthologue in A. nidulans, TcsB, has no significant alterations to its phenotype (15), yielding no obvious clue as to the function of the Sln1 homologue in filamentous fungi.
Osmotic stress in filamentous fungi also appears to be processed by the Hog1-related MAPK cascade. The os-2 and osc1 MAPK gene mutants, which are orthologues of S. cerevisiae HOG1 in N. crassa and C. lagenarium, respectively, were found to be sensitive to high osmolarity (23, 47). The Osc1p MAPK was phosphorylated and accumulated in the nucleus under high osmotic conditions, indicating the activation of the Osc1p by high osmotic stress (23). The mutants also showed increased resistance to phenylpyrrole and dicarboximide fungicides similar to that of the group III histidine kinase mutant (23, 47). Furthermore, these fungicides led to the improper activation of Hog1-related MAPKs in C. lagenarium, C. heterostrophus, and B. cinerea (23), which indicates that an osmotic stimulus is mediated by the group III histidine kinase and Hog1-related MAPK pathways in filamentous fungi and that the fungicides interfere with those osmoregulatory systems. However, the histidine kinases have not been proven to regulate the activation of the Hog1-related MAPK pathway, and the mechanism of the fungicide-induced, inappropriate activation of the Hog1-related MAPKs is unknown.
In this study, we determined a specific signature pattern of Hog1-related MAPK phosphorylation in C. heterostrophus wild-type and dic1-deficient strains after the cells were exposed to either osmotic stress or dicarboximide and phenylpyrrole fungicides. In addition to C. heterostrophus, N. crassa Os-2p MAPK activation in wild-type and os-1 strains was also assessed. The results revealed that the group III histidine kinases Dic1p and Os-1p were positive regulators of Hog1-related MAPK in each fungal species. This indicates that osmotic signal transmission in C. heterostrophus and N. crassa is regulated differently from that of S. cerevisiae because Sln1p, which is the sole histidine kinase in S. cerevisiae, negatively regulates the activation of the HOG pathway. Furthermore, we also demonstrated that these fungi possessed another upstream activator of Hog1-related MAPK.
MATERIALS AND METHODS
Strains and growth conditions.
The C. heterostrophus strain HITO7711 was used as the wild-type strain. The dicarboximide- and fludioxonil-resistant strains E4504, E4509, N9006, and N9010 were obtained in our previous work (45). Strains E4504, N9006, and N9010 had dic1 null mutations resulting from premature termination due to the presence of the stop codon at repeat 5 of the HAMP domain (E4504), or frameshifts within repeat 1 of the HAMP domain (E4504) or after the H-box of the kinase domain (N9010) (46).
The mutant DHOG101, which has a disrupted Hog1-related MAPK gene, BmHOG1 (GenBank accession no. AB208438), was generated as described following. The BmHOG1 was cloned using the degenerate PCR and the inverse PCR methods. The BmHOG1 of the wild-type strain was disrupted using the targeted insertion method by homologous recombination. The DNA fragment (from the 548th to the 1762nd nucleotide in the open reading frame), which not contained part of 5′ and 3′ coding region of BmHOG1, was amplified by PCR. The amplified fragment was introduced into the plasmid, pCB1004, containing the hph gene. Fungal transformation was performed using a method described previously (46). Genomic DNA from the resulting transformants was extracted and Southern analysis was performed to confirm occurring homologous recombination event (data not shown). One of the resulting disruptants, DHOG101, was used as the BmHOG1-disrupted mutant.
All C. heterostrophus strains were maintained on complete medium agar (CMA) at 25°C. CMA contains (per liter): 1.5 g Ca(NO3)2 · 4H2O, 0.5 g MgSO4 · 7H2O, 0.5 g KCl, 0.4 g KH2PO4, 30 mg K2HPO4, 10 g glucose, 1.0 g tryptone, 1.0 g yeast extract, and 15 g agar. All N. crassa strains were obtained from the Fungal Genetics Stock Center (University of Kansas Medical Center, Kansas City, KS). The wild-type strain 74-OR8-1a and the os mutants NM233t (os-1), Y256M209 (os-1), UCLA80 (os-2), and UCLA93 (os-2) were grown at 25°C on agar-solidified Vogel's medium N plus 1.5% (wt/vol) sucrose (43). To prepare the vegetative mycelia for analysis of the phosphorylation of Hog1-related MAPKs, C. heterostrophus and N. crassa were incubated in liquid complete medium (CM; CMA minus agar) and Vogel's liquid medium N plus sucrose, respectively. The fungicides used in this study were iprodione (Rovral WP, 50% active ingredient; Aventis Crop Sci.), and fludioxonil (Saphire flowable, 20% active ingredient; Novartis Agro) and were obtained commercially. The fungicides were added to the media from 1000-fold concentrated stock solutions in 70% ethanol (iprodione) or dimethyl sulfoxide (fludioxonil).
Southern blot and RT-PCR analysis.
The total DNA of C. heterostrophus was isolated using the method of Nakada et al. (29). DNA digestion and gel electrophoresis were performed according to standard methods (37). The gene fragments amplified by PCR using the primer sets ProbeA (5′-TTTGAGCAGGTGATTGAGAGTTTG-3′ as forward and 5′-TGCATGAATGTGCTCGTACCAAT-3′ as reverse) and ProbeB (5′-ACCACAGTTTAGCCTCCAATTCA-3′ as forward and 5′-ATACCGTTCACATTCTCAGTGAG-3′ as reverse) were used as DNA probes. DNA probes were labeled using the AlkPhos Direct labeling kit (Amersham Biosciences) according to the manufacturer's instructions. Hybridization and detection were carried out according to the manufacturer's instructions with the AlkPhos Direct detection system (Amersham Biosciences).
RNA isolation and first-strand cDNA synthesis were performed according to the previously described method (46). Three primer sets, PS-A (5′-AAGGAAATGGACCCTGAAATCACTCTCT-3′ as forward and 5′-GTTTACAGTGAGFTCATTCCACATG-3′ as reverse), PS-B (5′-AACCATATTCAACGCTCTCAAGTC-3′ as forward and 5′-CGAGATGCCATTTTCCAATGCAGAT-3′ as reverse), and GPD (5′-TACGTCGTCGAGTCTACCGGTGTCT-3′ as forward and 5′-GTCAGTGGAAACAATGTCATCCTCAGT-3′ as reverse), were used to amplify the fragments of cDNA from wild-type and dic1-mutant strains of C. heterostrophus.
Western blot analysis.
The phosphorylation of Hog1-related MAPKs in C. heterostrophus and N. crassa was examined using a Western blot analysis with an anti-dually phosphorylated p38 antibody (Cell Signaling Technology, Beverly, MA). Total protein samples were isolated as described below. Mycelia of each strain were incubated for 2 days in CM or liquid Vogel's medium at 25°C and were filtered through 4-layer gauze. The mycelia were then ground with a mortar and pestle to a fine powder under liquid nitrogen. Ice-chilled buffer containing protease and phosphatase inhibitors [50 mM Tris-HCl, pH 7.5, 1% sodium deoxycholate, 1% Triton X-100, 0.1% sodium dodecyl sulfate, 50 mM NaF, 5 mM sodium pyrophosphate, 0.1 mM sodium vanadate, protease inhibitor cocktail set (Roche, 1 tablet per 50 ml)] was added to the powder, and the suspensions were sonicated for 20 s to promote solubilization. After centrifuging the suspensions at 8,000 × g for 3 min, the resulting supernatants were separated on 10% polyacrylamide gels and blotted onto nitrocellulose membranes. The concentration of total proteins was calculated using a BCA protein assay reagent (Pierce), and 50 μg of proteins was loaded into each well. Anti-Hog1p antibody was purchased from Santa Cruz Biotechnology (C-terminal anti-Hog1), and antibody binding was visualized using an ECL Plus Western blotting detection regent (Amersham Biosciences) after the binding of a horseradish peroxidase-conjugated secondary antibody.
RESULTS
Histidine kinase gene dic1 was not expressed in the E4509 strain of C. heterostrophus.
To better understand the relationships between the group III histidine kinase Dic1p and Hog1-related MAPK, it was necessary to assess MAPK activation in the cells that did not produce the Dic1 protein. Therefore, we first attempted to find a mutant with a completely defective dic1 gene. We had previously isolated several dic1 mutant strains by chemical mutageneses (45). Among these, the E4509 strain was likely to have a deletion in the dic1 locus, since no fragment was amplified by PCR using the primer sets Dic1F and Dic1R designed for the DIC1 coding region (46). But it was not clear whether this was a null mutant. To determine if the Dic1 gene in the E4509 strain was null, Southern blot and RT-PCR analyses were performed.
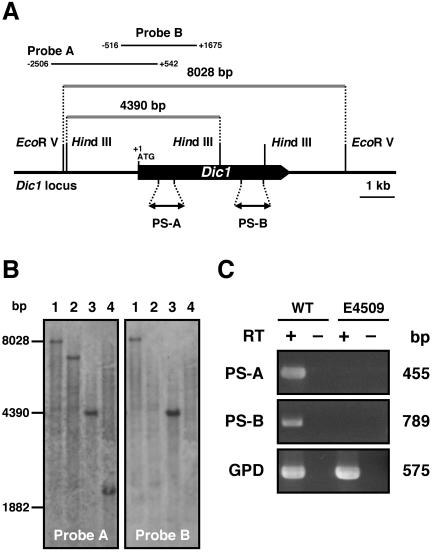
For the Southern blot analyses, DNA fragments containing parts of 5′-franking region and Dic1 open reading frame (probe A: 2,506 bp in the upstream sequence and 542 bp in the dic1 ORF, probe B: 516 bp in the upstream sequence and 1,675 bp in the DIC1 ORF) were constructed by PCR (Fig. 1A). Genomic DNA was isolated from the wild-type HITO7711 and the dic1-deficient strain E4509. The isolated genomic DNA was digested with EcoRV and HindIII, and hybridized with probe A. The results showed that HITO7711 contained the 8.0-kb EcoRV and 4.4-kb HindIII fragments (Fig. 1B left, lanes 1 and 3), but these fragments were not found in E4509. In contrast, the E4509 strain contained other truncated fragments (Fig. 1B left, lanes 2 and 4). In addition, no band was detected in the same membrane, using probe B in E4509 (Fig. 1B right, lanes 2 and 4). These gel blot analyses indicate that at least 2,191 bp fragment lying across the 5′ flanking region and Dic1 open reading frame is completely deleted in the E4509 strain.
FIG. 1.
dic1 deficiency in strain E4509 of C. heterostrophus. (A) Schematic representation of the chromosomal DIC1 locus in the wild-type strain HITO7711. The restriction enzyme sites, the point at which the probes hybridize, and the site amplified by RT-PCR are indicated. (B) Southern analysis of the dic1 locus in the wild-type and dic1 mutant strains. Chromosomal DNA of the wild-type strain HITO7711 (lanes 1 and 3) and of the dic1 mutant strain E4509 (lanes 2 and 4) was digested with EcoRV (lanes 1 and 2) or HindIII (lanes 3 and 4). An approximately 3-kb DNA fragment was first used as the probe (probe A), and, after reprobing, an approximately 2-kb DNA fragment (probe B) was hybridized. (C) Dic1 gene expression in the wild-type and dic1-deficient strains. RNA isolated from the wild-type strain HITO7711 (WT) and the dic1-deficient strain E4509 was used for cDNA synthesis. The cDNA synthesized (+) or not synthesized (−) with reverse transcriptase was used as the template for RT-PCR. The RT-PCR products amplified by two specific primer sets (PS-A and PS-B) and the primers for the C. heterostrophus GPD gene, a positive control, were separated by electrophoresis.
For RT-PCR analyses, two primer sets, PS-A and PS-B, were designed for the 5′ and 3′ ends of the DIC1 coding region, respectively. As shown in Fig. 1C, no band was detected in the E4509 strain, but the HITO7711 strain contained fragments that were consistent with the expected length. These data indicate that the histidine kinase gene dic1 was not expressed in the E4509 strain.
Phosphorylation of Hog1-type MAP kinase in wild-type and dic1-deficient C. heterostrophus.
To investigate the phosphorylation of Hog1-related MAPK in C. heterostrophus, we first cloned the Hog1-related MAP kinase gene (BmHOG1: GenBank accession no. AB208438) and disrupted the gene by the homologous recombination method (see Materials and Methods). The phosphorylation of the BmHog1p MAPK was analyzed by Western blotting using an antibody that specifically recognized the phosphorylated forms of p38-type kinase. A duplicate blot was incubated with an antibody raised against the C terminus of Hog1.
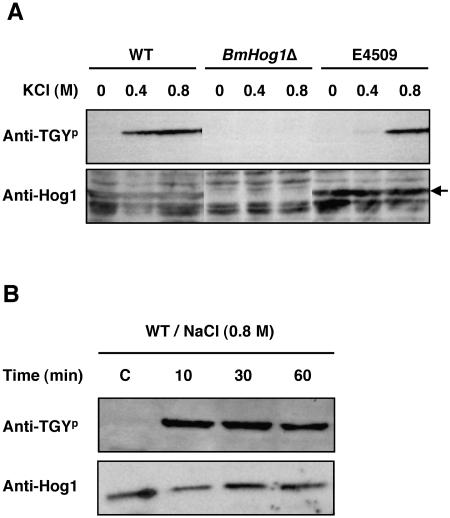
The Hog1 antibody detected several bands in the mycelia of C. heterostrophus. A 43-kDa band was missing in the mycelia of the BmHOG1-deleted mutant DHOG101, whereas this band was detected in the mycelia of the wild-type HITO7711, suggesting that this band represented the BmHog1p MAPK (Fig. 2A). The anti-phospho-p38 antibody clearly detected a band in HITO7711 mycelia after exposure to 0.4 or 0.8 M KCl for 10 min but not in the absence of osmotic stress. The size of the detected band was identical to the size of the band detected by the Hog1 antibody, indicating that the product detected by the anti-phospho-p38 antibody represented the phosphorylated BmHog1p MAPK. NaCl and sorbitol treatment also caused phosphorylation of BmHog1p to the same extent as did KCl treatment (data not shown), which suggests that BmHog1p MAPK was activated under high osmotic stress. The amount of phosphorylated BmHog1p was similar at 10, 30, and 60 min after the exposure to high osmotic stress was initiated (Fig. 2B). The phosphorylation of Hog1-related MAPK was analyzed after a 10-min exposure to the stress compounds.
FIG. 2.
Phosphorylation of C. heterostrophus BmHog1p MAPK induced by osmotic stress. (A) BmHog1p phosphorylation in the wild-type strain, the BmHOG1 disruption strain, and the E4509 strain. Mycelia of the strains HITO7711 (WT), DHOG101 (BmHOG1Δ), and E4509 were used for analysis. Prepared mycelia of the strain tested were incubated in CM with or without 0.4 and 0.8 M KCl for 10 min. The cells were harvested, and total protein extracts were prepared as described in Materials and Methods. Protein samples (50 μg) were subjected to SDS-PAGE and blotted onto nitrocellulose membranes for Western blot analysis. Phosphorylated BmHog1p was detected using anti-dually phosphorylated p38 antibody (indicated by anti-TGYp). The total amount of BmHog1p was measured using anti-Hog1 C terminus antibody (indicated by anti-Hog1). (B) Continuous activation of BmHog1p by NaCl in the wild-type strain. Mycelia of HITO7711 (WT) were incubated in CM with 0.8 M NaCl for 10, 30, and 60 min. Prepared mycelia were incubated in CM without NaCl as a control (indicated by C). The procedures for Western blot analysis were the same as those described above.
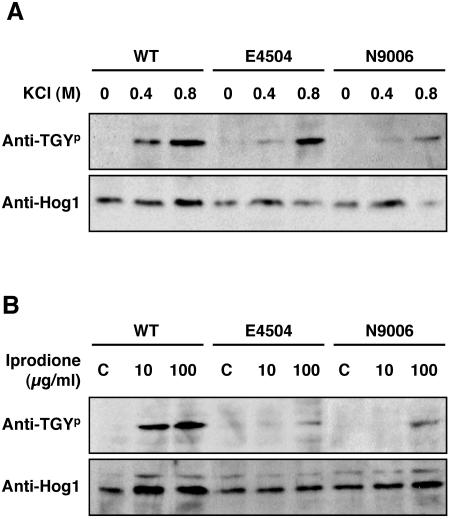
To investigate whether the group III histidine kinase Dic1p regulates the phosphorylation of BmHog1p MAPK, the phosphorylation of BmHog1p in the E4509 strain was also analyzed (Fig. 2A). As in the wild-type strain, phosphorylated BmHog1p was not detected in E4509 mycelia that were not exposed to osmotic stress. In contrast to the findings in the wild-type strain, some phosphorylated BmHog1p was detected in the mycelia of E4509 exposed to 0.4 M KCl. At the same condition, a trace of phosphorylated BmHog1p was detectable in other dic1 null strains E4504, N9006, and N9010 (Fig. 4A and data not shown). These data indicate that the histidine kinase Dic1p positively regulates the activation of BmHog1p MAPK. Of note, BmHog1p activation in the dic1-deficient strains was seen at a high level of osmotic stress (0.8 M KCl) (Fig. 2A and 4A), suggesting that BmHog1p activation was also controlled by another upstream activator.
FIG. 4.
Phosphorylation of C. heterostrophus BmHog1p MAPK in the wild-type strain and the dic1 mutants E4504 and N9006. (A) BmHog1p phosphorylation by osmotic stress. Mycelia of the strains HITO7711 (WT), E4504 and N9006 were used for analysis. Prepared mycelia of the strain tested were incubated in CM with or without 0.4 and 0.8 M KCl for 10 min. The cells were harvested, and total protein extracts were prepared as described in Materials and Methods. Protein samples (50 μg) were subjected to SDS-PAGE and blotted onto nitrocellulose membranes for Western blot analysis. Phosphorylated BmHog1p was detected using anti-dually phosphorylated p38 antibody (indicated by anti-TGYp). The total amount of BmHog1p was measured using anti-Hog1 C terminus antibody (indicated by anti-Hog1). (B) BmHog1p phosphorylation by iprodione in the wild-type strain and the dic1-deficient strains. Mycelia of HITO7711 (WT), E4504, and N9006 were used for analysis. Prepared mycelia were incubated in CM with 10 or 100 μg/ml iprodione for 10 min. Prepared mycelia of the strain tested were incubated in CM without iprodione as a control (indicated by C). The procedures for Western blot analysis were the same as those described above.
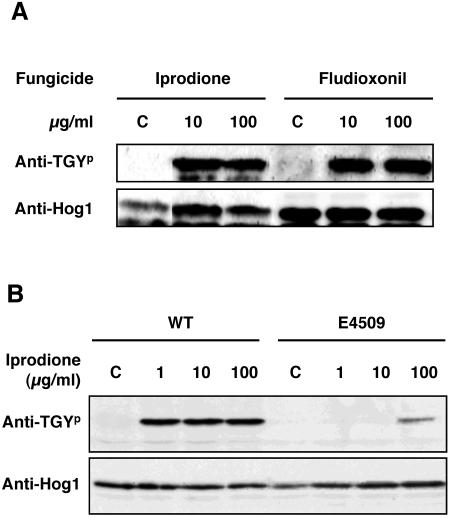
We previously reported that phosphorylation of a Hog1-related MAPK was induced by the phenylpyrrole fungicide fludioxonil in certain phytopathogenic fungi, such as C. heterostrophus (23). C. heterostrophus mutants that were resistant to dicarboximide showed a positive cross-resistance to phenylpyrrole fungicide (45). Thus, we examined whether phosphorylation of BmHog1p was stimulated by dicarboximide fungicides. As shown in Fig. 3A, the dicarboximide fungicide iprodione also led to the activation of BmHog1p MAPK in the wild-type strain, indicating that iprodione and fludioxonil had similar effects on the activation of Hog1-related MAPK.
FIG. 3.
Phosphorylation of the C. heterostrophus BmHog1p MAPK by iprodione and fludioxonil. (A) BmHog1p phosphorylation by iprodione and fludioxonil in the wild-type strain. Mycelia of HITO7711 (WT) were incubated in CM with 10 or 100 μg/ml iprodione or fludioxonil for 10 min. Prepared mycelia were incubated in CM with ethanol and dimethyl sulfoxide as a control (indicated by C). The cells were harvested, and total protein extracts were prepared as described in Materials and Methods. Protein samples (50 μg) were subjected to SDS-PAGE and blotted onto nitrocellulose membranes for Western blot analysis. Phosphorylated BmHog1p was detected using anti-dually phosphorylated p38 antibody (indicated by anti-TGYp). The total amount of BmHog1p was measured using anti-Hog1 C terminus antibody (indicated by anti-Hog1). (B) BmHog1p phosphorylation by iprodione in the wild-type strain and the dic1-deficient strain. Mycelia of HITO7711 (WT) and E4509 were used for analysis. Prepared mycelia were incubated in CM with 1, 10, or 100 μg/ml iprodione for 10 min. Prepared mycelia of the strain tested were incubated in CM without iprodione as a control (indicated by C). The procedures for Western blot analysis were the same as those described above.
Next, we compared the patterns of BmHog1p phosphorylation by iprodione in wild-type and dic1-defective strains (Fig. 3B and 4B). In the mycelia of the wild-type strain, BmHog1p was phosphorylated after exposure to iprodione at a concentration as low as 1 μg/ml. In the mycelia of dic1-deficient strains, phosphorylated BmHog1p was not observed following exposure to less 10 μg/ml of iprodione. This indicates that BmHog1p MAPK is activated through Dic1p by iprodione at a concentration of 1 μg/ml. In contrast, BmHog1p phosphorylation was detected in the mycelia of dic1-deficient strains that were treated with 100 μg/ml iprodione, demonstrating that high concentrations of iprodione can cause activation of BmHog1p in cells without Dic1p histidine kinase.
Phosphorylation of Os-2p MAP kinase in the wild-type and os-1 mutant of N. crassa.
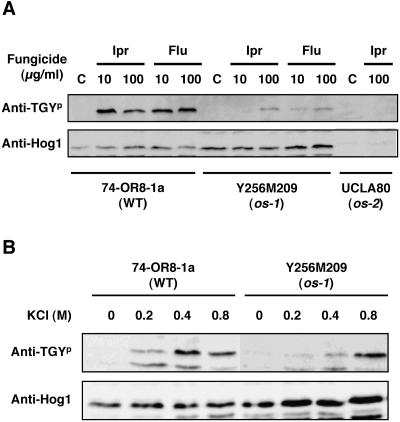
The N. crassa osmotic-sensitive mutants os-1, os-2, os-4, and os-5 are resistant to dicarboximide and phenylpyrrole fungicides (13, 14, 18). Schumacher et al. (36) reported that the os-1 gene encodes the putative histidine kinase, which was identical to the NIK1 gene product reported by Alex et al. (1). It was reported that the os-2 gene encodes a MAPK that is orthologous to S. cerevisiae Hog1p (47). To examine whether Os-2p MAPK activation was induced by the fungicides and was controlled by the histidine kinase Os-1p, we determined the levels of active, phosphorylated Os-2p in the wild-type and the os-1 mutant strains after exposure to the fungicides and osmotic stress (Fig. 5). A protein with a molecular weight similar to that of BmHog1p was clearly detected in the mycelia of the wild-type strain 74-OR8-1a after exposure to iprodione and fludioxonil, but was not present in the unexposed mycelia or in the UCLA80 strain (Fig. 5A). The UCLA80 strain was null with nonsense point mutations in the os-2 gene (47). The same results were obtained using another os-2 null mutant strain, UCLA93 (data not shown). These results suggest that the protein detected by the anti-phospho-p38 antibody represents the phosphorylated Os-2p MAPK in N. crassa.
FIG. 5.
Phosphorylation of Os-2p MAPK of N. crassa. (A) Treatment with fungicides. Mycelia of the wild-type strain 74-OR8-1a (WT) and the os-1 mutant Y256M209 (os-1) were used for analysis. Prepared mycelia of the strain tested were incubated in liquid Vogel's medium with 10 or 100 μg/ml iprodione (Ipr) for 10 min or with 10 or 100 μg/ml fludioxonil (Flu) for 10 min. Prepared mycelia were incubated in CM without iprodione and fludioxonil as a control. Mycelia of the os-2 mutant UCLA80 (os-2) were also exposed to liquid Vogel's medium with or without 100 μg/ml iprodione. The cells were harvested, and total protein extracts were prepared as described in Materials and Methods. Protein samples (50 μg) were subjected to SDS-PAGE and blotted onto nitrocellulose membranes for Western blot analysis. Phosphorylated Os-2p was detected using anti-dually phosphorylated p38 antibody (indicated by anti-TGYp). The total amount of BmHog1p was measured using anti-Hog1 C terminus antibody (indicated by anti-Hog1). Os-2p was detected using anti-dually phosphorylated p38 antibody (indicated by anti-TGYp). The total amount of BmHog1p was measured using anti-Hog1 C terminus antibody (indicated by anti-Hog1). (B) Under osmotic stress. Mycelia of the wild-type strain 74-OR8-1a (WT) and the os-1 mutant Y256M209 (os-1) were used for analysis. Prepared mycelia of the strain tested were incubated in liquid Vogel's medium with 0.2, 0.4, or 0.8 M KCl for 10 min. Prepared mycelia were incubated in liquid Vogel's medium without KCl as a control. The procedures for Western blot analysis were the same as those described above.
In the os-1 strain Y256M209, which has a null mutation at the os-1 locus (30), the phosphorylated Os-2p was not or barely detectable following exposure to 10 μg/ml iprodione and fludioxonil. Phosphorylated Os-2p was detectable in the os-1 strain after exposure to 100 μg/ml of the fungicides, but the amount of phosphorylated Os-2p was significantly reduced in the mutant strain compared with that in the wild-type strain. Similarly, phosphorylated Os-2p was clearly detected in the mycelia of the wild-type strain after exposure to 0.4 M KCl (Fig. 5B) and was also detected in the mycelia of the os-1 strain under the KCl treatment, but the amount of phosphorylated protein detected in the mutant strain was significantly reduced compared with that in the wild type. Similar results were obtained using the os-1 null mutant strain NM233t (data not shown). These results suggest that the activation of Os-2p is positively regulated by the histidine kinase Os-1p/Nik1p.
DISCUSSION
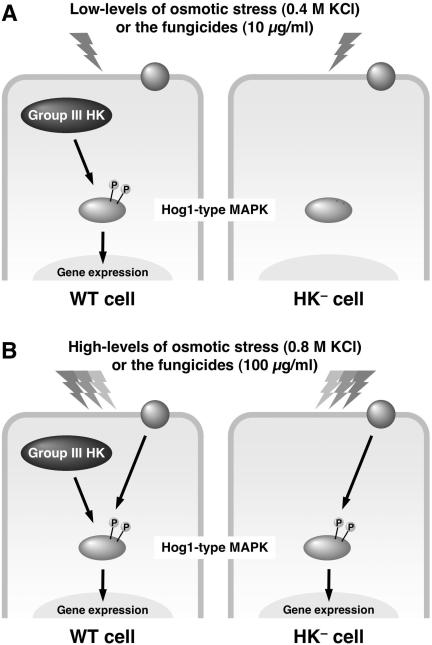
We analyzed the phosphorylation pattern of the Hog1-type MAPK BmHog1p in C. heterostrophus wild-type and dic1-deficient strains after exposure to osmotic stress or dicarboximide and phenylpyrrole fungicides. The results revealed that the activation of BmHog1p-MAPK was positively regulated by the protein encoded by DIC1 (Fig. 6A). Similar results were obtained from the analyses of phosphorylated Os-2p MAPK in the wild-type strain and in the os-1 mutant of N. crassa.
FIG. 6.
Models of Hog1-related MAPK phosphorylation. (A) Phosphorylation patterns of Hog1-related MAPK in a wild-type cell and a cell without group III histidine kinase (HK) at low levels of osmotic stress or fungicide. (B) Phosphorylation patterns of Hog1-related MAPK in a wild-type cell and a cell without group III histidine kinase at high levels of osmotic stress or fungicide.
The DIC1 and os-1 genes each encode a putative two-component histidine kinase that consists of HAMP domain repeats, a phosphoacceptor histidine kinase domain, and a response regulator domain (1, 38, 46). Filamentous ascomycetes, including N. crassa, C. heterostrophus, G. moniliformis, and B. fuckeliana, have been found to possess several histidine kinase genes that were subsequently classified into 11 groups (5, 16). The histidine kinase genes that were homologous to S. cerevisiae SLN1 were categorized into group VI, and both the dic1 and os-1 genes were categorized into group III. Group VI histidine kinases are thought to be putative membrane proteins because they possess two potential transmembrane helices in their N-termini. In contrast, no membrane-spanning domain is predicted in the group III proteins, which suggests that Dic1p and Os-1p regulate osmotic signal transmission in a manner different from that mediated by Sln1p in S. cerevisiae. Indeed, Dic1p and Os-1p are positive regulators of Hog1-related MAPK in C. heterostrophus and N. crassa, whereas S. cerevisiae Sln1p negatively regulates the activation of Hog1 MAPK.
Group III histidine kinase genes were also found in other filamentous fungi such as A. nidulans, Candida albicans, and M. grisea (Table 1). In addition, mutations of the group III histidine kinase genes were responsible for osmotic sensitivity in several fungi (8, 26, 27, 30, 31, 46). These observations suggest that the group III histidine kinase in filamentous fungi commonly functions in osmotic sensing.
TABLE 1.
Known group III histidine kinases and Hog1-related MAP kinases in filamentous fungi
| Fungal species | Gene name | % Amino acid sequence identitya | GenBank accession no. | Reference(s) |
|---|---|---|---|---|
| Group III histidine kinase | ||||
| Neurospora crassa | Nik1/os-1 | U53289 | 1, 37 | |
| Cochliobolus heterostrophus | DIC1 | 63 | AB095748 | 46 |
| Botrytis cinerea | BcOS1/daf1 | 78 | AF435964 | 8, 31 |
| Magnaporthe grisea | HIK1 | 83 | AB041647 | 27 |
| Candida albicans | CaNIK1/COS1 | 51 | AB006363 | 2, 28 |
| Gibberella moniliformis | GmNIK1 | 79 | AY456038 | 5 |
| Aspergillus nidulans | Not named | 67 | Sequencing Projectb | |
| Hogl-related MAP kinase | ||||
| Neurospora crassa | os-2 | AF297032 | 47 | |
| Cochliobolus heterostrophus | BmHog1 | 90 | AB208438 | Unpublished |
| Colletotrichum lagenarium | OSC1 | 95 | AF184980 | 23 |
| Magnaporthe grisea | OSM1 | 95 | AB162820 | 9 |
| Aspergillus nidulans | SAKA | 80 | AF282891 | 21 |
| Saccharomyces cerevisiae | HOG1 | 77 | L06279 | 4 |
| Schizosaccharomyces pombe | SPC1/STY1 | 82 | X89262 | 39 |
Amino acid sequence identities were calculated by DNAsis Maximum Matching program for Os-1p (histidine kinase) and Os-2p (MAP kinase).
There are several reports concerning the functional analysis of Hog1-related MAPK in filamentous fungi. In N. crassa, one of the osmotic-sensitive genes, os-2, encodes a Hog1-related MAPK and could complement the osmotic-sensitive phenotype of an S. cerevisiae hog1 mutant (47). The C. lagenarium Hog1-related MAPK, Osc1p, is involved in responses to hyperosmotic stress and is translocated to the nucleus after exposure to osmotic stress (23). In addition, C. heterostrophus BmHog1p MAPK was activated by hyperosmotic stress. In these filamentous fungi, the amino acid sequences of the Hog1-related MAPKs have been well deduced (Table 1). Therefore, it is feasible that the Hog1-type MAPK of filamentous fungi generally functions to transmit extracellular osmotic signals to the nucleus. As mentioned above, our data showed that Hog1-type MAPK activation was controlled by the group III histidine kinase in both C. heterostrophus and N. crassa (Fig. 5A). Therefore, we believe that filamentous fungi have a common mechanism to sense and respond to extracellular osmotic changes, namely, the group III histidine kinase acting as a positive regulator of Hog1-type MAPK in filamentous fungi.
The results obtained in this study also revealed that C. heterostrophus and N. crassa might possess another upstream activator of Hog1-type MAPK because even the dic1-deficient strains and the os-1 mutants showed MAPK phosphorylation following exposure to high levels of the fungicides and osmotic stress (Fig. 6B). The genomes of C. heterostrophus and N. crassa have been found to contain 21 and 11 histidine kinase genes, respectively (5, 16). Although Dic1p and Os-1p are involved in fungicide resistance and osmotic adaptation in each fungus, the functions of the other histidine kinases remain unknown. In the opportunistic pathogen C. albicans, CaNIK1/COS1 is a homologue of C. heterostrophus DIC1 and is required for osmotic tolerance (2, 40). In addition to CaNIK1/COS1, C. albicans CaSLN1, which is related to S. cerevisiae SLN1, causes slight growth retardation at high osmolarity (28). These findings suggest that C. albicans employs these histidine kinases to adapt to extracellular osmotic changes. Therefore, it is possible that another histidine kinase, such as an Sln1p homologue in C. heterostrophus and N. crassa, can act as an alternative activator of the Hog1-type MAPK for the group III histidine kinases.
In S. cerevisiae, the upstream part of the HOG pathway is thought to have two branches, both of which regulate Pbs2p MAPKK (25). One branch contains the putative membrane protein Sho1p and the MAPKKK Ste11p. The other upstream branch contains a His-Asp phosphorelay signaling complex composed of Sln1p, Ypd1p, and Ssk1p (24). It was reported that these two-branches have different response characteristics to high osmolarity (25). Our results show that only a high concentration of the osmoticum and the fungicides led to the activation of Hog1-related MAPK in the cells without the histidine kinase. This suggests that the response of the histidine kinase to osmolarity is different from that of another activator of Hog1-related MAPK and raised the possibility that upstream mechanisms similar to the Sho1p branch might be used to respond to environmental osmotic changes in filamentous fungi. We are planning to conduct functional analyses of the candidate activators in C. heterostrophus and N. crassa.
Dicarboximide and phenylpyrrole fungicides are used to control a variety of important plant pathogenic fungi (17, 35). Despite intensive study (8, 10, 11, 13, 14, 26, 45, 46, 47), the primary targets of these fungicides have not been identified. In N. crassa, osmotic-sensitive mutants such as os-1, os-2, os-4, and os-5 show increased resistance to dicarboximides and phenylpyrroles, and these osmotic loci encode the group III histidine kinase, Hog1-related MAPK, Ssk22-related MAPKKK, and Pbs2-related MAPKK, respectively. In several phytopathogenic fungi, resistance against dicarboximide and phenylpyrrole fungicides in the field and/or laboratory was caused by mutations of the group III histidine kinase genes, and the gene mutants often showed osmolarity-sensitive phenotypes. Moreover, the Hog1-type MAPK of some filamentous fungi was abnormally activated by exposure to phenylpyrrole fungicide (23). Therefore, it is now thought that these fungicides interact with the protein(s) related to the osmotic signaling pathway.
Our data obtained in this study are consistent with this hypothesis because C. heterostrophus BmHog1p and N. crassa Os-2p MAPK were also improperly activated by the fungicides, and the amount of phosphorylated MAPK was significantly reduced in the fungicide-resistant mutants of C. heterostrophus and N. crassa when the cells were exposed to low-levels of the fungicides (Fig. 6A). However, MAPK phosphorylation was detected even in the resistant mutants upon exposure to high levels of the fungicides (100 μg/ml). This suggests that the fungicides can lead to abnormal activation of the MAPKs even when the histidine kinases do not act as intermediates (Fig. 6B).
Currently, we have no evidence regarding the potential presence of another effector(s), different from the effector for osmotic stress, which leads to the activation of the Hog1-related MAPK in response to fungicide exposure. The hyphal growth rate of the C. heterostrophus dic1 strains used in this study was not reduced by 50% at the fungicide concentration of 100 μg/ml (median effective concentration > 100 μg/ml) (45). Nevertheless, the MAPK was improperly activated by the fungicides at this concentration, which suggests that the antifungal activity of the fungicides is not caused only by the abnormal activation of Hog1-related MAPK. Further studies are needed to elucidate the relationship between the fungicide activity and the signal transduction pathway of osmotic sensing.
Acknowledgments
This work was supported in part by grants-in-aid (11760035 and 15380104) from JSPS and the Center of Excellence for Microbial Process Development Pioneering Future Production Systems (COE program of the Ministry of Education, Culture, Sports, Science and Technology of Japan).
REFERENCES
- 1.Alex, L. A., K. A. Borkovich, and M. I. Simon. 1996. Hyphal development in Neurospora crassa: Involvement of a two-component histidine kinase. Proc. Natl. Acad. Sci. USA 93:3416-3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alex, L. A., C. Korch, C. P. Selitrennikoff, and M. I. Simon. 1998. COS1, a two component histidine kinase that is involved in hyphal development in the opportunistic pathogen Candida albicans. Proc. Natl. Acad. Sci. USA 95:7069-7073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bourret, R. B., K. A. Borkovich, and M. I. Simon. 1991. Signal transduction pathways involving protein phosphorylation in prokaryotes. Annu. Rev. Biochem. 60:401-441. [DOI] [PubMed] [Google Scholar]
- 4.Brewster, J. L., T. de Valoir, N. D. Dwyer, E. Winter, and M. C. Gustin. 1993. An osmosensing signal transduction pathway in yeast. Science 259:1760-1763. [DOI] [PubMed] [Google Scholar]
- 5.Catlett, N. L., O. C. Yoder, and B. G. Turgeon. 2003. Whole-genome analysis of two-component signal transduction genes in fungal pathogens. Eukaryot. Cell 2:1151-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang, C., S. F. Kwok, A. B. Bleecker, and E. M. Meyerowitz. 1993. Arabidopsis ethylene-response gene ETR1: similarity of product to two-component regulator. Science 262:539-544. [DOI] [PubMed] [Google Scholar]
- 7.Cobb, M. H., and E. J. Goldsmith. 1995. How MAP kinases are regulated. J. Biol. Chem. 270:14843-14846. [DOI] [PubMed] [Google Scholar]
- 8.Cui, W., R. E. Beever, S. L. Parkes, P. L. Weeds, and M. D. Templeton. 2002. An osmosensing histidine kinase mediates dicarboximide fungicide resistance in Botryotinia fuckeliana (Botrytis cinerea). Fungal Genet. Biol. 36:187-198. [DOI] [PubMed] [Google Scholar]
- 9.Dixon, K. P., J.-R. Xu, N. Smirnoff, and N. J. Talbot. 1999. Independent signaling pathways regulate cellular turgor during hyperosmotic stress and appressorium-mediated plant infection by Magnaporthe grisea. Plant Cell 11:2045-2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edlich, W., and H. Lyr. 1995. Mechanism of action of dicarboximide fungicides. p. 119-131 In H. Lyr (ed.), Modern selective fungicides-properties, applications, mechanisms of action. Gustav Fischer Verlag, New York, NY.
- 11.Faretra, F., and S. Pollastro. 1991. Genetic basis of resistance to benzimidazole and dicarboximide fungicides in Botryotinia fuckeliana (Botrytis cinerea). Mycol. Res. 95:943-951. [Google Scholar]
- 12.Fassler, J. S., W. M. Gray, C. L. Malone, W. Tao, H. Lin, and R. J. Deschenes. 1997. Activated alleles of yeast SLN1 increase Mcm1-dependent reporter gene expression and diminish signaling trough the Hog1 osmosensing pathway. J. Biol. Chem. 272:13365-13371. [DOI] [PubMed] [Google Scholar]
- 13.Fujimura, M., N. Ochiai, A. Ichiishi, R. Usami, K. Horikoshi, and I. Yamaguchi. 2000. Sensitivity to phenylpyrrole fungicides and abnormal glycerol accumulation in os and cut mutant strains of Neurospora crassa. J. Pestic. Sci. 25:31-36. [Google Scholar]
- 14.Fujimura, M., N. Ochiai, A. Ichiishi, R. Usami, K. Horikoshi, and I. Yamaguchi. 2000. Fungicide resistance and osmotic stress sensitivity in os mutants of Neurospora crassa. Pestic. Biochem. Physiol. 67:125-133. [Google Scholar]
- 15.Furukawa, K., Y. Katsuno, T. Urao, T. Yabe, T. Yamada-Okabe, H. Yamada-Okabe, Y. Yamagata, K. Abe, and T. Nakajima. 2002. Isolation and functional analysis of a gene, tcsB, encoding a transmembrane hybrid-type histidine kinase from Aspergillus nidulans. Appl. Environ. Microbiol. 68:5304-5310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galagan, J. E., S. E. Calvo, K. A. Borkovich, E. U. Selker, N. D. Read, D. Jaffe, W. FitzHugh, L.-J. Ma, S. Smirnov, S. Purcell, B. Rehman, T. Elkins, R. Engels, S. Wang, C. B. Nielsen, J. Butler, M. Endrizzi, D. Qui, P. Ianakiev, D. Bell-Pedersen, M. A. Nelson, M. Werner-Washburne, C. P. Selitrennikoff, J. A. Kinsey, E. L. Braun, A. Zelter, U. Schulte, G. O. Kothe, G. Jedd, W. Mewes, C. Staben, E. Marcotte, D. Greenberg, A. Roy, K. Foley, J. Naylor, N. Stange-Thomann, R. Barrett, S. Gnerre, M. Kamal, M. Kamvysselis, E. Mauceli, C. Bielke, S. Rudd, D. Frishman, S. Krystofova, C. Rasmussen, R. L. Metzenberg, D. D. Perkins, S. Kroken, C. Cogoni, G. Macino, D. Catcheside, W. Li, R. J. Pratt, S. A. Osmani, C. P. C. DeSouza, L. Glass, M. J. Orbach, J. A. Berglund, R. Voelker, O. Yarden, M. Plamann, S. Seiler, J. Dunlap, A. Radford, R. Aramayo, D. O. Natvig, L. A. Alex, G. Mannhaupt, D. J. Ebbole, M. Freitag, I. Paulsen, M. S. Sachs, E. S. Lander, C. Nusbaum, and B. Birren. 2003. The genome sequence of the filamentous fungus Neurospora crassa. Nature 422:859-868. [DOI] [PubMed] [Google Scholar]
- 17.Gehmann, K., R. Nyfeler, A. J. Leadbeater, D. Nevill, and D. Sozzi. 1990. CGA 173506: a new phenylpyrrole fungicide for broad-spectrum disease control. Brighton Crop Prot. Conf. Pests Dis. 2:399-406. [Google Scholar]
- 18.Grindle, M., and W. Temple. 1982. Fungicide-resistant of os mutants of Neurospora crassa. Neurospora Newsl. 29:16-17. [Google Scholar]
- 19.Gustin, M. C., J. Albertyn, M. Alexander, and K. Davenport. 1998. MAP kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 62:1264-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hohmann, S. 2002. Osmotic stress signaling and osmoadaptation in yeast. Microbiol. Mol. Biol. Rev. 66:300-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawasaki, L., O. Sanchez, K. Shiozaki, and J. Aguirre. 2002. SakA MAP kinase is involved in stress signal transduction, sexual development and spore viability in Aspergillus nidulans. Mol. Microbiol. 45:1153-1163. [DOI] [PubMed] [Google Scholar]
- 22.Kojima, K., T. Kikuchi, Y. Takano, E. Oshiro, and T. Okuno. 2002. The mitogen-activated protein kinase gene MAF1 is essential for the early differentiation phase of appressorium formation in Colletotrichum lagenarium. Mol. Plant-Microbe Interact. 15:1268-1276. [DOI] [PubMed] [Google Scholar]
- 23.Kojima, K., Y. Takano, A. Yoshimi, C. Tanaka, T. Kikuchi, and T. Okuno. 2004. Fungicide activity through activation of a fungal signaling pathway. Mol. Microbiol. 53:1785-1796. [DOI] [PubMed] [Google Scholar]
- 24.Maeda, T., S. M. Wurgler-Murphy, and H. Saito. 1994. A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature 369:242-245. [DOI] [PubMed] [Google Scholar]
- 25.Maeda, T., M. Takekawa, and H. Saito. 1995. Activation of yeast PBS2 MAPKK by MAPKKKs or by binding of an SH3-containing osmosensor. Science 269:554-558. [DOI] [PubMed] [Google Scholar]
- 26.Millar, T. K., S. Renault, and C. P. Selitrennikoff. 2002. Molecular dissection of alleles of the osmotic-1 locus of Neurospora crassa. Fungal Genet. Biol. 35:147-155. [DOI] [PubMed] [Google Scholar]
- 27.Motoyama, T., K. Kodama, T. Ohira, A. Ichiishi, M. Fujimura, I. Yamaguchi, and T. Kubo. 2005. A two-component histidine kinase of the rice blast fungus is involved in osmotic stress response and fungicide action. Fungal Genet. Biol. 42:200-212. [DOI] [PubMed] [Google Scholar]
- 28.Nagahashi, S., T. Mio, N. Ono, T. Yamada-Okabe, M. Arisawa, H. Bussey, and H. Yamada-Okabe. 1998. Isolation of CaSLN1 and CaNIK1, the genes for osmosensing histidine kinase homologues, from pathogenic fungus Candida albicans. Microbiology 144:425-432. [DOI] [PubMed] [Google Scholar]
- 29.Nakada, M., C. Tanaka, K. Tsunewaki, and M. Tsuda. 1994. RFLP analysis for species separation in genera Bipolaris and Curvularia. Mycoscience 35:271-278. [Google Scholar]
- 30.Ochiai, N., M. Fujimura, T. Motoyama, A. Ichiishi, R. Usami, K. Horikoshi, and I. Yamaguchi. 2001. Characterization of mutants in the two-component histidine kinase gene that confer fludioxonil resistance and osmotic sensitivity in the os-1 mutants of Neurospora crassa. Pest Manag. Sci. 57:437-442. [DOI] [PubMed] [Google Scholar]
- 31.Oshima, M., M. Fujimura, S. Banno, C. Hashimoto, T. Motoyama, A. Ichiishi, and I. Yamaguchi. 2002. A point mutation in two-component histidine kinase BcOS1 gene confers dicarboximide resistance in field isolates of Botrytis cinerea. Phytopathology 92:75-80. [DOI] [PubMed] [Google Scholar]
- 32.Ota, I. M., and A. Varshavsky. 1993. A yeast protein similar to bacterial two-component regulator. Science 262:566-569. [DOI] [PubMed] [Google Scholar]
- 33.Pandey, A., M. G. Roca, N. D. Read, and N. L. Glass. 2004. Role of a mitogen-activated protein kinase pathway during conidial germination and hyphal fusion in Neurospora crassa. Eukaryot. Cell 3:348-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parkinson, J. S., and E. C. Kofoid. 1992. Communication modules in bacterial signaling proteins. Annu. Rev. Genet. 26:71-112. [DOI] [PubMed] [Google Scholar]
- 35.Pommer, E. H., and G. Lorenz. 1995. Dicarboximide fungicides, p. 119-131. In H. Lyr (ed.), Modern selective fungicides-properties, applications, mechanisms of action. Gustav Fischer Verlag, New York, NY.
- 36.Robinson, M. J., and M. H. Cobb. 1997. Mitogen-activated protein kinase pathways. Curr. Opin. Cell Biol. 9:180-186. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Schumacher, M. M., C. S. Enderlin, and C. P. Selitrennikoff. 1997. The osmotic-1 locus of Neurospora crassa encodes a putative histidine kinase similar to osmosensors of bacteria and yeast. Curr. Microbiol. 34:340-347. [DOI] [PubMed] [Google Scholar]
- 39.Shiozaki, K., M. Shiozaki, and P. Russell. 1997. Mcs4 mitotic catastrophe suppressor regulates the fission yeast cell cycle through the Wik1-Wis1-Spc1 kinase cascade. Mol. Biol. Cell 8:409-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Srikantha, T., L. Tsai, K. Daniels, L. Enger, K. Highley, and D. R. Soll. 1998. The two-component hybrid kinase regulator CaNIK1 of Candida albicans. Microbiology 144:2715-2729. [DOI] [PubMed] [Google Scholar]
- 41.Stock, A. M., V. L. Robinson, and P. N. Gaudreau. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69:183-215. [DOI] [PubMed] [Google Scholar]
- 42.Takano, Y., T. Kikuchi, Y. Kubo, J. E. Hamer, K. Mise, and I. Furusawa. 2000. The Colletotrichum lagenarium MAP kinase gene CMK1 regulates diverse aspects of fungal pathogen. Mol. Plant-Microbe Interact. 13:374-383. [DOI] [PubMed] [Google Scholar]
- 43.Vogel, H. J. 1964. Distribution of lysine pathways among fungi: evolutionary implications. Am. Nat. 98:435. [Google Scholar]
- 44.Xu, J.-R. 2000. MAP kinase in fungal pathogens. Fungal Genet. Biol. 31:137-152. [DOI] [PubMed] [Google Scholar]
- 45.Yoshimi, A., J. Imanishi, A. Gafur, C. Tanaka, and M. Tsuda. 2003. Characterization and genetic analysis of laboratory mutants of Cochliobolus heterostrophus resistant to dicarboximide and phenylpyrrole fungicides. J. Gen. Plant Pathol. 69:101-108. [Google Scholar]
- 46.Yoshimi, A., M. Tsuda, and C. Tanaka. 2004. Cloning and characterization of the histidine kinase gene Dic1 from Cochliobolus heterostrophus that confers dicarboximide resistance and osmotic adaptation. Mol. Genet. Genomics 271:228-236. [DOI] [PubMed] [Google Scholar]
- 47.Zhang, Y., R. Lamm, C. Pillonel, S. Lam, and J.-R. Xu. 2002. Osmoregulation and fungicide resistance: the Neurospora crassa os-2 gene encodes a Hog1 mitogen-activated protein kinase homologue. Appl. Environ. Microbiol. 68:532-538. [DOI] [PMC free article] [PubMed] [Google Scholar]