Abstract
An inverted repeat corresponding to parts of the centrin gene of Chlamydomonas reinhardtii was placed downstream of the NIT1 promoter, which is induced by ammonium starvation. After induction, transformants developed centrin deficiency as assayed by immunofluorescence, Western blotting, and Northern blotting. The effect was reversible, demonstrating that the NIT1 promoter allowed controlled RNA interference in Chlamydomonas reinhardtii.
Sequencing of the genome of the unicellular green alga Chlamydomonas reinhardtii has led to the discovery of numerous functionally uncharacterized genes (JGI website: http://genome.jgi-psf.org/chlre2/chlre2.home.html). Because it allows targeting of specific genes, RNA interference (RNAi) is a useful technique to investigate the role of gene products (1). In C. reinhardtii, a variety of genes have been silenced using vectors that contain constitutive promoters and inverted repeats (IR) corresponding to the gene of interest (4, 7, 9). To achieve temporal and spatial control of gene silencing, vectors based on inducible promoters have been developed for animals, plants, and some protists, e.g., Trypanosoma brucei (2, 6, 11). Here we report a vector for inducible RNAi in Chlamydomonas based on the NIT1 promoter.
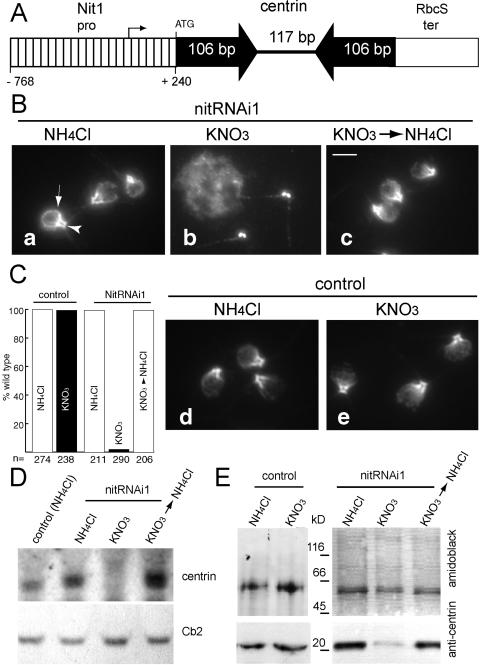
The NIT1 structural gene encodes nitrate reductase (NR; EC1.6.6.2); its expression is repressed by ammonium and induced by ammonium starvation (3, 8). An IR consisting of parts of the vfl2 gene encoding the cytoskeletal protein centrin (7) was fused between the NIT1 promoter and the RbcS terminator (Fig. 1A) and transformed into C. reinhardtii CC3395 (arg7-8 cwd mt−). Strains nitRNAi1 and nitRNAi2 were chosen for further analysis. When maintained in Tris-acetate-phosphate (TAP) medium containing NH4Cl (5), most nitRNAi1 cells (98.2%, n = 211, Fig. 1Ba) had a wild-type centrin system consisting of the distal connecting fiber (dCF) and the nucleus-basal body connectors (NBBCs) (12) as revealed by indirect immunofluorescence using monoclonal anticentrin.
FIG. 1.
Induction and repression of centrin RNAi. A. Schematic presentation of the NIT1-centrin-IR construct. The putative start of transcription is based on data available at http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?val=AF203033.1. B-E. Control and nitRNAi1 cells were maintained in either NH4Cl medium (NH4Cl) or NH4Cl-free medium (KNO3) or returned to NH4Cl medium after being maintained in ammonium-free medium for several weeks (KNO3 → NH4Cl). B. Indirect immunofluorescence for centrin. Arrowhead, dCF; arrow, NBBC. Bar, 5 μm. C. The % of cells with wild-type centrin fibers for control and nitRNAi1 cells. D. Northern blot probed with genomic [32P]ATP-labeled DNA encoding centrin or, as a loading control, a biotin-labeled probe to Cb2 (13); 25 μg of total RNA was loaded per lane. E. Western blot of detergent-extracted control and nitRNAi1 cells. The lower part of the membrane was stained with anticentrin, and the upper part was stained with amido black. Similar amounts of protein were loaded per lane.
Cells were collected by centrifugation (800 × g, 5 min), washed twice, and transferred to ammonium-free TAP medium with KNO3 as a nitrogen source. After several days of ammonium starvation, centrin fibers were largely absent (97.9% of cells, n = 290) and residual centrin accumulated at the basal bodies (Fig. 1Bb, C). Transcripts encoding centrin were greatly reduced (Fig. 1D) and, in Western blots, only traces of centrin were detected (Fig. 1E).
Severe centrin deficiency interferes with basal body assembly and control of basal body number (7, 12). nitRNAi1 cells with abnormal numbers of basal bodies were rare (<1%) during the first week of induction of the RNAi transgene (0.5% on day 7), but increased to 51.5% of cells during the following 2 weeks (not shown). When cells were returned to NH4Cl medium for an extended period of time (see below), they lost the centrin-deficient phenotype (Fig. 1Bc, C) and returned to wild-type levels of centrin transcripts and protein (Fig. 1D and E). A control strain transformed with the selectable marker alone displayed a wild-type centrin cytoskeleton and wild-type amounts of centrin under both conditions (Fig. 1Bd, e, C, and E).
As previously reported for transformants expressing the centrin IR from the strong constitutive HSP70A/RbcS2 fusion promoter, nitRNAi1 lost the centrin-deficient phenotype after 15 to 20 weeks of constant induction (not shown) and it was not possible to knock down centrin expression again even after maintaining the cells in NH4Cl medium for 90 days (not shown). PCR and Southern blotting revealed that the NIT1-centrin-IR transgene was still present (not shown). Stock cultures of nitRNAi1 maintained permanently in medium supplemented with NH4Cl were still inducible at this time point, suggesting that inactivation of the transgene initially required expression of the construct.
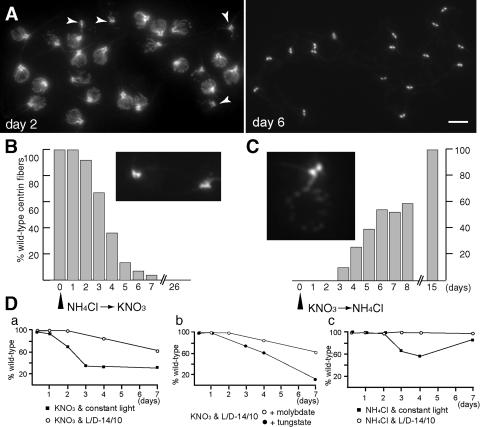
After transfer to NH4Cl-free medium, it took several days for the cells to respond, e.g., 91.6% of cells retained wild-type centrin fibers 2 days after induction, but more than 90% of the cells had lost centrin fibers after 6 days (Fig. 2A and B). Similarly, the mutant phenotype was lost gradually after repression of the transgene (Fig. 2C). The NIT1 promoter was previously used in reporter constructs and the gene products were detected 2 hours after induction and plateaued within 24 hours (8, 10, 14). To explain the slow development of the centrin deficient phenotype, we assume that some centrin is carried over from the mother into the daughter cells each division and that several days were required to turn the degradation of mRNAs on and off.
FIG. 2.
Culture conditions for inducible centrin RNAi. A. Anticentrin staining of methanol-fixed nitRNAi1 cells 2 and 6 days after transfer to NH4Cl-free medium. Arrowheads (day 2), cells with short NBBCs. Bar, 5 μm. B. The % of nitRNAi1 cells displaying wild-type centrin fibers on different days after transfer to ammonium-free medium. Inset: Cells from day 4 lacking NBBCs but retaining dCFs. C. Time course showing the % of nitRNAi2 cells displaying a wild-type phenotype after return to NH4Cl medium. Inset: Cells examined at intermediate times (day 4) frequently displayed single NBBCs and a punctate staining on the cell nucleus but lacked dCFs. D. nitRNAi1 cells initially maintained in ammonium-containing medium with a light-dark (L/D) cycle of 14 hours/10 hours were transferred to the medium and conditions indicated. The % of cells with a wild-type phenotype was determined by anticentrin indirect immunofluorescence at different time points. a, KNO3 as a nitrogen source and constant light at 25°C (▪) or a light-dark cycle of 14 h/10 h at 22°C (○). b, KNO3 as a nitrogen source with a light-dark cycle of 14 h/10 h in medium supplemented with 1 μm Na2MoO4 (○) or Na2WO4 (•). c, NH4Cl as a nitrogen source and constant light (▪) or a light-dark cycle of 14 h/10 h (○).
Loppes et al. (8) showed that the NIT1 promoter is activated by light and repressed by active NR, which is inhibited by tungstate. Indeed, the centrin-deficient phenotype developed faster in constant bright light than with a light/dark cycle of 14 h/10 h and when tungstate was substituted for molybdate in NH4Cl-free TAP. However, in the presence of NH4Cl, constant bright light induced defective centrin fibers (up to 43.9%, n = 214), whereas wild-type fibers were present in a light-dark cycle of 14 h/10 h (98%, n = 232). Tungstate had no effect on cells maintained in NH4Cl medium and did not enhance the phenotype when added to cells fully induced by ammonium starvation and constant light. In general, we used constant light and NH4Cl-free medium for induction and a light-dark cycle of 14 h/10 h in NH4Cl medium for repression.
Analysis of several transformants showed that the phenotypes were weaker than those observed when the same centrin IR was expressed from the strong constitutive HSP70A/RbcS2 fusion promoter (fully induced nitRNAi2 had 1.1 basal bodies/cell, compared to 0.53 basal bodies/cell for the fusion promoter) (7). Using a NIT1::ARS reporter construct, the transgene was induced up to 30-fold higher when expressed in an NR− background compared to NR+ strains (8). Here, an NR+ strain was used; we expect that better expression of the NIT1-centrin-IR transgene could be obtained in an NR− background.
In summary, the NIT1-centrin-IR construct allowed inducible and reversible generation of a centrin-deficient phenotype simply by transferring cells from NH4Cl-containing to NH4Cl-free medium. Inducible RNAi of centrin enabled us to analyze the phenotype during the course of development. The NIT1 promoter is controlled tightly enough to maintain the wild-type phenotype under repressive conditions. This feature might be especially useful for the analysis of essential genes and others involved in cell growth and division which, when constitutively repressed, would affect cell viability. Controlled RNAi using the NIT1 promoter can facilitate the functional characterization of genes in C. reinhardtii.
Acknowledgments
We thank George Witman (Department of Cell Biology, University of Massachusetts Medical School) for critical reading of the manuscript and helpful suggestions. We are grateful to Gottfried Weissenböck (Botanisches Institut, Universität zu Köln, Cologne, Germany) for providing laboratory space during the last month of this study.
This study was supported by the Deutsche Forschungsgemeinschaft (DFG, Le806/3-4 and 4-2).
REFERENCES
- 1.Baulcombe, D. 2004. RNA silencing in plants. Nature 431:356-363. [DOI] [PubMed] [Google Scholar]
- 2.Chen, S., D. Hofius, U. Sonnewald, and F. Bornke. 2003. Temporal and spatial control of gene silencing in transgenic plants by inducible expression of double-stranded RNA. Plant J. 36:731-740. [DOI] [PubMed] [Google Scholar]
- 3.Fernández, E., R. Schnell, L. P. Ranum, S. C. Hussey, C. D. Silflow, and P. A. Lefebvre. 1989. Isolation and characterization of the nitrate reductase structural gene of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 86:6449-6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fuhrmann, M., A. Stahlberg, E. Govorunova, S. Rank, and P. Hegemann. 2001. The abundant retinal protein of the Chlamydomonas eye is not the photoreceptor for phototaxis and photophobic responses. J. Cell Sci. 114:3857-3863. [DOI] [PubMed] [Google Scholar]
- 5.Gorman, D. S., and R. P. Levine. 1965. Cytochrome f and plastocyanin: their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardi. Proc. Natl. Acad. Sci. USA 54:1665-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gupta, S., R. A. Schoer, J. E. Egan, G. J. Hannon, and V. Mittal. 2004. Inducible, reversible, and stable RNA interference in mammalian cells. Proc. Natl. Acad. Sci. USA 101:1927-1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koblenz, B., J. Schoppmeier, A. Grunow, and K. F. Lechtreck. 2003. Centrin deficiency in Chlamydomonas causes defects in basal body replication, segregation and maturation. J. Cell Sci. 116:2635-2646. [DOI] [PubMed] [Google Scholar]
- 8.Loppes, R., M. Radoux, M. C. Ohresser, and R. F. Matagne. 1999. Transcriptional regulation of the Nit1 gene encoding nitrate reductase in Chlamydomonas reinhardtii: effects of various environmental factors on the expression of a reporter gene under the control of the Nit1 promoter. Plant Mol. Biol. 41:701-711. [DOI] [PubMed] [Google Scholar]
- 9.Rohr, J., N. Sarkar, S. Balenger, B. R. Jeong, and H. Cerutti. 2004. Tandem inverted repeat system for selection of effective transgenic RNAi strains in Chlamydomonas. Plant J. 40:611-621. [DOI] [PubMed] [Google Scholar]
- 10.Schoppmeier, J., W. Mages, and K. F. Lechtreck. 2005. GFP as a tool for the analysis of proteins in the flagellar basal apparatus of Chlamydomonas. Cell Motil. Cytoskeleton 61:189-200. [DOI] [PubMed] [Google Scholar]
- 11.Shi, H., A. Djikeng, T. Mark, E. Wirtz, C. Tschudi, and E. Ullu. 2000. Genetic interference in Trypanosoma brucei by heritable and inducible double-stranded RNA. RNA 6:1069-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taillon, B. E., S. A. Adler, J. P. Suhan, and J. W. Jarvik. 1992. Mutational analysis of centrin: an EF-hand protein associated with three distinct contractile fibers in the basal body apparatus of Chlamydomonas. J. Cell Biol. 119:1613-1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Von Kampen, J., U. Nielander, and M. Wettern. 1994. Stress-dependent transcription of a gene encoding a g-beta-like polypeptide from Chlamydomonas reinhardtii. J. Plant Physiol. 143:756-758. [Google Scholar]
- 14.Zhang, D., and P. A. Lefebvre. 1997. A negative regulatory locus required for the repression of the nitrate reductase gene in Chlamydomonas reinhardtii. Genetics 146:121-133. [DOI] [PMC free article] [PubMed] [Google Scholar]