Abstract
The stress-activated protein kinase (SAPK) pathway plays a central role in coordinating gene expression in response to diverse environmental stress stimuli. We examined the role of this pathway in the translational response to stress in Schizosaccharomyces pombe. Exposing wild-type cells to osmotic stress (KCl) resulted in a rapid but transient reduction in protein synthesis. Protein synthesis was further reduced in mutants disrupting the SAPK pathway, including the mitogen-activated protein kinase Wis1 or the mitogen-activated protein kinase Spc1/Sty1, suggesting a role for these stress response factors in this translational control. Further polysome analyses revealed a role for Spc1 in supporting translation initiation during osmotic stress, and additionally in facilitating translational adaptation. Exposure to oxidative stress (H2O2) resulted in a striking reduction in translation initiation in wild-type cells, which was further reduced in spc1− cells. Reduced translation initiation correlated with phosphorylation of the α subunit of eukaryotic initiation factor 2 (eIF2α) in wild-type cells. Disruption of Wis1 or Spc1 kinase or the downstream bZip transcription factors Atf1 and Pap1 resulted in a marked increase in eIF2α phosphorylation which was dependent on the eIF2α kinases Hri2 and Gcn2. These findings suggest a role for the SAPK pathway in supporting translation initiation and facilitating adaptation to environmental stress in part through reducing eIF2α phosphorylation in fission yeast.
Cells prevent and repair damage in response to environmental stress conditions by regulating gene expression at the levels of both transcription and translation. Mitogen-activated protein kinase signaling pathways are highly conserved through evolution and play an important role in mediating the transduction of the signals from the cell surface to the nucleus, resulting in altered gene expression and modulation of protein activity (22). In mammalian cells, the stress-activated protein kinases (SAPKs) p38/ RK/CSBP kinase and c-Jun N-terminal kinase (JNK) play a key role in response to a variety of environmental stresses, including osmotic and oxidative stresses, heat shock, UV irradiation, protein synthesis inhibitors, and DNA-damaging agents, and upon exposure to proinflammatory cytokines and vasoactive neuropeptides. Upon activation, these stress-related protein kinases can phosphorylate and activate the transcription factors c-Jun, ATF2, and Elk-1, which coordinate gene expression responses to environmental stimuli (34).
A highly conserved SAPK pathway is present in the fission yeast Schizosaccharomyces pombe that is comprised of a central kinase cascade, including the mitogen-activated protein kinase kinases Wak1 (also known as Wis4 or Wik1) and Win1, the mitogen-activated protein kinase kinase Wis1, and the mitogen-activated protein kinase Spc1 (also known as Sty1 or Phh1). The Spc1 mitogen-activated protein kinase in S. pombe is structurally and functionally related to its mammalian counterparts and is activated by a wide range of cellular insults (35). Activation of Spc1 stimulates a transcriptional response to stress through the bZip transcription factors Atf1 (also known as Gad7 or Mts1), Pcr1, and Pap1. Atf1 is constitutively nuclear and binds to and is phosphorylated by the Spc1 mitogen-activated protein kinase (9, 30, 32, 43, 45). Atf1 is required for mating and for survival following exposure to osmotic stress, and acute high levels of oxidative stress (9, 28, 45). Pcr1 forms a heterodimer with Atf1 and functions similarly to Atf1 (43). Pap1 accumulates in the nucleus in a stress- and Spc1-dependent manner and is required for survival under low levels of oxidative stress (25, 28, 36). Although the precise mechanisms by which Spc1 activates Atf1 or Pap1 dependent transcription are unclear, a number of target stress response genes have now been identified which are dependent on Spc1, Atf1, or Pap1 (2,27, 30, 35, 44, 45).
Protein synthesis is also regulated in response to environmental stress, resulting in the translational down-regulation to protect cells from the generation of misfolded or toxic proteins, and in the expression of specific genes to mitigate cell injury (5, 11, 19). In mammalian cells, stress-induced activation of the p38 mitogen-activated protein kinase phosphorylates and activates the protein kinases Mnk1 and Mnk2 (8, 41). The translation initiation factor 4E (eIF4E) is a substrate of Mnk1 (38, 41, 42) and phosphorylation enhances the binding of eIF4E to the 5′ cap structure, thus enhancing cap-dependent translation initiation (31).
Both mammals and yeasts regulate general translation initiation in response to a variety of environmental stresses through reversible phosphorylation of the α subunit of eukaryotic initiation factor 2 (eIF2α) (5). Phosphorylation of eIF2α on serine 51 inhibits the guanine nucleotide exchange factor eIF2B and leads to a reduction of the levels of the active eIF2-GTP complex required for delivery of aminoacylated initiator tRNA to the translational machinery, thus inhibiting general protein synthesis (5). Four distinct eIF2α kinases have been identified in mammals: double-stranded RNA-dependent protein kinase (PKR), pancreatic eIF2α kinase (PEK), GCN2, and heme-regulated inhibitor kinase (HRI), which are activated and phosphorylate eIF2α under different stress conditions (5). In yeast, the number of eIF2α kinases is fewer, with S. pombe containing only GCN2 and two HRI-related protein kinases (48). eIF2α kinases are thought to reduce global protein synthesis, allowing cells to conserve resources as they alter their gene expression program to block or alleviate stress damage.
Previous reports have suggested a possible link between the SAPK pathway and translation in fission yeast. Inhibition of the SAPK pathway, and overexpression of translation-related genes sum1+ and sum3+/ded1+, were found to suppress S-M checkpoint mutants and to inhibit the osmotic stress cell cycle response in fission yeast (7, 16). Sum1 was found to be an essential component of the eIF3 translation initiation complex (6). In addition, sum3+/ded1+ encodes a general translation factor analogous to Ded1 in Saccharomyces cerevisiae (10). Since reduced protein synthesis through overexpression of these genes and inhibition of the SAPK exhibited similar phenotypes under stress, these observations strongly suggested a role for the SAPK pathway in modulating translation in response to stress. We therefore sought to examine the role of the SAPK pathway in the translational response to stress in fission yeast. These studies provide evidence for a general translational response to osmotic and oxidative stresses and further define a role for the SAPK pathway in supporting translation initiation and translational adaptation in response to these environmental stresses in fission yeast.
MATERIALS AND METHODS
Yeast strains and stress sensitivity assays.
The following S. pombe strains were used in this study: TH9 (wild-type h−), TH123 (spc1-m13 ura4-D18 leu1-32 h−),TH393 (atf1::ura4+ leu1-32 ura4-D18 his7.366 h+), TH454 (pap1::ura4+ leu1-32 h−),TH815 (wis1::ura4+ leu1-32 ura4-D18 h+), TH474 (atf1::ura4 pap1::ura4 leu1-32 ura4-D18, his7.366 h−), TH1740 (sty1-1 leu1-32 ura4-D18 his3-D1 ade6-D1 h+), TH2033 (hri1::ura4+ his3-D ade6-m216 leu1-32 ura4-D18 h+), TH2036 (hri2::leu1 ura4-D18 his3-D h+), TH2025 (hri1::ura4+ sty1-1 ura4-D18 ade6-m216 leu1-32 his3D h−), TH2028 (hri2::leu1 sty1-1 ade6-m216 ura4-D18 leu1-32 his3D h+), TH2019 (hri1::ura4+ hri2::leu1 ade6-m216 leu1-32 ura4-D18 h−), TH2026 (hri1::ura4+ hri2::leu1 sty1-1 ade6-m216 leu1-32 his3D ura4-D18 h−), TH2038 (gcn2::ura4+ ura4-D18 ade6-m216 leu1-32 his3-D h−), TH2029 (gcn2::ura4+ sty1-1 ade6-m216 ura4-D18 leu1-32 his3D h−), TH2021 (hri2::leu1 gcn2::ura4+ ade6-m216 leu1-32 ura4-D18 h−), and TH2031 (hri2::leu1 gcn2::ura4+ sty1-1 ade6-m216 ura4-D18 leu1-32 h−).
The growth media and general methods for studying fission yeast have been previously described (23). Cells were grown to early logarithmic phase in YE5S medium at 30°C and treated with 0.6 M KCl (osmotic stress) or 1 mM H2O2 (oxidative stress) for the times indicated. For viability studies, cells were subjected to stress as described above, diluted at times indicated, and plated on YE5S plates. Numbers of colonies were determined after incubation for 3 days at 30°C. For the serial dilution colony-spotting assay, cells were serially diluted from 107 to 103 cells/ml and 5 μl was spotted onto YE5S plates with or without 2 mM H2O2 and incubated for 5 days at 30°C.
Radiolabel incorporation assay.
For [35S]methionine and [35S]cysteine incorporation experiments, 100 μCi of Pro-mix l-[35S] in vitro cell labeling mix (Anachem) was added to 6 × 107 cells and incubated for 20 min at 30°C. Labeled cells were harvested by centrifugation and stored at −80°C. Thawed cells were lysed with acid-washed glass beads (425 to 600 μm, Sigma) in a Bio-Savant Fast Prep 120 machine and protein extracts prepared in 2x sample buffer (20). Samples were analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) (12%). Gels were stained with Coomassie blue, dried, and autoradiography performed using a PhosphorImager (Bio-Rad). Quantification of radiolabeled proteins of three independent experiments was performed using the Quantity One software and results were normalized by quantification of the total amount of proteins loaded and stained by Coomassie blue. A representative gel is presented.
Polysome profiles.
Polysomes were obtained using a protocol modified from Tzamarias et al. (37). To preserve polysomes, cycloheximide was added to stressed or nonstressed cells as indicated at a final concentration of 50 μg/ml just prior to harvesting. Cells were chilled rapidly, washed once in breaking buffer solution (10 mM Tris-HCl pH 7.0, 100 mM NaCl, 30 mM MgCl2, 50 μg/ml cycloheximide and 200 μg/ml heparin) and harvested by centrifugation. 50 ml of cells grown logarithmically to an optical density (595 nm) of 0.5, or the equivalent, were lysed with acid-washed glass beads (425 to 600 μm, Sigma) in 200 μl of breaking buffer. Cell lysates were clarified by centrifugation at 13,000 × g for 2 min. Supernatants were fractionated on 7 to 47% sucrose gradients prepared in a solution containing 50 mM Tris-acetate, pH 7.0, 50 mM NH4Cl and 12 mM MgCl2 for 105 min at 40,000 rpm using a SW40-Ti rotor in a Beckman L70 centrifuge. Polysome profiles were obtained by monitoring the absorbance at 254 nm along the gradient using an LKB 2238 Uvicord SII and a Picolog analog to digital converter and data logging software (Pico Technology, Cambridge). The polysome/monosome ratio was calculated as the ratio of the estimated area of the two to four polysomes to that of the 80 S monosomes. Experiments were performed three times and representative polysome profiles for each strain and time point are shown.
SDS-PAGE and immunoblotting.
Following exposure to osmotic or oxidative stress, cells were lysed with acid-washed glass beads (425 to 600 μm, Sigma) and vortexing, clarified by centrifugation, and 20 μg of each protein sample was separated by electrophoresis on SDS-PAGE (12.5%). Proteins were transferred electrophoretically to nitrocellulose membrane and were subsequently immunoblotted with affinity-purified antibody that specifically recognizes eIF2α phosphorylated at serine-51 (Biosource) or antiserum that recognizes total eIF2α. Detection was performed using peroxidase-conjugated anti-rabbit immunoglobulin G and visualized by chemiluminescence.
RESULTS
The Spc1 mitogen-activated protein kinase supports protein synthesis under conditions of environmental stress.
To address whether protein synthesis in fission yeast is modulated in response to stress, wild-type and spc1-m13 cells, in which the Spc1 mitogen-activated protein kinase is disrupted, were exposed to a pulse of [35S]methionine-cysteine under conditions of osmotic or oxidative stress. Levels of de novo protein synthesis were determined by SDS-PAGE analysis of radiolabeled proteins. Following 20 min exposure of wild-type cells to osmotic stress (0.6 M KCl), translation was reduced to 58% of unstressed levels. Protein synthesis then increased to 86% after 40 min and returned to unstressed wild-type levels after 60 min of KCl exposure (Fig. 1A and 1B). These data indicate that protein synthesis was rapidly but transiently reduced in wild-type cells following exposure to osmotic stress. They also demonstrate that wild-type cells have the ability to restore normal global protein synthesis after being subjected to osmotic stress.
FIG. 1.
Effects of osmotic and oxidative stress on protein synthesis in wild-type (wt) and spc1-m13 cells. (A) [35S]methionine incorporation in wild-type (TH9) and spc1-m13 (TH123) cells under osmotic stress. Cells were treated with 0.6 M KCl for times indicated and were labeled for 20 min prior to harvesting. Radiolabeled proteins were visualized by SDS-PAGE, followed by autoradiography (top panel), and quantitated using the Quantity One software. Coomassie stain of above gel showed the total protein levels loaded in each lane (bottom panel). (B) Graph representing the quantification of three independent experiments as described above. (C) [35S] methionine incorporation in wild-type (TH9) and spc1-m13 (TH123) cells under oxidative stress. Cells were treated with 1 mM H2O2 for times indicated and were labeled for 20 min prior to harvesting. Radiolabeled proteins were visualized by SDS-PAGE, followed by autoradiography (top panel), and quantitated using the Quantity One software. Coomassie stain of above gel showed the total protein levels loaded in each lane (bottom panel). (D) Graph representing the quantification of three independent experiments as described above. (E) Cell viability of wild-type (black squares) and spc1-m13 (gray triangles) cells following exposure to 1 mM H2O2 for the times indicated. Values represent an average of three independent experiments.
Interestingly, translation of spc1-m13 cells was further reduced to 25% of unstressed levels after 20 min of osmotic stress. Translation recovered only partially in spc1-m13 cells to 39% of unstressed levels after 40 min and 60 min of the KCl treatment (Fig. 1A and 1B). No loss of viability was observed in wild-type or spc1-m13 cells under these conditions (our unpublished data). These findings show that the level of general protein synthesis was rapidly and significantly reduced in spc1-m13 mutants compared to wild-type cells following exposure to osmotic stress. These results indicate a role for the Spc1 kinase in maintaining protein synthesis immediately following exposure to osmotic stress and a further role in facilitating translational adaptation to these conditions at later time points.
The effects of oxidative stress on protein synthesis were also examined. Following exposure of wild-type cells to oxidative stress (1 mM H2O2), the level of radiolabel incorporation was reduced to 54% of unstressed levels after 20 min, and was decreased further to 21% after 40 and 60 min (Fig. 1C and 1D). No loss of viability was observed following exposure to 1 mM H2O2 for 60 min (Fig. 1E). These data indicate that the rate of protein synthesis was rapidly reduced in wild-type cells under oxidative stress conditions. Following exposure of spc1-m13 cells to 1 mM H2O2, translation levels were significantly reduced compared to wild-type cells. In spc1-m13 cells, there was a 4.5-fold decrease in protein synthesis following 20 min of oxidative stress, and a ∼12-fold reduction after 60 min of H2O2 treatment compared to unstressed conditions (Fig. 1C and 1D). These results show that protein synthesis was rapidly and significantly reduced in spc1-m13 cells compared to wild-type cells following exposure to oxidative stress. Furthermore, in contrast to wild-type cells, continued exposure of spc1-m13 cells to 1 mM H2O2 resulted in loss of viability, with about 20% viability after H2O2 exposure for 90 min (Fig. 1E). As protein synthesis in spc1-m13 cells was reduced disproportionately compared to loss of viability at the earlier time points (compare Fig. 1D and 1E), these data indicate a role for the Spc1 mitogen-activated protein kinase in maintaining general levels of protein synthesis under conditions of oxidative stress.
The SAP kinase cascade is involved in the translational stress response.
Wis1, a mitogen-activated protein kinase kinase, is required for the activation of the Spc1 mitogen-activated protein kinase under osmotic, oxidative and thermal stresses (3,29). We therefore measured translation in wis1− cells following osmotic and oxidative stress. Under nonstressed conditions, there were no significant differences in protein synthesis between wild-type, spc1-m13 and wis1− cells (Fig. 2A, lanes 1, 4, and 7). Following a 20 min exposure of wis1− cells to either osmotic stress (Fig. 2A, lane 5) or oxidative stress (Fig. 2A, lane 6), a significant reduction in de novo protein synthesis was observed compared to wild-type cells (Fig. 2A, lanes 2 and 3; Fig. 2B). No loss of viability in wis1− cells was observed at 20 min after exposure to KCl (our unpublished data) or to H2O2 (Fig. 2C), indicating that the reduced translation level is not the result of loss of viability. However, with longer periods of oxidative stress, wis1− cells displayed lower viability that was not detected in wild-type cells (Fig. 2C). Levels of radiolabel incorporation into proteins and viability of wis1− cells correlated closely with those observed in spc1-m13 cells (Fig. 2A, lanes 8and 9). These data strongly suggest that the Wis1-Spc1 kinase cascade is important for maintaining general levels of protein synthesis under conditions of stress.
FIG. 2.
Effects of osmotic and oxidative stresses on protein synthesis and viability in wis1− cells. (A) [35S] methionine incorporation levels in wis1− (TH815) cells. Cells were subjected to 0.6 M KCl or 1 mM H2O2 and labeled for 20 min prior to harvesting. [35S]methionine incorporation levels were determined by SDS-PAGE analysis (top panel). Coomassie stain of above gel showing total protein levels is presented (bottom panel). (B) Graph representing the quantification of above gel. Standard deviations do not exceed 10% of the mean value. (C) Cell viability of wild-type (black squares), and wis1− (black circles) cells following exposure to 1 mM H2O2 for the times indicated. Values represent an average of three independent experiments.
The SAPK pathway supports general translation initiation during exposure to osmotic or oxidative stress.
To address which step in protein synthesis is impeded in response to stress in fission yeast, polysome profiles were examined using lysates prepared from wild-type and spc1-m13 cells grown under conditions of stress. Following exposure of wild-type cells to 0.6 M KCl, there was a minor reduction in the ratio of polysomes to monosomes after 10 min of exposure, and polysome profiles appeared to also resemble those of unstressed cells at later time points (Fig. 3A, left panels and graph). These data indicate that translation initiation was mildly reduced in wild-type cells under these conditions. In contrast to wild-type cells, a significant reduction in the polysome-to-monosome ratio was observed in spc1-m13 cells after just 10 min incubation in 0.6 M KCl (Fig. 3A, right panels and graph). Decreased polysomes continued to be observed following 20 and 40 min of osmotic stress compared to those of wild-type cells. These findings indicate that translation initiation was rapidly reduced in spc1-m13 cells compared to wild-type cells following osmotic stress, and identify a role for the Spc1 mitogen-activated protein kinase in maintaining efficient translation initiation under conditions of osmotic stress.
FIG. 3.
Polysome profile analysis of wild-type (wild-type) and spc1-m13 cells under stress conditions. Exponentially growing cultures of wild-type (TH9) and spc1-m13 (TH123) cells in YE5S were incubated in 0.6 M KCl (A) or 1 mM H2O2 (B) for the time indicated. Samples were collected and polysome profile analysis performed following velocity sedimentation of whole cell extracts on sucrose gradients (7 to 47%). Fractions were scanned at 254 nm and absorbance profiles are shown (from 0 to 1.0) with sucrose concentrations increasing from left to right, as shown (gray gradients). The positions of 80S ribosomes and polysomes are indicated. The graphs represent the quantification of the polysome-to-monosome ratio of wild-type and spc1-m13 cells after exposure to 0.6 M KCl (left graph) and to 1 mM H2O2 (right graph). Standard deviations do not exceed 10% of the mean value.
Following exposure of wild-type cells to oxidative stress, a striking reduction in the polysomes was observed within 20 min (Fig. 3B, left panel, 20 min and graph). Large monosome peaks were maintained throughout the 90 min time course (Fig. 3B, left panels and graph). These results indicated a significant reduction in translation initiation in wild-type cells following oxidative stress, and correlated well with the reduced protein synthesis levels observed under these conditions (Fig. 1C and 1D). Thus, in contrast to osmotic stress, there is a striking down-regulation of translation initiation in response to oxidative stress in wild-type cells. Following exposure of spc1-m13 cells to oxidative stress, a sharp increase in the monosomes, and reduction in polysomes was observed after just 10 min incubation in 1 mM H2O2 (Fig. 3B, right panel, 10 min, and graph), and this lowered translation initiation continued following up to 90 min of stress. This decrease in polysomes was greater in magnitude and occurred more rapidly in the spc1-m13 cells compared to the wild-type strain. These results correlated well with reduced protein synthesis levels observed in spc1-m13 under these conditions (Fig. 1C and 1D), showing a central role for the Spc1 mitogen-activated protein kinase in maintaining general translation initiation under oxidative stress.
SAPK pathway modulates eIF2α phosphorylation levels in response to oxidative stress.
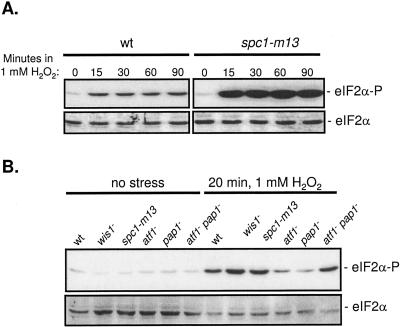
Phosphorylation of eIF2α in S. pombe has recently been observed in response to environmental stresses (48, 49). We therefore examined eIF2α phosphorylation levels in response to osmotic and oxidative stress in both wild-type cells and mutants in which the SAPK pathway was disrupted. No significant induction of eIF2α phosphorylation occurred in wild-type or spc1-m13 cells following exposure to osmotic stress (our unpublished results). In contrast, exposure of wild-type cells to 1 mM H2O2 resulted in enhanced eIF2α phosphorylation levels within 15 min, which was sustained for at least 90 min (Fig. 4A, left panel). These results are consistent with significantly reduced translation initiation observed under these conditions.
FIG. 4.
Elevated eIF2α phosphorylation occurs in spc1-m13 cells during oxidative stress. (A) Wild-type (TH9) or spc1-m13 (TH123) cells were incubated at 30°C in the presence of 1 mM H2O2 for the times indicated. Total protein extracts were analyzed by immunoblotting using an antibody that specifically recognizes phosphorylated eIF2α(eIF2α-P) or antiserum that recognizes total eIF2 (eIF2α). (B) Wild-type (TH9), wis1− (TH 815), spc1-m13 (TH123), atf1− (TH393), pap1− (TH 454), and atf1− pap1− (TH474) cells were incubated for 20 min at 30°C in the absence (left panel) or presence of 1 mM H2O2 (right panel). Western blotting was performed as described above.
Under unstressed conditions, eIF2α phosphorylation levels in spc1-m13, wis1−, atf1−, and pap1− cells were similar to those of unstressed wild-type cells (Fig. 4A and B). These findings are consistent with the findings that basal translation is unaffected following disruption of the SAPK pathway under unstressed conditions (Fig. 1 and 3). However, following exposure to oxidative stress, analysis of spc1-m13 cells revealed a striking further increase in the eIF2α phosphorylation levels in comparison to those of wild-type cells at all time points examined (Fig. 4A, right panel). These results correlated with the further reduction in translation initiation observed in spc1-m13 cells under these conditions. Increased eIF2α phosphorylation levels were also observed in wis1− cells (Fig. 4B, right panel). These results identify a link between kinases Wis1 and Spc1 and eIF2α phosphorylation in response to oxidative stress.
The bZIP transcription factors Atf1 and Pap1 are regulated by the mitogen-activated protein kinase pathways in response to environmental stress. We wished to address whether loss of Atf1 and Pap1 also perturbed translational control in response to oxidative stress. Individual atf1− or pap1− mutations led to a reduction in eIF2α phosphorylation during oxidative stress compared to wild-type cells. Interestingly, when both Atf1 and Pap1 were disrupted, there was a return to high levels of eIF2α phosphorylation in response to oxidative stress (Fig. 4B, right panel). These results identify a role for the downstream target bZip transcription factors Atf1 and Pap1 in modulating eIF2α phosphorylation levels under oxidative stress conditions.
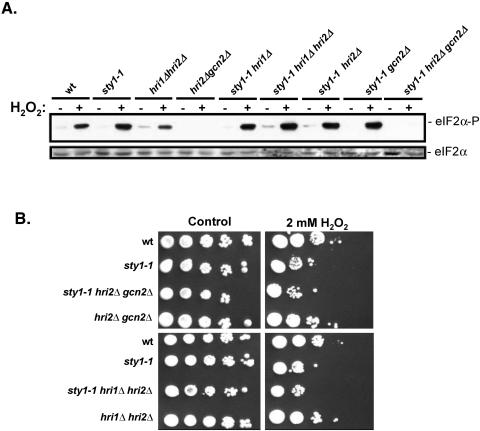
The eIF2 kinases Hri2 and Gcn2 have recently been shown to phosphorylate eIF2α under conditions of oxidative stress in fission yeast (48). Further experiments were therefore performed to determine whether these kinases were also required for the increased eIF2α phosphorylation observed following disruption of the SAPK pathway under oxidative stress conditions. Analysis of strains in which there were disruptions in Spc1 and different combinations of each of the S. pombe eIF2α kinases Hri1, Hri2, and Gcn2 revealed that high levels of eIF2α phosphorylation were still observed in sty1-1 (functionally equivalent to spc1-m13), sty1-1 hri1Δ, sty1-1 hri2Δ, sty1-1 gcn2Δ, and sty1-1 hri1Δ hri2Δ compared to that of wild-type cells following exposure to oxidative stress (Fig. 5A). In contrast, eIF2α phosphorylation was abrogated in hri2Δ gcn2Δ and sty1-1 hri2Δ gcn2Δ cells in either the absence or presence of oxidative stress. These results indicate that Hri2 and Gcn2 are both required for eIF2α phosphorylation in both wild-type and Spc1-disrupted cells under conditions of oxidative stress.
FIG. 5.
Effects of eIF2 kinase deletion on eIF2α phosphorylation and viability in wild-type and sty1-1 cells during oxidative stress. (A) eIF2α phosphorylation in wild-type (wt) eIF2 kinase deleted and sty1-1 cells during oxidative stress. Wild-type (TH9), sty1-1 (TH 1740), hri1Δ hri2Δ (TH2019), hri2Δ gcn2Δ (TH2021), sty1-1 hri1Δ (TH2025), sty1-1 hri1Δ hri2Δ (TH2026), sty1-1 hri2Δ (TH2028), sty1-1 gcn2Δ (TH2029), and sty1-1 hri2Δ gcn2Δ (TH2031) were incubated at 30°C in the absence or presence of 1 mM H2O2. Total protein extracts were analyzed by immunoblotting using an antibody that specifically recognizes phosphorylated eIF2α (eIF2α-P) or antiserum that recognizes total eIF2 (eIF2α). (B) Sensitivity of eIF2 kinases and Spc1/Sty1 mutants to oxidative stress. Tenfold serial dilutions of wild-type (TH9), sty1-1 (TH1740), sty1-1 hri2Δ gcn2Δ (TH2031), hri2Δ gcn2Δ (TH2021), hri1Δ hri2Δ (TH2019), and sty1-1 hri1Δ hri2Δ (TH2026) were plated on YE5S medium (control) and YE5S medium containing 2 mM H2O2 (as indicated) and incubated for 5 days at 30°C.
We further wished to test whether increased eIF2α phosphorylation contributed to loss of viability observed in Spc1-disrupted cells when grown under oxidative stress conditions. sty1-1 cells exhibited a significant loss of viability when spotted onto plates containing 2 mM H2O2 compared to both wild-type and hri2Δ gcn2Δ cells (Fig. 5B). However, sty1-1 hri2Δ gcn2Δ cells, in which eIF2α phosphorylation is not observed (Fig. 5A), exhibited no difference in viability compared to sty1-1 alone under these conditions (Fig. 5B). Furthermore, no difference in sty1-1 viability was observed when combined with disruption of other eIF2α kinase combinations tested under these conditions (Fig. 5B, our unpublished results).
DISCUSSION
The results presented here provide important insights into the translational responses of fission yeast to osmotic and oxidative stresses. Following exposure of wild-type fission yeast to osmotic stress, protein synthesis was reduced rapidly, but translation initiation appeared only mildly reduced and no significant changes in eIF2α phosphorylation were observed (Fig. 1, 3, and 4). These findings raise the possibility that another translation step, possibly translation elongation, may be transiently lowered following osmotic stress in fission yeast. Following exposure to oxidative stress, translation initiation was rapidly reduced, and correlated with elevated levels of eIF2α phosphorylation (Fig. 1 and 4). These results strongly suggest that translation initiation is repressed following exposure to oxidative stress through eIF2α phosphorylation in fission yeast. This conclusion is strongly supported by the findings presented here, and those of Zhan et al. (49), that the eIF2α kinases Hri2 and Gcn2 are required to phosphorylate eIF2α in response to H2O2 in fission yeast (48).
In mammalian cells, heme-regulated inhibitor (HRI) has also been found to phosphorylate eIF2α in response to oxidative stress (21), indicating that this stress response is evolutionarily conserved. Reduced translation would allow cells additional time to repair cell damage induced by oxidative stress prior to synthesizing new proteins. Furthermore, lowered protein synthesis would be expected to result in down-regulation of some regulatory proteins with short half-lives as a result of a rapid turnover. Indeed, such regulation has been observed in mammalian cells for IκBα, the inhibitor of NF-κB regulation (4, 17), and for CReP, a constitutive repressor of eIF2α phosphorylation (18). eIF2α phosphorylation can promote changes in gene expression through preferential translation of stress response genes (5).
One example of such a response comes from yeast studies in which GCN4 was found to be translationally induced by stress signals such as amino acid or glucose deprivation, or exposure to methylmethane sulfonate, allowing the transcriptional activation of stress response mRNAs (5, 14, 15, 24, 47). In mammalian cells, the transcription factor ATF4, the cationic amino acid transporter CAT-1, and the transcription factors derived from CCAAT/enhancer binding protein C/EBPβ have also been shown to be selectively translated under stress conditions (1, 5, 12, 40, 46). These responses collectively promote survival and adaptative responses to environmental stresses. Consistent with this, eIF2α phosphorylation has been demonstrated to promote resistance to oxidative stress (13, 33).
Our studies identified an important role for the SAPK pathway in supporting translation initiation immediately following exposure to either osmotic or oxidative stresses (Fig. 3A and 3B; 10 min). In response to oxidative stress, the SAPK pathway was found to maintain eIF2 activity (i.e., reduce eIF2α phosphorylation) through the concerted activities of both Atf1 and Pap1 bZip transcription factors (Fig. 4). Intriguingly, deletion of either Atf1 or Pap1 individually resulted in reduced eIF2α phosphorylation levels compared to the wild type, suggesting that downstream components of the SAPK pathway can contribute both positively and negatively to the modulation of the eIF2α kinase stress pathway.
Further analysis indicated that the increase in eIF2α phosphorylation levels observed following disruption of Spc1 was found to be dependent on both Hri2 and Gcn2 kinases under oxidative stress conditions. These results indicate that there is regulatory coordination between the SAPK and eIF2 kinase stress pathways. Our findings suggest a model in which the SAPK pathway functions to negatively regulate the Hri2 and Gcn2 kinases under conditions of oxidative stress in fission yeast. This regulation could be either through direct interaction between downstream components of the SAPK pathway and both Hri2 and Gcn2, or indirect through reducing levels of reactive oxygen species that may activate Hri2 and Gcn2. Alternatively, it is possible that increased eIF2α phosphorylation levels result from reduced eIF2α phosphatase activity following disruption of the SAPK pathway in fission yeast. Loss of such phosphatase activity would not be detectable in an hri2Δ gcn2Δ background, where eIF2α phosphorylation is abrogated under oxidative stress conditions. There is precedent for such a model, where mammalian cell GADD34 encodes a stress-inducible regulatory subunit of a holophosphatase complex that dephosphorylates eIF2α, and its inactivation prevents eIF2α dephosphorylation and recovery of protein synthesis, normally observed late in the stress response (26). However, analysis of the global transcriptional responses to environmental stress, including both oxidative and osmotic stresses, did not identify any known translation factors, including potential GADD34 orthologs or ribosomal genes, whose expression was up-regulated in response to stress in fission yeast (2). Studies in mammalian cells have previously identified a role for the Spc1 homolog p38 mitogen-activated protein kinase in facilitating translation initiation through enhancing cap-dependent translation initiation (8, 31, 38, 41, 42). Thus, the findings presented here suggest a distinct mechanism by which the SAPK pathway may support translation initiation in eukaryotes.
Recent studies in the evolutionarily divergent budding yeast Saccharomyces cerevisiae suggested a role for the homologous Hog mitogen-activated protein kinase pathway in the adaptation of translation initiation after inhibition by osmotic stress (39). Our studies also identified a role for the SAPK pathway in facilitating translational adaptation following exposure to either osmotic or oxidative stress conditions in fission yeast. Although protein synthesis had largely recovered within an hour in wild-type cells following exposure to 0.6 M KCl, protein synthesis in spc1-m13 and atf1− cells had not (Fig. 1; our unpublished data). Moreover, in contrast to wild-type cells, spc1-m13 cells underwent a rapid collapse in translation initiation following exposure to oxidative stress after which loss of viability was observed (Fig. 1 and 3). Our data indicated that loss of viability could occur in the absence of eIF2α phosphorylation following disruption of the SAPK pathway in an hri2Δ gcn2Δ background (Fig. 5A and 5B). These findings are consistent with an essential role for the SAPK pathway in transcriptional regulation under oxidative stress conditions (25, 28, 36) but may mask an important role for the SAPK pathway in translational adaptation under these conditions. Further analysis of the relationship between the highly conserved SAPK pathway and the translation machinery in fission yeast is therefore likely to provide important insights into the underlying mechanisms of homeostasis in eukaryotic cells.
Acknowledgments
We are grateful to Jonathan Millar and to Paul Russell for supplying the strains used in this study.
I.D.-S., C.W., A.P., and T.H. were supported by the Medical Research Council. R.W. and J.N. were supported by NIH grant RO1 GM49164.
REFERENCES
- 1.Calkhoven, C. F., C. Muller, and A. Leutz. 2000. Translational control of C/EBPalpha and C/EBPbeta isoform expression. Genes Dev. 14:1920-1932. [PMC free article] [PubMed] [Google Scholar]
- 2.Chen, D., W. M. Toone, J. Mata, R. Lyne, G. Burns, K. Kivinen, A. Brazma, N. Jones, and J. Bahler. 2003. Global transcriptional responses of fission yeast to environmental stress. Mol. Biol. Cell 14:214-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Degols, G., K. Shiozaki, and P. Russell. 1996. Activation and regulation of the Spc1 stress-activated protein kinase in Schizosaccharomyces pombe. Mol. Cell. Biol. 16:2870-2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deng, J., P. D. Lu, Y. Zhang, D. Scheuner, R. J. Kaufman, N. Sonenberg, H. P. Harding, and D. Ron. 2004. Translational repression mediates activation of nuclear factor κB by phosphorylated translation initiation factor 2. Mol. Cell. Biol. 24:10161-10168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dever, T. E. 2002. Gene-specific regulation by general translation factors. Cell 108:545-556. [DOI] [PubMed] [Google Scholar]
- 6.Dunand-Sauthier, I., C. Walker, C. Wilkinson, C. Gordon, R. Crane, C. Norbury, and T. Humphrey. 2002. Sum1, a component of the fission yeast eIF3 translation initiation complex, is rapidly relocalized during environmental stress and interacts with components of the 26S proteasome. Mol. Biol. Cell 13:1626-1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Forbes, K. C., T. Humphrey, and T. Enoch. 1998. Suppressors of cdc25p overexpression identify two pathways that influence the G2/M checkpoint in fission yeast. Genetics 150:1361-1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fukunaga, R., and T. Hunter. 1997. MNK1, a new mitogen-activated protein kinase-activated protein kinase, isolated by a novel expression screening method for identifying protein kinase substrates. EMBO J. 16:1921-1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaits, F., G. Degols, K. Shiozaki, and P. Russell. 1998. Phosphorylation and association with the transcription factor Atf1 regulate localization of Spc1/Sty1 stress-activated kinase in fission yeast. Genes Dev. 12:1464-1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grallert, B., S. E. Kearsey, M. Lenhard, C. R. Carlson, P. Nurse, E. Boye, and K. Labib. 2000. A fission yeast general translation factor reveals links between protein synthesis and cell cycle controls. J. Cell Sci. 113:1447-1458. [DOI] [PubMed] [Google Scholar]
- 11.Harding, H. P., M. Calfon, F. Urano, I. Novoa, and D. Ron. 2002. Transcriptional and translational control in the Mammalian unfolded protein response. Annu. Rev. Cell Dev. Biol. 18:575-599. [DOI] [PubMed] [Google Scholar]
- 12.Harding, H. P., I. Novoa, Y. Zhang, H. Zeng, R. Wek, M. Schapira, and D. Ron. 2000. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell 6:1099-1108. [DOI] [PubMed] [Google Scholar]
- 13.Harding, H. P., Y. Zhang, H. Zeng, I. Novoa, P. D. Lu, M. Calfon, N. Sadri, C. Yun, B. Popko, R. Paules, D. F. Stojdl, J. C. Bell, T. Hettmann, J. M. Leiden, and D. Ron. 2003. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell 11:619-633. [DOI] [PubMed] [Google Scholar]
- 14.Hinnebusch, A. G. 1994. Translational control of GCN4: an in vivo barometer of initiation-factor activity. Trends Biochem. Sci. 19:409-414. [DOI] [PubMed] [Google Scholar]
- 15.Hinnebusch, A. G., and K. Natarajan. 2002. Gcn4p, a master regulator of gene expression, is controlled at multiple levels by diverse signals of starvation and stress. Eukaryot. Cell 1:22-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Humphrey, T., and T. Enoch. 1998. Sum1, a highly conserved WD-repeat protein, suppresses S-M checkpoint mutants and inhibits the osmotic stress cell cycle response in fission yeast. Genetics 148:1731-1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang, H. Y., and R. C. Wek. 2005. GCN2 phosphorylation of eIF2alpha activates NF-kappaB in response to UV irradiation. Biochem. J. 385:371-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jousse, C., S. Oyadomari, I. Novoa, P. Lu, Y. Zhang, H. P. Harding, and D. Ron. 2003. Inhibition of a constitutive translation initiation factor 2alpha phosphatase, CReP, promotes survival of stressed cells. J. Cell Biol. 163:767-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaufman, R. J., D. Scheuner, M. Schroder, X. Shen, K. Lee, C. Y. Liu, and S. M. Arnold. 2002. The unfolded protein response in nutrient sensing and differentiation. Nat. Rev. Mol. Cell. Biol. 3:411-421. [DOI] [PubMed] [Google Scholar]
- 20.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 21.Lu, L., A. P. Han, and J. J. Chen. 2001. Translation initiation control by heme-regulated eukaryotic initiation factor 2alpha kinase in erythroid cells under cytoplasmic stresses. Mol. Cell. Biol. 21:7971-7980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marshall, C. J. 1994. MAP kinase kinase kinase, MAP kinase kinase and MAP kinase. Curr. Opin. Genet. Dev. 4:82-89. [DOI] [PubMed] [Google Scholar]
- 23.Moreno, S., A. Klar, and P. Nurse. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194:795-823. [DOI] [PubMed] [Google Scholar]
- 24.Natarajan, K., M. R. Meyer, B. M. Jackson, D. Slade, C. Roberts, A. G. Hinnebusch, and M. J. Marton. 2001. Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol. Cell. Biol. 21:4347-4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nguyen, A. N., A. Lee, W. Place, and K. Shiozaki. 2000. Multistep phosphorelay proteins transmit oxidative stress signals to the fission yeast stress-activated protein kinase. Mol. Biol. Cell 11:1169-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Novoa, I., Y. Zhang, H. Zeng, R. Jungreis, H. P. Harding, and D. Ron. 2003. Stress-induced gene expression requires programmed recovery from translational repression. EMBO J. 22:1180-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paredes, V., A. Franco, M. Madrid, T. Soto, J. Vicente-Soler, M. Gacto, and J. Cansado. 2004. Transcriptional and post-translational regulation of neutral trehalase in Schizosaccharomyces pombe during thermal stress. Yeast 21:593-603. [DOI] [PubMed] [Google Scholar]
- 28.Quinn, J., V. J. Findlay, K. Dawson, J. B. Millar, N. Jones, B. A. Morgan, and W. M. Toone. 2002. Distinct regulatory proteins control the graded transcriptional response to increasing H2O2 levels in fission yeast Schizosaccharomyces pombe. Mol. Biol. Cell 13:805-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Samejima, I., S. Mackie, and P. A. Fantes. 1997. Multiple modes of activation of the stress-responsive MAP kinase pathway in fission yeast. EMBO J. 16:6162-6170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shiozaki, K., and P. Russell. 1996. Conjugation, meiosis, and the osmotic stress response are regulated by Spc1 kinase through Atf1 transcription factor in fission yeast. Genes Dev. 10:2276-2288. [DOI] [PubMed] [Google Scholar]
- 31.Sonenberg, N. 1996. mRNA 5′ cap-binding protein eIF4E and control of cell growth, p. 245-269. In N. Mathews and N. Sonenberg (ed.), Translational control. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 32.Takeda, T., T. Toda, K. Kominami, A. Kohnosu, M. Yanagida, and N. Jones. 1995. Schizosaccharomyces pombe atf1+ encodes a transcription factor required for sexual development and entry into stationary phase. EMBO J. 14:6193-6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tan, S., N. Somia, P. Maher, and D. Schubert. 2001. Regulation of antioxidant metabolism by translation initiation factor 2alpha. J. Cell Biol. 152:997-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tibbles, L. A., and J. R. Woodgett. 1999. The stress-activated protein kinase pathways. Cell. Mol. Life Sci. 55:1230-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Toone, W. M., and N. Jones. 1998. Stress-activated signalling pathways in yeast. Genes Cells 3:485-498. [DOI] [PubMed] [Google Scholar]
- 36.Toone, W. M., S. Kuge, M. Samuels, B. A. Morgan, T. Toda, and N. Jones. 1998. Regulation of the fission yeast transcription factor Pap1 by oxidative stress: requirement for the nuclear export factor Crm1 (Exportin) and the stress-activated MAP kinase Sty1/Spc1. Genes Dev. 12:1453-1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tzamarias, D., I. Roussou, and G. Thireos. 1989. Coupling of GCN4 mRNA translational activation with decreased rates of polypeptide chain initiation. Cell 57:947-954. [DOI] [PubMed] [Google Scholar]
- 38.Ueda, T., R. Watanabe-Fukunaga, H. Fukuyama, S. Nagata, and R. Fukunaga. 2004. Mnk2 and Mnk1 are essential for constitutive and inducible phosphorylation of eukaryotic initiation factor 4E but not for cell growth or development. Mol. Cell. Biol. 24:6539-6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uesono, Y., and Toh−E. A. 2002. Transient inhibition of translation initiation by osmotic stress. J. Biol. Chem. 277:13848-13855. [DOI] [PubMed] [Google Scholar]
- 40.Vattem, K. M., and R. C. Wek. 2004. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc. Natl. Acad. Sci. USA 101:11269-11274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Waskiewicz, A. J., A. Flynn, C. G. Proud, and J. A. Cooper. 1997. Mitogen-activated protein kinases activate the serine/threonine kinases Mnk1 and Mnk2. EMBO J. 16:1909-1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Waskiewicz, A. J., J. C. Johnson, B. Penn, M. Mahalingam, S. R. Kimball, and J. A. Cooper. 1999. Phosphorylation of the cap-binding protein eukaryotic translation initiation factor 4E by protein kinase Mnk1 in vivo. Mol. Cell. Biol. 19:1871-1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Watanabe, Y., and M. Yamamoto. 1996. Schizosaccharomyces pombe pcr1+ encodes a CREB/ATF protein involved in regulation of gene expression for sexual development. Mol. Cell. Biol. 16:704-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wilkinson, M. G., and J. B. Millar. 1998. SAPKs and transcription factors do the nucleocytoplasmic tango. Genes Dev. 12:1391-1397. [DOI] [PubMed] [Google Scholar]
- 45.Wilkinson, M. G., M. Samuels, T. Takeda, W. M. Toone, J. C. Shieh, T. Toda, J. B. Millar, and N. Jones. 1996. The Atf1 transcription factor is a target for the Sty1 stress-activated MAP kinase pathway in fission yeast. Genes Dev. 10:2289-2301. [DOI] [PubMed] [Google Scholar]
- 46.Yaman, I., J. Fernandez, H. Liu, M. Caprara, A. A. Komar, A. E. Koromilas, L. Zhou, M. D. Snider, D. Scheuner, R. J. Kaufman, and M. Hatzoglou. 2003. The zipper model of translational control: a small upstream ORF is the switch that controls structural remodeling of an mRNA leader. Cell 113:519-531. [DOI] [PubMed] [Google Scholar]
- 47.Yang, R., S. A. Wek, and R. C. Wek. 2000. Glucose limitation induces GCN4 translation by activation of Gcn2 protein kinase. Mol. Cell. Biol. 20:2706-2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhan, K., J. Narasimhan, and R. C. Wek. 2004. Differential activation of eIF2 kinases in response to cellular stresses in Schizosaccharomyces pombe. Genetics 168:1867-1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhan, K., K. M. Vattem, B. N. Bauer, T. E. Dever, J. J. Chen, and R. C. Wek. 2002. Phosphorylation of eukaryotic initiation factor 2 by heme-regulated inhibitor kinase-related protein kinases in Schizosaccharomyces pombe is important for resistance to environmental stresses. Mol. Cell. Biol. 22:7134-7146. [DOI] [PMC free article] [PubMed] [Google Scholar]