Abstract
Several models for specific excision of micronucleus-specific DNA sequences during macronuclear development in ciliates exist. While the template-guided recombination model suggests recombination events resulting in specific DNA excision and reordering of macronucleus-destined sequences (MDS) guided by a template, there is evidence that an RNA interference-related mechanism is involved in DNA elimination in holotrichous ciliates. We describe that in the stichotrichous ciliate Stylonychia, snRNAs homologous to micronucleus-specific sequences are synthesized during macronuclear differentiation. Western and in situ analyses demonstrate that histone H3 becomes methylated at K9 de novo during macronuclear differentiation, and chromatin immunoprecipitation revealed that micronucleus-specific sequences are associated with methylated H3. To link both observations, expression of a PIWI homolog, member of the RNA-induced silencing complex, was silenced. In these cells, the methylated micronucleus-specific histone H3 variant “X” is still present in macronuclear anlagen and no K9 methylation of histone H3 is observed. We suggest that snRNA recruits chromatin-modifying enzymes to sequences to be excised. Based on our and earlier observations, we believe that this mechanism is not sufficient for specific excision of sequences and reordering of MDS in the developing macronucleus and propose a model for internal eliminated sequence excision and MDS reordering in stichotrichous ciliates.
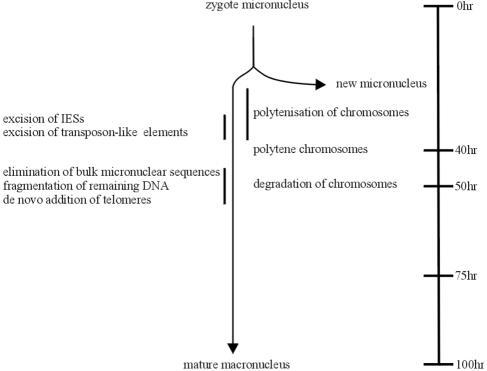
The diploid micronucleus and the DNA-rich macronucleus of ciliated protozoa are derived from the same zygote nucleus formed by two haploid micronuclei during sexual reproduction. In all ciliates, DNA rearrangement processes during macronuclear development are observed from which the most extensive forms occur in stichotrichous ciliates, such as Stylonychia or Oxytricha. In these organisms, up to over 90% of micronuclear DNA sequences are eliminated during macronuclear differentiation, resulting in a macronucleus with a highly reduced kinetic complexity and macronuclear DNA organized in small DNA molecules, so-called “nanochromosomes” (3, 2), which range in size from 0.4 to over 20 kbp. In Stylonychia lemnae, each macronucleus contains about 15,000 different nanochromosomes amplified to specific copy numbers ranging from several hundred to 106 copies per macronucleus (9, 25). A schematic diagram showing macronuclear differentiation in the stichotrichous ciliate Stylonychia lemnae is shown in Fig. 1. Sequence reduction occurs at different stages during macronuclear development. In Stylonychia, only about 30% of micronuclear chromosomes enter the first DNA synthesis phase, which leads to the formation of polytene chromosomes, while the other chromosomes become pycnotic (1, 19). Macronucleus-destined sequences (MDSs) are interrupted by small internal eliminated sequences (IES), ranging in size from about 1 bp to 100 bp in stichotrichous ciliates and up to several kbps in length in the holotrichous ciliates Tetrahymena and Paramecium (25), and by transposable elements (12). Both sequence elements are excised in stichotrichous ciliates during polytene chromosome formation, followed by rejoining of MDSs. About 30% of the MDSs studied so far are in a scrambled disorder and have to be reordered in the course of IES excision (26). Finally, bulk DNA containing repetitive and unique sequences are eliminated during polytene chromosome breakdown, and at this stage, macronuclear DNA becomes fragmented and telomeres are added de novo to the macronuclear nanochromosomes (25).
FIG. 1.
Schematic diagram showing the processes occurring during macronuclear differentiation in Stylonychia lemnae. The timescale starts when the two conjugants have separated (modified from reference 1 with permission of the publisher; see also reference 25).
Several models for DNA excision during macronuclear differentiation have been proposed (14, 20, 21, 27). An elegant model for IES excision and unscrambling of genes in stichotrichous ciliates is the “template-guided recombination model” (27). According to that model, pairs of short repeats (pointers) at the ends of the MDSs undergo homologous recombination to excise IESs and splice MDSs. But since the repeat sequences are too short to guide their own alignment, a template-guided recombination is proposed in which the template comes from the old macronucleus. While this model solves the problem of the correct pairing of pointers, precisely identifies MDS-IES junctions, and explains how unscrambling of macronuclear precursor sequences can be achieved, the presence of such a template has yet to be demonstrated. Very different models for DNA excision have been proposed for the holotrichous ciliates Tetrahymena and Paramecium (8, 14, 20, 21). According to these models, the complete micronuclear genome is transcribed bidirectionally and these transcripts form double-stranded RNAs (dsRNAs). The dsRNAs are processed into small dsRNAs (snRNAs, scanRNAs) and scanRNAs homologous to micronucleus-specific sequences are selected. They are then transferred to the developing macronucleus (macronuclear anlage) where they target sequences to be eliminated. As a consequence, a histone H3 histone methyltransferase methylates histone H3 at the lysine 9 (K9) and chromodomain proteins can then bind, resulting in heterochromatization of the sequences to be eliminated. In fact, all the components proposed in this model have been identified in Tetrahymena, and it is now widely accepted that genome rearrangement in this and possibly other ciliates involves an RNA interference (RNAi)-related mechanism (for a review, see reference 21). Still, such a mechanism does not explain the unscrambling of MDSs, a phenomenon not observed outside stichotrichous ciliates (26), nor their exact excision.
In this study, we asked whether an RNAi-related mechanism for DNA elimination also occurs in stichotrichous ciliates. We demonstrate that snRNAs homologous to micronuclear sequences are present during macronuclear development in Stylonychia and that histone H3 becomes methylated de novo in the macronuclear anlage and that this modified histone is associated with sequences to be eliminated. Inhibition of histone methylation results in a defective macronuclear differentiation, and upon inhibition of the expression of one component in the RNAi pathway, the highly methylated micronuclear histone H3 variant “X” (28) is still observed in the macronuclear anlage, while de novo methylation of macronuclear anlagen histone H3 does not take place. Based on our earlier, present, and unpublished observations, we conclude that snRNAs are required for targeting sequences to be eliminated but are not sufficient for correct DNA excision and reordering.
MATERIALS AND METHODS
Growth of Stylonychia, isolation of nuclei.
The growth of Stylonychia lemnae and isolation of macronuclei, micronuclei, and macronuclear anlagen were performed as described before (1). The cell density in vegetative cultures ranges between 300 and 400 cells per ml. For mating, cells of two different mating types were mixed and allowed to conjugate. The conjugation efficiency was between 80 and 90%. Exconjugant cells at different stages of macronuclear development were used to isolate RNA and nuclear proteins.
Isolation of DNA, RNA, and nuclear proteins.
The isolation of DNA followed the procedure described previously (6). Total RNA was isolated from approximately 5 × 104 vegetative and exconjugant cells at different stages of macronuclear development. Cells were collected on a 30-μm gauze, and RNA was isolated with Trizol LS reagent (Invitrogen) according to the manufacturer's protocol. These RNA preparations were then digested with RNase-free DNase (Invitrogen). As controls, the RNA preparation was also digested with RNase (Applichem). The RNA concentration was determined by UV spectroscopy. RNA was separated on a 15% polyacrylamide urea gel. snRNA was eluted from the gel, end labeled with [γ-32P]ATP, and hybridized to Southern or slot blots. For extraction of nuclear proteins, the various nuclei were isolated (approximately 5 × 104), collected by centrifugation, resuspended in loading buffer (13), heated for 10 min at 100°C, and separated on 15% sodium dodecyl sulfate (SDS)-polyacrylamide gels.
Southern, slot blot, and Western analyses.
For Southern analyses, DNA was transferred to nitrocellulose filters and hybridized with labeled snRNA at 40°C as described previously (20). For slot blot analyses, 1 μg DNA was blotted and hybridized under conditions similar to those of the Southern analyses. Proteins were transferred to a nylon membrane and probed with a polyclonal rabbit anti-H3 (trimethyl K9) antibody (catalogue no. ab8898; Abcam). Detection was done using the digoxigenin system (Roche).
Immunodetection of H3 trimethyl K9.
Isolated nuclei were fixed in 2% paraformaldehyde for 20 min at room temperature, washed twice with phosphate-buffered saline (PBS), and immobilized on poly-l-lysine-coated coverslips. Nuclei were permeabilized with 0.5% Triton X-100 and PBS for 20 min. Blocking was performed in 3% bovine serum albumin, 0.1% Triton X-100, and PBS for 20 min at room temperature. Antibodies were diluted in blocking solution according to the manufacturer's recommendation. Between applications of primary and secondary antibodies, washing steps were performed in PBS for 20 min. Incubations with both primary and secondary antibodies were performed for 1.5 h at 37°C. Histone H3 trimethylated at lysine 9 was detected using polyclonal rabbit anti-H3 (trimethyl K9) antibody (24) and then goat anti-rabbit Alexa Fluor 488 conjugate (Molecular Probes). Nuclei were counterstained with 0.1 μg/ml 4′,6′-diamidino-2-phenylindole (DAPI) (Sigma) and 1 μM To-Pro-3 (Molecular Probes) in PBS. Slides were mounted in Vectashield antifade medium (Vector Laboratories) and sealed with nail polish. Nuclei were analyzed by laser scanning confocal microscopy. Acquisition of light optical serial sections was done with a Leica TCP SP confocal laser scanning microscope (Leica Microsystems) equipped with an oil immersion PlanApochromat 100×/1.4 NA objective lens. Fluorochromes were visualized with an argon laser with an excitation wavelength of 488 nm for Alexa Fluor 488 and with a helium-neon laser with an excitation wavelength of 633 nm for To-Pro-3. Fluorochrome images were scanned sequentially, generating 8-bit grayscale images. The image resolution was 512 by 512 pixels, with a pixel size ranging from 195 to 49 nm, depending on the selected zoom factor. The axial distance between light optical serial sections was 250 nm. To obtain an improved signal-to-noise ratio, each section image was averaged from four successive scans. The 8-bit grayscale single-channel images were overlaid to an RGB image, assigning false colors to each channel, and then assembled into tables using the open source software ImageJ (version 1.31; Wayne Rasband, NIH) and Corel Photo Paint, version 10, software.
Inhibition of histone H3 methylation by trichostatin A.
Five micrograms of trichostatin A (stock solution of 1 mg/ml)/milliliter was added to conjugating cells.
PCR amplifications.
PCR conditions were as follows: initial denaturation at 94°C for 5 min; 35 cycles of 94°C for 30 s, 53°C for 30 s, and 74°C for 45 s; and final extension at 74°C for 1 min. The primers used for the amplification of the transposon-like element MaA81 were P4 (5′-GCGGGTACCATCAGATAACTCGCAAC-3′) and P23 (5′-GCATTTACCGATGGATCAATGA-3′). The primers used for the amplification of the 1.3-kbp nanochromosome were either P20 (5′-CCGCAGGATCCTTGAGAGTCTGCCATTTAAC-3′) and P28 (5′-GATTAGCTAGCTTGATCGTAATCGTAG-3′) or P9 (5′-GGCTCGAGTTGCTACTCTCAGATATTC-3′) and P23 (5′-GTTAAATGGCAGACTCTCAAGAAGAAATGC-3′) (see Fig. 5B, C, and D for primer positions).
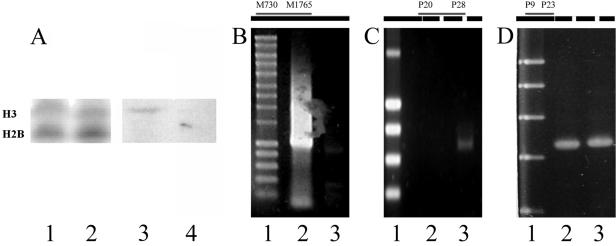
FIG. 5.
Effect of trichostatin A on DNA processing. (A) Inhibition of histone H3 methylation by trichostatin A; histone H3 from macronuclear anlagen from control cells or trichostatin A-treated cells. Lanes 1 and 2, Coomassie-stained gel (lane 1, control cells; lane 2, trichostatin A-treated cells); lanes 3 and 4, Western analyses using antibodies directed against trimethylated histone H3 (lane 3, control cells; lane 4, trichostatin A-treated cells). (B) Effect of trichostatin A on excision of the transposon-like element MaA81. Lane 1, molecular size marker (0.2 kbp to 10 kbp, Smart ladder; Eurogentec); lane 2, PCR product from the repetitive element of cells treated with trichostatin A at 40 h postconjugation; lane 3, control cells at 40 h postconjugation. At the top, the positions of the primers used and the amplified region (gray) are given in the schematic drawing of the repetitive element (black box). (C) Effect of trichostatin A on excision of the IESs from the 1.3-kbp MDS. Lane 1, molecular size marker; lane 2, control cells at 40 h postconjugation; lane 3, PCR product of cells treated with trichostatin A at 40 h postconjugation. At the top, the positions of the primers used and the amplified region (gray) are given in the schematic drawing of the pCE7 gene cluster (black box; IESs are indicated by white boxes within). (D) Control PCR using primers derived from the 5′ region of the 1.3-kbp nanochromosome showing that equal amounts of template DNA were present in the PCRs. Lane 1, molecular size marker; lane 2, control cells at 40 h postconjugation; lane 3, PCR product of cells treated with trichostatin A at 40 h postconjugation. At the top, the positions of the primers used and the amplified region (gray) are given in the schematic drawing of the pCE7 gene cluster (black box; IESs are indicated by white boxes within).
Chromatin immunoprecipitation (ChIP).
Isolated nuclei were cross-linked with PBS-1% formaldehyde (15 min, 4°C). After washing twice in PBS, the nuclei were resuspended in lysis buffer 1 (0.25% Triton X-100, 10 mM EDTA, 0.5 mM EGTA, 10 mM HEPES, pH 7.5). After incubation (15 min, 4°C), the nuclei were collected by centrifugation and resuspended in lysis buffer 2 (0.2 M NaCl, 1 mM EDTA, 0.5 mM EGTA, 10 mM HEPES, pH 7.5) and incubated (10 min, 4°C). They were then resuspended in sonication buffer (150 mM NaCl, 25 mM Tris-Cl, pH 7.5, 5 mM EDTA, 1% Triton X-100, 0.1% SDS, 0.5% sodium deoxycholate). Sonication parameters were as follows: intensity, 5; 20-s bursts, 2× for 3 min, 1-min pause, 4°C (Branson Sonifier, Microtip). The sonicated DNA-protein solution was cleared by centrifugation (6,000 × g, 15 min, 4°C), and the supernatant was incubated (12 h, 4°C) with polyclonal rabbit anti-histone H3 trimethyl K9 antibody (1 μg/ml; Abcam). The antibody-histone-DNA complex was then isolated with protein G Dynabeads (Dynal) according to the manufacturer's protocol. The cross-link was reversed by heating (30 min, 65°C), and the proteins were digested with proteinase K (20 μg/ml, 14 h, 37°C). The DNA was purified by phenol extraction, precipitated with ethanol, and resuspended in 10 mM Tris (pH 7.4).
Inhibition of PIWI expression by RNAi.
Silencing of PIWI expression by RNAi was achieved by feeding Escherichia coli cells expressing various regions of the Stylonychia PIWI macronuclear gene as described earlier (23).
RESULTS
Small nuclear RNAs homologous to micronucleus-specific sequences are synthesized during macronuclear development.
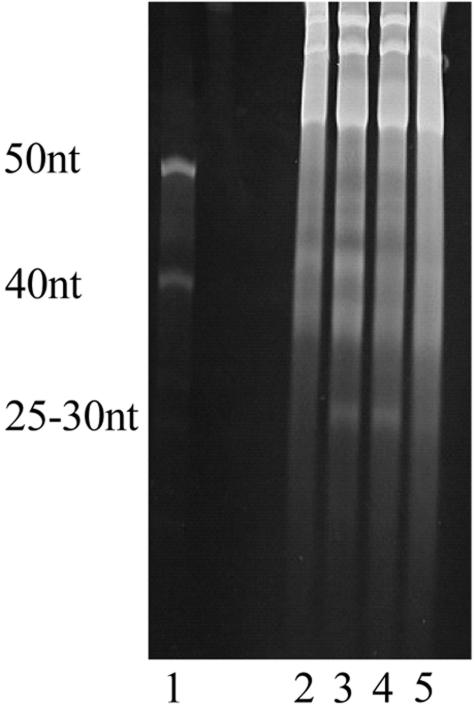
RNA was isolated from vegetative Stylonychia cells and from exconjugant cells at different stages of macronuclear development and separated on denaturing polyacrylamide gels. As shown in Fig. 2, an RNA species with an approximate size between 25 and 30 nucleotides (nt) appears about 10 h postconjugation (i.e., when the two conjugating cells have separated) and can no longer be observed in exconjugants in the polytene chromosome stage. This 25- to 30-nt band was also visible after DNase I digestion, demonstrating that it represents no short DNA sequences which could occur during IES excision. Furthermore, after RNase digestion, no ethidium bromide-stained material could be observed on the gels. To identify whether these RNAs show homology to micronucleus-specific sequences, they were isolated from the gel, end labeled, and hybridized either to total DNA or to restriction-digested micronuclear DNA (Fig. 3A). Figure 3B shows a Southern hybridization to total DNA (lane 1) and to restriction-digested micronuclear DNA (lane 2) using snRNAs as a probe. In these analyses, snRNA hybridizes only to undigested high-molecular-weight micronuclear DNA in total DNA preparations but not to macronuclear DNA (lane 1), while a strong hybridization to restriction-digested micronuclear DNA is observed (lane 2). To further demonstrate that this anlagen-specific snRNA is homologous to micronucleus-specific DNA sequences, slot blot hybridizations to various sequence classes were made (Fig. 3C). The following sequences were analyzed: a, the macronuclear nanochromosome encoding histone H4 (Fig. 3C, a); b, the macronuclear nanochromosome encoding calmodulin (Fig. 3C, b); c, macronuclear rRNA genes (Fig. 3C, c); d, total DNA (macronuclear and micronuclear DNA) of vegetative cells (Fig. 3C, d); e, total macronuclear anlagen DNA isolated from the early polytene chromosome stage about 30 h postconjugation (Fig. 3C, e); f, the macronuclear 1.3-kbp nanochromosome (Fig. 3C, f); g, a middle repetitive 800-bp DNA sequence (pLJ01) eliminated after the breakdown of polytene chromosomes (Fig. 3C, g); h, a cluster of macronuclear precursor sequences (MDSs) derived from pCE7 by PCR amplification (this PCR fragment encodes the 1.1-kbp and 1.3-kbp MDS; in the micronuclear genome, these MDSs are interrupted by 5 IESs with sizes between 10 bp and 68 bp and separated by an 11-bp spacer sequence) (5, 11) (Fig, 3C, h; see also Fig. 5C and D, top); i, the XbaI fragment of the transposon-like element MaA81 (19) (Fig. 3C, i); j, the complete transposon-like element MaA81 (Fig. 3C, j; see also Fig. 5B, top). As shown in this figure, the snRNA isolated from exconjugants does not hybridize to macronuclear DNA sequences (Fig. 3C, a, b, c, f) but hybridizes to macronuclear anlagen DNA (Fig. 3C, e) and to repetitious eliminated DNA such as MaA81 (Fig. 3C, i, j) and pLJ01 (Fig. 3C, g) and also to the macronuclear precursor sequences amplified from pCE7 (Fig. 3C, h), showing its homology to the IESs and spacer present in this sequence.
FIG. 2.
Isolation of RNA from vegetative and exconjugant cells at different stages of macronuclear differentiation. The RNA was separated on a denaturing polyacrylamide gel as described in Materials and Methods. The size of the snRNA synthesized during macronuclear development was estimated by using 40- to 50-nt oligonucleotides as markers. Lane 1, oligonucleotides (40 nt and 50 nt); lane 2, total RNA from vegetative cells; lanes 3 to 5, RNA isolated from exconjugants at 10 h (lane 3), 25 h (lane 4), and 50 h (lane 5) postconjugation. The RNA was isolated from approximately 108 cells.
FIG. 3.
Southern and slot blot analyses using snRNA as a probe. (A) Ethidium bromide-stained agarose gel with total DNA (lane 1) and XbaI-digested micronuclear DNA (lane2). (B) Southern hybridization to total DNA (macronuclear and micronuclear DNA) from vegetative cells (lane 1) and to XbaI-digested micronuclear DNA (lane 2) using snRNAs from exconjugant cells at 10 h postconjugation as a probe. snRNA was isolated from the gel and end labeled as described previously (20). Hybridization was performed as described in Materials and Methods. (C) Slot blot of radiolabeled snRNAs of 10-h-postconjugation cells hybridized on different DNA sequences from Stylonychia: a, histone H4; b, calmodulin; c, rRNA genes; d, total DNA of vegetative cells; e, DNA isolated from polytene chromosomes; f, 1.3-kbp nanochromosome isolated from vegetative cells; g, pLJ01; h, 1.1-kbp and 1.3-kbp nanochromosome contained in pCE7; i, XbaI fragment of the repetitive element; j, MaA81. Each slot contains 1 μg DNA.
H3K9 methylation in the nuclei.
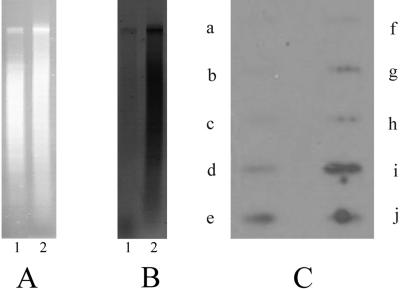
Since it is supposed that the RNAi machinery can recruit histone deacetylases and histone methyltransferases to the target sequences (16), we analyzed H3K9 methylation in the macronuclei, micronuclei, and macronuclear anlagen using antibodies directed against trimethylated H3K9. The methylation pattern in these nuclei was studied both by Western analysis and by in situ antibody staining. In the micronucleus of Stylonychia, the 16-kDa histone H3 is replaced by a histone H3 variant “X” with a size of about 21 kDa. In the course of macronuclear differentiation, this histone variant is replaced with normal histone H3 (28). For Western analyses, proteins from macronuclei, micronuclei, and anlagen nuclei in the first stage of DNA amplification (i.e., polytenization) were separated on SDS-polyacrylamide gels, blotted onto nylon membranes, and probed with an antibody directed against trimethylated H3K9 (Fig. 4A). No staining of the macronuclear histone H3 was observed (Fig. 4A, lane 5), but a strong signal was obtained with the micronuclear histone H3 variant “X” (Fig. 4A, lane 4) and with histone H3 in anlagen nuclei (Fig. 4A, lane 6), suggesting that H3 becomes de novo methylated in the course of macronuclear differentiation. A similar picture was obtained by in situ analyses (Fig. 4B). The antibody strongly reacts with the micronucleus of vegetative cells and the macronuclear anlagen but not with the macronucleus (Fig. 4B, a). In light optical sections, it becomes obvious that mainly the banded regions of the polytene chromosomes are stained (Fig. 4B, b), which are supposed to be specifically eliminated in the form of heterochromatic chromatin rings (18). In the course of polytene chromosome degradation, the staining of anlagen nuclei becomes weaker and can no longer be observed in nuclei in the DNA-poor stage (Fig. 4B, c) or in developing macronuclei in the second DNA amplification phase.
FIG. 4.
Analysis of histone H3K9 methylation in the different nuclei. (A) Western analyses using antibodies directed against histone H3 trimethylated at K9. Macronuclei and micronuclei were isolated from vegetative cells and macronuclear anlagen were isolated from exconjugant cells at 20 h postconjugation. They were separated on a 15% SDS-polyacrylamide gel, and either the gel was stained with Coomassie blue or the proteins were blotted onto a nylon membrane. Western analyses were performed as described in Materials and Methods. Lanes 1 to 3, Coomassie-stained proteins; lane 1, micronuclear proteins; lane 2, macronuclear proteins; lane 3, proteins of the macronuclear anlagen (20 h postconjugation); lanes 4 to 6, Western analysis using anti-histone H3 trimethyl K9 antibodies; lane 4, micronuclear proteins; lane 5, macronuclear proteins; lane 6, proteins of the macronuclear anlagen. Arrows point to the micronuclear histone H3 variant “X” and to histone H3. (B) Immunolocalization of histone H3 trimethyl K9 in Stylonychia nuclei at successive stages of macronuclear anlagen development. Fluorochrome channels were scanned sequentially, generating 8-bit grayscale images. False colors were assigned to each channel (histone H3 trimethyl K9, green; To-Pro-3, red) before being overlaid. (a) Maximum-intensity projections of 24 light optical serial sections showing two micronuclei (m), a macronucleus (M), and a macronuclear anlage containing polytene chromosomes (A). Histone H3 trimethyl K9 is found in micronuclei as well as in distinct areas of the macronuclear anlage but is absent in the macronucleus. (b) Mid-light optical sections of a macronuclear anlage containing polytene chromosomes (A). Histone H3 trimethyl K9 is localized in distinct areas of the macronuclear anlage (green and green-red areas). Discrete amorphous areas, possibly representing DNA already processed, contain no histone H3 trimethyl K9 (red areas). The graticule frames a segment of a polytene chromosome. In this segment, histone H3 trimethyl K9 appears to be localized preferentially in the chromosomal bands. (c) A late macronuclear anlage in the DNA-poor stage (late A). No histone H3 trimethyl K9 is found in late macronuclear anlagen in the DNA-poor stage. (C) Slot blot analysis of digoxigenin-labeled immunoprecipitated DNA isolated from exconjugants at 20 h postconjugation hybridized to different DNA sequence classes. Slots: a, macronuclear 1.1-kbp nanochromosome; b, macronuclear 1.3-kbp nanochromosome; c, macronuclear DNA; d, XbaI fragment of the transposon-like element MaA81; e, complete transposon-like element MaA81; f, repetitive sequence pLJ01; g, macronuclear nanochromosome encoding histone H4. Each slot contains 1 μg DNA.
To identify sequences of the developing macronucleus associated with H3K9-methylated H3, ChIP analyses were performed. Chromatin was solubilized and precipitated with an antibody directed against trimethylated histone H3K9. DNA from the immunoprecipitate was isolated, labeled, and hybridized to selected sequence classes, i.e., the 1.1-kbp macronuclear nanochromosome (Fig. 4C, a), the macronuclear 1.3-kbp nanochromosome (Fig. 4C, b), total macronuclear DNA (Fig. 4C, c), XbaI-digested total DNA isolated from the macronuclear anlagen in the polytene chromosome stage (Fig. 4C, d), the transposon-like element MaA81 (Fig. 4C, e), the middle repetitive DNA sequence pLJ01 (Fig. 4C, f), and the macronuclear nanochromosome encoding histone H4 (Fig. 4C, g). No hybridization to macronuclear DNA or macronucleus-specific sequences was observed (Fig. 4C, a, b, c, g), but strong signals were obtained with anlagen DNA (Fig. 4C, d), the transposon-like element MaA81 (Fig. 4C, e), and the repetitive sequence pLJ01 (Fig. 4C, f). In these hybridization analyses, no macronuclear precursor sequences were included, since due to kinetic reasons, it seemed impossible to detect specific hybridization to small IESs when using total immunoprecipitated DNA as a probe.
Histone deacetylation and subsequent de novo histone methylation can be efficiently inhibited by the drug trichostatin A (4, 29). Upon application of trichostatin A to conjugating and exconjugant cells, no H3 methylation can be detected by Western analysis in the macronuclear anlage (Fig. 5A), although production of snRNAs was not influenced by treatment with this drug. These exconjugants stop macronuclear development before reaching the polytene chromosome stage. To see whether this drug affects elimination of micronucleus-specific sequences, the excision of the IESs from the 1.3k-bp macronuclear precursor sequence encoded on pCE7 and the transposon-like element MaA81 in control cells and trichostatin A-treated cells was studied. PCR amplification was made from 5 cells of the same stage of macronuclear development. As demonstrated in Fig. 5B, the transposon-like element MaA81 is almost completely excised in control cells at 40 h postconjugation (Fig. 5B, lane 3), but a strong PCR amplification product is still obtained at that time from trichostatin A-treated cells (Fig. 5B, lane 2). Similarly, a PCR fragment containing 3 IESs of the 1.3-kbp MDS could be amplified from trichostatin A-treated cells at 40 h postconjugation (Fig. 5C, lane 3), while these IESs were already excised in control cells about 15 to 20 h postconjugation (Fig. 5C, lane 2). To demonstrate that equal amounts of template were used in these experiments, a control PCR was done from the 5′ region of the 1.3-kbp nanochromosome (Fig. 5D). This strongly suggests that H3K9 methylation is a necessary prerequisite not only for subsequent excision of IESs and transposon-like elements but also for elimination of bulk DNA.
PIWI is required for H3K9 methylation in macronuclear anlagen.
To analyze whether the expression of snRNA is correlated to de novo histone H3 methylation, we silenced PIWI expression by RNAi. This does not affect the production of snRNAs, since PIWI is a member of the RNA-induced silencing complex required to regulate downstream effects in the RNAi pathway (22). We have recently shown that a homolog of this protein is specifically expressed during macronuclear development in Stylonychia (7). When the expression of PIWI is knocked down in Stylonychia, the old macronuclei are not degraded, the macronuclear anlagen development arrests in the polytene chromosome stage, and eventually the developing macronucleus disintegrates (23).
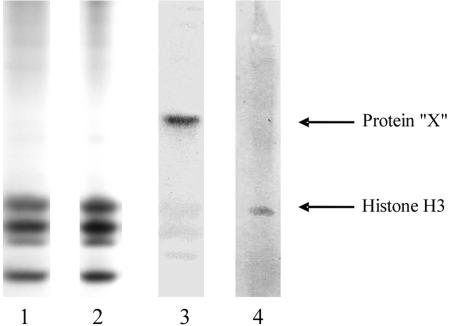
The effect of PIWI expression on histone H3 methylation in the macronuclear anlagen was analyzed by Western analysis. While in control cells fed with E. coli but without the RNAi construct the methylated micronucleus-specific histone H3 variant “X” is replaced by methylated histone H3 during macronuclear development (Fig. 6, lane 4), this methylated micronuclear H3 variant is still observed in exconjugant cells in which PIWI expression was silenced and macronuclear anlagen-specific H3 is visible on Coomassie-stained gels but it does not become methylated de novo (Fig. 6, lane 3).
FIG. 6.
Silencing of PIWI expression by RNAi in exconjugant cells and Western analysis using an antibody directed against histone H3 trimethyl K9. Isolation of nuclear proteins and Western analyses were performed as described in Materials and Methods and the legend to Fig. 4. Proteins of the macronuclear anlagen were isolated at 20 h postconjugation. Lanes 1 and 2, Coomassie-stained gel used for Western analysis; lane 1, proteins from cells in which PIWI expression was silenced by RNAi; lane 2, proteins from control cells; lanes 3 and 4, Western analysis using anti-histone H3 trimethyl K9 antibodies on PIWI-silenced cells and control cells; lane 3, proteins from exconjugant cells in which PIWI expression was silenced by RNAi; lane 4, proteins from control cells. Arrows point to the micronuclear histone H3 variant “X” and to histone H3.
DISCUSSION
Small nuclear RNAs have been shown to play a crucial role in the control of gene activity. Originally, it was described that binding of these RNAs to homologous regions of a transcript leads to its degradation or to a translational stop (17). More recently, it became clear that snRNAs can act as mediators for the formation of heterochromatin by the recruitment of histone deacetylases and histone methylases (15). It also now seems clear that snRNA is involved in the specific elimination of DNA sequences in the ciliate Tetrahymena, in which binding of these RNAs leads to the creation of a specific chromatin structure of sequences to be eliminated during macronuclear differentiation. The models proposed (20, 21, 14) do explain how sequences to be eliminated are discriminated from sequences to be retained in the course of macronuclear differentiation, although it is not yet clear how the final excision process takes place.
The most dramatic DNA reorganization processes are observed in stichotrichous ciliates, such as Stylonychia or Oxytricha. Similar to Tetrahymena and Paramecium, IESs are excised during the first DNA amplification process before bulk DNA becomes eliminated (25) (Fig. 1). While IESs in Tetrahymena vary in size between about 0.6 and 22 kbp, thus allowing them to be incorporated into a distinct chromatin structure, over 95% of the IESs in stichotrichous ciliates are smaller than 100 bp, and a significant number is even smaller than 10 bp (26). Additionally, about 30% of the MDSs are in a scrambled disorder in the micronuclear genome and have to be reordered in the course of IES excision. For this reason, a template-guided model to excise IESs and reorder MDSs in stichotrichous ciliates has been proposed (27).
In this report, we describe that snRNAs homologous to micronucleus-specific DNA sequences are transcribed early during macronuclear differentiation. Their homology to micronucleus-specific sequences was clearly demonstrated by Southern analysis of macronuclear and micronuclear DNA and by slot blot analyses to selected sequence classes having different destinies during macronuclear differentiation. These analyses imply that snRNAs homologous to IESs exist, with the snRNA being larger than the IES to be targeted. In a parallel set of experiments, we show that the micronuclear histone H3 variant “X” occurs in a methylated form during vegetative growth, and in the course of macronuclear development, this histone variant is replaced by or processed to conventional histone H3, which occurs methylated during the first DNA amplification stage in macronuclear development. In the course of DNA elimination, this methylated H3 is lost from the nucleus and no methylated H3 is observed in the mature macronucleus. In situ analyses revealed that this modified H3 colocalizes with the banded region of the polytene chromosomes which are supposed to contain DNA sequences to be eliminated in the form of heterochromatic chromatin rings (18). ChIP experiments supported this idea, showing that preferentially micronucleus-specific DNA sequences become precipitated by an antibody directed against H3K9. Inhibition experiments using the drug trichostatin A implied that histone H3 methylation is a necessary prerequisite for excision of both a transposon-like element and IESs. PCR amplifications of the transposon-like element MaA81 and of an MDS revealed that excision of these sequences is delayed or may not even occur at all after treatment with trichostatin A. Furthermore, trichostatin A leads to a developmental arrest in the polytene chromosome stage, suggesting that histone H3 methylation is also required for elimination of bulk DNA. To correlate both observations, the transcription of snRNAs homologous to micronucleus-specific DNA sequences and the association of micronucleus-specific DNA sequences with methylated histone H3, the expression of PIWI, which is a component of the RNAi pathway, was silenced. Interestingly, the micronucleus-specific histone H3 variant “X” was retained in the macronuclear anlage, and no methylation of macronuclear anlagen H3 could be observed.
Taking all these data together, a scenario very similar to that described for holotrichous ciliates (8, 14, 20, 21) appears to also be true for stichotrichous ciliates: scanRNAs specify the sequences to be removed and recruit chromatin-modifying enzymes to induce heterochromatin formation at these sequences. Several problems regarding this model arise in stichotrichous ciliates: (i) it does not explain MDS reordering, (ii) many of the IESs are smaller than the corresponding scanRNA and therefore cannot target IES precisely, and (iii) since the majority of IESs in stichotrichous ciliates are smaller than the nucleosome core particle, no specific chromatin structure can form specifically above them. This suggests that additional mechanisms are required for correct processing of MDSs. Transfection experiments (10) showed that truncated MDSs become corrected in the course of macronuclear development, and therefore, a “proofreading mechanism” using a template from the old macronucleus was postulated. More recently, we observed numerous cases of imprecise IES excision during the process of macronuclear development (M. Möllenbeck, A. Cavalcanti, W. J. Chang, F. Jönsson, H. J. Lipps, and L. Landweber, unpublished data). This numerous imprecise excision of IESs should lead to the death of exconjugant cells if there would be no correction mechanism. We therefore propose the following model for MDS processing in stichotrichous ciliates. scanRNAs specify DNA sequences to be eliminated or excised, and a specific chromatin structure is formed. But due to the small size of most of the IESs, chromatin structure would not be sufficient to allow correct excision of sequences and also would not explain the reordering of MDSs. Thus, we suggest that a template from the old macronucleus guides and corrects excision and reordering. This macronuclear template could be used to correct imprecise IES excision by filling the gaps created by incorrect IES removal as suggested earlier (10). Alternatively, such gaps could be corrected by recombination events, and in this case, MDS reordering could occur simultaneously according to the original template-guided model.
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft and the National Science Foundation (grant no. EIA-0121422).
We appreciate the discussions we had about this work with L. Landweber, Princeton, N.J.; G. Rozenberg, Leiden, The Netherlands; and D. M. Prescott, Boulder, Colo. We are grateful to T. Cremer, Munich, Germany, for allowing us to use microscopic facilities and to A. Peters, Basel, Switzerland, for donating antibodies.
REFERENCES
- 1.Ammermann, D., G. Steinbruck, L. von Berger, and W. Hennig. 1974. The development of the macronucleus in the ciliated protozoan Stylonychia mytilus. Chromosoma 45:401-429. [DOI] [PubMed] [Google Scholar]
- 2.Cavalcanti, A. R., D. M. Dunn, R. Weiss, G. Herrick, L. F. Landweber, and T. G. Doak. 2004. Sequence features of Oxytricha trifallax (class Spirotrichea) macronuclear telomeric and subtelomeric sequences. Protist 155:311-322. [DOI] [PubMed] [Google Scholar]
- 3.Cavalcanti, A. R., N. A. Stover, L. Orecchia, T. G. Doak, and L. F. Landweber. 2004. Coding properties of Oxytricha trifallax (Sterkiella histriomuscorum) macronuclear chromosomes: analysis of a pilot genome project. Chromosoma 113:69-76. [DOI] [PubMed] [Google Scholar]
- 4.Duharcourt, S., and M. C. Yao. 2002. Role of histone deacetylation in developmentally programmed DNA rearrangements in Tetrahymena thermophila. Eukaryot. Cell 1:293-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eder, C., C. Maercker, J. Meyer, and H. J. Lipps. 1993. The processing of macronuclear DNA sequences during macronuclear development of the hypotrichous ciliate Stylonychia lemnae. Int. J. Dev. Biol. 37:473-477. [PubMed] [Google Scholar]
- 6.Elsevier, S. M., H. J. Lipps, and G. Steinbruck. 1978. Histone genes in macronuclear DNA of the ciliate Stylonychia mytilus. Chromosoma 69:291-306. [DOI] [PubMed] [Google Scholar]
- 7.Fetzer, C. P., D. J. Hogan, and H. J. Lipps. 2002. A PIWI homolog is one of the proteins expressed exclusively during macronuclear development in the ciliate Stylonychia lemnae. Nucleic Acids Res. 30:4380-4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garnier, O., V. Serrano, S. Duharcourt, and E. Meyer. 2004. RNA-mediated programming of developmental genome rearrangements in Paramecium tetraurelia. Mol. Cell. Biol. 24:7370-7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jonsson, F., J. Postberg, C. Schaffitzel, and H. J. Lipps. 2002. Organization of the macronuclear gene-sized pieces of stichotrichous ciliates into a higher order structure via telomere-matrix interactions. Chromosome Res. 10:445-453. [DOI] [PubMed] [Google Scholar]
- 10.Jonsson, F., G. Steinbruck, and H. J. Lipps. 25. January 2001, posting date. Both subtelomeric regions are required and sufficient for specific DNA fragmentation during macronuclear development in Stylonychia lemnae. Genome Biol. 2 [Online.] http://genomebiology.com/2001/2/2/RESEARCH/0005. [DOI] [PMC free article] [PubMed]
- 11.Jonsson, F., J. P. Wen, C. P. Fetzer, and H. J. Lipps. 1999. A subtelomeric DNA sequence is required for correct processing of the macronuclear DNA sequences during macronuclear development in the hypotrichous ciliate Stylonychia lemnae. Nucleic Acids Res. 27:2832-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klobutcher, L. A., and C. L. Jahn. 1991. Developmentally controlled genomic rearrangements in ciliated protozoa. Curr. Opin. Genet. Dev. 1:397-403. [DOI] [PubMed] [Google Scholar]
- 13.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 14.Le Mouel, A., A. Butler, F. Caron, and E. Meyer. 2003. Developmentally regulated chromosome fragmentation linked to imprecise elimination of repeated sequences in paramecia. Eukaryot. Cell 2:1076-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lippman, Z., and R. Martienssen. 2004. The role of RNA interference in heterochromatic silencing. Nature 431:364-370. [DOI] [PubMed] [Google Scholar]
- 16.Matzke, M. A., and J. A. Birchler. 2005. RNAi-mediated pathways in the nucleus. Nat. Rev. Genet. 6:24-35. [DOI] [PubMed] [Google Scholar]
- 17.Mello, C. C., and D. Conte, Jr. 2004. Revealing the world of RNA interference. Nature 431:338-342. [DOI] [PubMed] [Google Scholar]
- 18.Meyer, G. F., and H. J. Lipps. 1980. Chromatin elimination in the hypotrichous ciliate Stylonychia mytilus. Chromosoma 77:285-297. [DOI] [PubMed] [Google Scholar]
- 19.Meyer, G. F., and H. J. Lipps. 1981. The formation of polytene chromosomes during macronuclear development of the hypotrichous ciliate Stylonychia mytilus. Chromosoma 82:309-314. [DOI] [PubMed] [Google Scholar]
- 20.Mochizuki, K., N. A. Fine, T. Fujisawa, and M. A. Gorovsky. 2002. Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in Tetrahymena. Cell 110:689-699. [DOI] [PubMed] [Google Scholar]
- 21.Mochizuki, K., and M. A. Gorovsky. 2004. Small RNAs in genome rearrangement in Tetrahymena. Curr. Opin. Genet. Dev. 14:181-187. [DOI] [PubMed] [Google Scholar]
- 22.Parker, J. S., S. M. Roe, and D. Barford. 2004. Crystal structure of a PIWI protein suggests mechanisms for siRNA recognition and slicer activity. EMBO J. 23:4727-4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paschka, A. G., F. Jönsson, V. Maier, M. Möllenbeck, K. Paeschke, J. Postberg, S. Rupprecht, and H. J. Lipps. 2003. The use of RNAi to analyze gene function in spirotichous ciliates. Eur. J. Protistol. 39:449-454. [Google Scholar]
- 24.Peters, A. H., S. Kubicek, K. Mechtler, R. J. O'Sullivan, A. A. Derijck, L. Perez-Burgos, A. Kohlmaier, S. Opravil, M. Tachibana, Y. Shinkai, J. H. Martens, and T. Jenuwein. 2003. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol. Cell 12:1577-1589. [DOI] [PubMed] [Google Scholar]
- 25.Prescott, D. M. 1994. The DNA of ciliated protozoa. Microbiol. Rev. 58:233-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prescott, D. M. 2000. Genome gymnastics: unique modes of DNA evolution and processing in ciliates. Nat. Rev. Genet. 1:191-198. [DOI] [PubMed] [Google Scholar]
- 27.Prescott, D. M., A. Ehrenfeucht, and G. Rozenberg. 2003. Template-guided recombination for IES elimination and unscrambling of genes in stichotrichous ciliates. J. Theor. Biol. 222:323-330. [DOI] [PubMed] [Google Scholar]
- 28.Schlegel, M., S. Muller, F. Ruder, and W. Büsen. 1990. Transcriptionally inactive micronuclei, macronuclear anlagen and transcriptionally active macronuclei differ in histone composition in the hypotrichous ciliate Stylonychia lemnae. Chromosoma 99:401-406. [Google Scholar]
- 29.Selker, E. U. 1998. Trichostatin A causes selective loss of DNA methylation in Neurospora. Proc. Natl. Acad. Sci. USA 95:9430-9435. [DOI] [PMC free article] [PubMed] [Google Scholar]