Abstract
Polyketides are a class of secondary metabolites that exhibit a vast diversity of form and function. In fungi, these compounds are produced by large, multidomain enzymes classified as type I polyketide synthases (PKSs). In this study we identified and functionally disrupted 15 PKS genes from the genome of the filamentous fungus Gibberella zeae. Five of these genes are responsible for producing the mycotoxins zearalenone, aurofusarin, and fusarin C and the black perithecial pigment. A comprehensive expression analysis of the 15 genes revealed diverse expression patterns during grain colonization, plant colonization, sexual development, and mycelial growth. Expression of one of the PKS genes was not detected under any of 18 conditions tested. This is the first study to genetically characterize a complete set of PKS genes from a single organism.
Secondary metabolites are a remarkably diverse class of cellular products that often exhibit taxonomic specificity. Secondary metabolites are generally considered “nonessential” for organismal growth under culture conditions (5). In microbes, these unique biochemicals are frequently produced in culture after a period of active growth has depleted the substrate (6, 9) and biosynthesis often coincides with differentiation, especially sporulation (5). However, the circumstances of production and their role in life cycles in the natural environment are not well understood.
Polyketides (derived from polyketone) are a class of secondary metabolites produced by most organisms, but they have been most extensively examined in bacteria and fungi. In fungi, polyketides include a range of compounds such as the mycotoxins aurofusarin (48), aflatoxin (7), and zearalenone (57) and spore pigments (38, 59). Many functions have been proposed for polyketides (11, 23, 60). However, the most plausible explanation for the diversity of secondary metabolites is that the ability to produce a wide variety of compounds is ecologically and evolutionarily advantageous as a resource for bioactive compounds (15, 22). For example, several actinomycetes synthesize two chemically unrelated metabolites that act against the same target organism (15, 17). Thus, unrelated compounds may be selected for the same activity when important to organism survival.
The study of fungal polyketides has been limited by the difficulty of detecting and characterizing the polyketide itself. Complete analysis of the genetic potential of fungi to produce polyketides became possible only recently when the genomic sequences of several fungal species became available. Now, the selective cloning of genes encoding polyketide synthases (PKSs) can precede identification of a product and contribute to the overall analysis of polyketide diversity and function in an organism.
Polyketides are synthesized by adding successive acetate groups to form a chain of C2 units with alternating ketones and the resulting chain can be either linear or aromatic. The minimal PKS consists of three domains: a ketosynthase domain, an acyltransferase domain, and an acyl carrier protein domain. Such an enzyme is said to be nonreducing and the resulting polyketide is aromatic with unmodified ketone groups. In addition to the three requisite domains, PKSs may also have β-ketoreductase and dehydratase and, occasionally, enoyl reductase domains to reduce the ketone groups to various extents (24, 43). The resulting enzymes are said to be reducing and produce linear polyketide compounds. Reducing PKSs often also have a methyltransferase domain. Variability in the type, number, and activity of these domains after each condensation cycle contributes further to diversity of the metabolites generated by fungal PKSs.
Sequence comparisons of the ketosynthase domains also support the existence of the two major groups of PKSs, the reducing PKSs and the nonreducing PKSs (8, 42). Recently, Kroken et al. (34) conducted a thorough phylogenetic analysis of putative PKS genes from the genomes of Neurospora crassa, Cochliobolus heterostrophus, Gibberella moniliformis, Botryotinia fuckeliana, Saccharomyces cerevisiae, Eremothecium gossypii, Schizosaccharomyces pombe, Gibberella zeae, and previously characterized fungal and bacterial PKS genes. This analysis suggests that the filamentous fungi rival the actinomycetes in polyketide number and diversity.
The filamentous fungus Gibberella zeae [Schwein.] Petch (anamorph Fusarium graminearum Schwabe) is a major causal agent of fusarium head blight of wheat and barley worldwide and produces the trichothecene mycotoxin deoxynivalenol, which is a significant contaminant of human food and animal feed (16). The fungus also produces three polyketide-derived mycotoxins, zearalenone (57), aurofusarin (48), and fusarin C (49). Zearalenone is an important source of environmental estrogens when contaminated grain is consumed by humans (35) and may also have effects on sexual development in fungi (41, 61). The toxicological effects of aurofusarin and fusarin C have not been studied extensively. Aurofusarin is known to have adverse effects on avian species; it affects the antioxidant composition and the fatty acid profile of the eggs (21). Fusarin C has been shown to have mutagenic properties in the Ames test. It has also been shown to cause chromosomal aberrations in mammalian cell cultures (26).
We initiated the work described here to identify the genes responsible for production of zearalenone in G. zeae (25). During the investigation, the genomic sequence of G. zeae was released, allowing us to identify 15 PKS genes. To determine the functions of the products of these PKS genes in the life cycle of G. zeae, we have genetically disrupted each of the 15 PKS genes and characterized the mutant isolates. A complete analysis of the phenotypes of the individual mutants suggests a variety of roles, some obvious and others more subtle, for these polyketides in the growth and development of the organism. We also analyzed the expression of each of the genes under a range of developmental, nutritional and pathogenic conditions. This is the first study to genetically characterize a complete set of PKS genes from a single organism.
MATERIALS AND METHODS
Fungal strains and growth conditions.
The strain of G. zeae used for this study was a Michigan field isolate, PH-1 (FGSC 9075, NRRL 31084). All strains were maintained as mycelia in 30% glycerol at −80°C and subcultured on V8 juice medium (56) as needed. PH-1 was also maintained on sterile soil at −20°C.
Identification of PKS genes.
To identify the set of putative PKS genes from the G. zeae genome, we analyzed the first version of the sequence, before automated gene calling had been done. The genomic sequence of G. zeae was obtained from the Broad Institute at the Massachusetts Institute of Technology (http://www.broad.mit.edu/). Ab initio gene identification was accomplished using FGENESH (http://www.softberry.com) with organism-specific parameters for Neurospora crassa (12). The local F. graminearum genome database, predicted open reading frame database, predicted protein database, and expressed sequence tag database were set up using FORMATDB in the NCBI stand-alone BLAST tool (ftp://ftp.ncbi.nih.gov/BLAST/executables/). Sequence similarity searches were applied to the local F. graminearum database using BLASTN, BLASTP, and TBLASTN in the NCBI stand-alone BLAST tool. Default parameters were used but the cutoff E-value was set at 1 instead of 10. The initial queries for the searches used the nucleotide sequences for F. graminearum PKS clones (generated previously using degenerate primers) to perform the BLASTN search against the F. graminearum genome database.
For each PKS gene identified, the predicted protein sequence was used to perform the TBLASTN search against the F. graminearum genome database to identify additional related sequences. Similarly, 12 previously characterized fungal PKS sequences (Aspergillus nidulans wAp, GenBank Q03149; Aspergillus fumigatus ALB1p, GenBank AAC39471; Aspergillus terreus LOVBp, GenBank AAD39830; Aspergillus terreus MSASp, GenBank AAC49814; Aspergillus parasiticus PKSL2p, GenBank AAC23536; Cochliobolus heterostrophus PKS1p, GenBank AAB08104; Colletotrichum lagenarium PKS1p, GenBank BAA18956; Emericella nidulans PKSSTp GenBank Q12397; Gibberella moniliformis FUM1p, GenBank AAD43562; Gibberella fujikuroi PKS4p, GenBank CAD19100; Nodulisporium sp. strain ATCC 74245 PKS1p, GenBank AAD38786; and Penicillium patulum MSASp, GenBank P22367) obtained from NCBI were used to perform TBLASTN and BLASTP, searching against the F. graminearum genome database and predicted protein database.
The 12 known fungal PKS sequences were aligned using the CLUSTAL W (53) default parameters in slow/accurate mode. The multiple alignment was used as input for the HMMBUILD module of HMMER 2.2g (20) to build the profile hidden markov model (HMM). The profile HMM was used to search the F. graminearum predicted protein database using HMMSEARCH. The default cutoff values were used to filter the query results. The 12 PKS sequences were used as input for the motif discovery tool Multiple Expectation-maximization for Motif Elicitation (MEME) to build PKS motif profiles (2). The log-odds matrix of the motif(s) was used to search the F. graminearum predicted protein database by using the Motif Alignment Search Tool (MAST) (3) with default parameters. The search results were also curated manually as described above.
For each putative gene identified, and for known fungal PKS genes used in this study, BLASTP, CDART, and RPS_BLAST searches were applied to the GenBank nr database and the conserved domain database, respectively, using NCBI web services (http://www.ncbi.nlm.nih.gov/BLAST/) to obtain and compare domain structures of these genes. Each putative PKS gene was also used to perform a TBLASTN search against the F. graminearum EST database to investigate the transcription of the predicted genes. At that time, the database consisted of expressed sequence tags (ESTs) from three libraries: perithecium forming, carbon starved, and nitrogen starved (55). Amino acid sequences were analyzed by the PILEUP and PRETTY programs of the Wisconsin Package version 10.3-UNIX (19).
Fungal transformation and gene disruption.
To determine the functions of each of the 15 PKS genes, we genetically disrupted them individually and subsequently analyzed the disrupted transformants for changes in phenotype under a variety of conditions. Disruption vectors were generated by PCR amplification of a 500- to 1750-bp fragment located downstream of the start codon of each predicted gene (Table 1). The amplified fragment was then ligated into the SmaI site of pHYG4, a vector carrying the selectable marker gene for hygromycin B phosphotransferase (hph) (14). These vectors were used to transform the wild-type PH-1 as described below. Genes were disrupted by single-crossover integration of the disruption vector into the homologous portion of the genome.
TABLE 1.
Structures of the predicted polyketide synthase genes in Gibberella zeae
| Gene designationa | Other gene designationb | Kroken designationb | Disruption fragment (contig)a | Reducing or nonreducing | No. of amino acids | Domainsc | Product |
|---|---|---|---|---|---|---|---|
| PLSP1 | FG04488 | PKS15 | 103263-103805 (193) | Nonreducing | 2,172 | KS-AT-ACP (choline/carnitine O-acyltransferase) | Unknown |
| PKS6 | FG12109* | PKS6 | 88677-89734 (329) | Reducing | 2,554 | KS-AT-DH-ME-ER-KR-ACP | Unknown |
| GzFUS1 | FG12100* | PKS10 | 71008-72251 (320) | Reducing | 3,920 | KS-AT-DH-ME-ER-KR-ACP (nonribosomyl peptide synthase) | Fusarin C |
| AUR1 | FG12040* | PKS12 | 134173-134960 (116) | Nonreducing | 2,072 | KS-AT-ACP-TE | Aurofusarin |
| GRS1 | FG03964 | PKS14, 16 | 228126-229359 (168) | Nonreducing | 2,029 | KS-AT-ACP-TE | Unknown |
| PGL1 | FG12125* | PKS3 | 48922-50045 (371) | Nonreducing | 2,287 | KS-AT-ACP-ACP (fatty acyl-CoA reductase; AR) | Perithecium pigment |
| ZEA2 | FG12055* | PKS13 | 6149-7355 (119) | Reducing | 2,345 | KS-AT-DH-ER-KR-ACP | Zearalenone (Gaffoor et al.) |
| ZEA1 | FG02395 | PKS4 | 2628-3760 (118) | Nonreducing | 1,995 | KS-AT-ACP | Zearalenone (Gaffoor et al.) |
| PKS1 | FG10548 | PKS1, 8 | 180638-181700 (441) | Reducing | 2,463 | KS-AT-DH-ER-KR-ACP | Unknown |
| PKS9 | FG12121* | PKS9 | 4984-6117 (436) | Reducing | 2,642 | KS-AT-DH-ME-KR-ACP | Unknown |
| PKS11 | FG01790 | PKS11 | 52197-53594 (93) | Reducing | 2,465 | KS-AT-DH-ME-ER-KR-ACP | Unknown |
| PKS7 | FG08795 | PKS7 | 76385-77657 (355) | Reducing | 2,350 | KS-AT-DH-ER-KR-ACP | Unknown |
| PKS17 | FG03340 | 31914-32951 (152) | Reducing | 2,529 | KS-AT-DH-ME-ER-KR-ACP | Unknown | |
| PKS5 | FG05794 | PKS5 | 303919-311398 (233) | Reducing | 3,177 | KS-AT-DH-ME-ER-KR-ACP (unknown conserved domain) | Unknown |
| PKS2 | FG04694 | PKS2 | 14355-1606 (196) | Reducing | 2,563 | KS-AT-DH-ME-ER-KR-ACP | Unknown |
| KSA1 | FG07226 | 117170-118209 (303) | 427 | KS | Unknown |
Based on the Broad Institute assigned gene designation (www.broad.mit.edu/cgi-bin/annotation/fusarium) and sequence.
The asterisk (*) indicates name revision based on MIPS annotation (mips.gsf.de/genre/proj/fusarium); Kroken designations refer to reference 34.
Domains not usually associated with PKS genes are indicated in parentheses and domains that may be inactive due to mutations in the sequence are italicized. Domain abbreviations: KS, ketosynthase; AT, acyl transferase; DH, dehydratase; ER, enoyl reductase; KR, ketoreductase; ACP, acyl carrier protein; ME, methyl transferase; TE, thioesterase.
Transformations were performed on germinated conidia using a previously published protocol (46) with the following modifications. Approximately 0.3 g soil stock was used to inoculate carboxymethylcellulose (Sigma Chemical Co., St. Louis, MO) medium which was then incubated for 72 h at 25°C at 250 rpm (13). Conidia were harvested by centrifugation and germinated in YEPD (0.3% yeast extract, 1% Bacto peptone, and 2% d-glucose) broth for 12 to 14 h at room temperature at 175 rpm. Isolation of protoplasts occurred in 25 mg/ml driselase (InterSpex Products, Inc. San Mateo, CA), 0.05 mg/ml chitinase (Sigma Chemical Co., St. Louis, MO), and 5 mg/ml lysing enzyme (Sigma Chemical Co., St. Louis, MO) in a 1.2 M KCl buffer. Protoplasts were collected by filtration through a 30-mm Nitex nylon membrane (Tetko Inc., Kansas City, MO) and washed three times in STC buffer (1.2 M sorbitol; 10 mM Tris-HCl, 50 mM CaCl2, pH 8.0).
Transformation was performed in a 500-μl volume with 106 to 108 protoplasts, 25 μg of the disruption vector, and was mediated by 30% polyethylene glycol solution (30% polyethylene glycol 8000, 10 mM Tris-HCl, 50 mM CaCl2, pH 8.0) in STC buffer. Protoplasts were transferred to 250 ml regeneration medium (0.1% yeast extract, 0.1% casein enzyme hydrolysate, 0.8 M sucrose, and 0.75% agarose) and distributed to petri plates (15 ml each). Cultures were incubated for 15 h and then overlaid with 10 ml RM amended with 150 μg/ml hygromycin B (Calbiochem-Novabiochem Corp., San Diego, CA) to recover transformants. Putative transformants were selected within 4 to 7 days and hygromycin resistance was confirmed by growth on V8 juice medium amended with 450 μg/ml hygromycin B. Hygromycin-resistant (Hygr) colonies were transferred to a 2% water agar medium and genetically pure single spore isolates were obtained for further analysis.
Single-spore isolates of putative transformants were screened for gene disruption using Southern blot analysis. Genomic DNA was isolated from putative transformants as previously described (28, 30) and cut independently with two different restriction enzymes. The restriction enzymes chosen for each analysis were paired such that one recognized a unique site within the disruption vector and one recognized sites only outside the vector. Restriction-cut DNA was separated by electrophoresis and Southern analysis was performed for each of the digested samples. Probes were based on the specific amplicon used for gene disruption and a portion of hph (588 bp). Based on Southern data, we selected, for further analysis, two independent disruption mutants for each PKS gene and one transformant that resulted from ectopic integration of each vector. All nucleic acid manipulations and molecular procedures were carried out according to standard methods (47).
Characterization of growth and pathogenicity of PKS mutants.
Transformants were characterized for their ability to produce perithecia and to discharge normal ascospores and their mycelial growth compared to the parent PH-1. Both PKS mutants and the ectopic transformant for each target PKS were center inoculated onto carrot agar (32) in 100-mm petri dishes, and two diameters were measured per plate after 3.5 days of growth to determine the area of the growing colony. The experiment was performed in triplicate for each isolate. A mixed model analysis of variance (SAS PROC UNIVARIATE) was used to compare the growth rates between the mutants and the wild-type. Differences in growth rate were compared using a Tukey's comparison (33). Once the mycelia grew to the edge of the plate, mature perithecia were generated as previously described (32). On day 10 postinoculation, the gross morphology of the perithecia was observed and ascospore discharge was characterized as previously described (54).
Transformants were screened for changes in pathogenicity on wheat. Five greenhouse virulence tests were conducted on the wheat cultivar Wheaton essentially as described (18). Inocula were generated by growth on mung bean liquid medium for 4 days at 28°C with shaking at 200 rpm (1). The macroconidia were filtered, concentrated by centrifugation, rinsed, and suspended in water. Wheat heads were inoculated by injecting a drop of water containing approximately 103 macroconidia into one floret of a spikelet located in the lower third of each head. Ten heads were injected for each treatment and control heads were injected with water. Injected heads were enclosed in plastic bags for three days. Visual disease assessments were recorded at 2- to 3-day intervals 18 to 21 days after inoculation and overall disease severity was calculated as a percentage of blighted spikelets in each head. The wheat heads were allowed to mature, then were harvested and individually threshed to collect seeds.
Chemical analysis of PKS mutants.
Transformants were tested to detect the loss of ability to produce zearalenone, fusarin C, and aurofusarin. To test whether zea was accumulating in the disrupted transformants, we used thin-layer chromatography (TLC). Transformants were grown on Uncle Ben's Original Converted Rice (MasterFoodServices, San Antonio, TX), a vitamin-supplemented parboiled rice found to stimulate zearalenone production (44, 50). Moist, sterile rice (25 g) was inoculated with 106 to 108 conidiospores in petri dishes (100 mm by 25 mm), taped with Tenderskin hypoallergenic paper tape (The Kendall Company, Mansfield, MA), and incubated at room temperature (23 to 25°C) for 1 week. During this period, cultures were monitored to confirm uniform colonization of rice across all samples. After 1 week, the cultures were transferred to an incubator at 11°C for 2 more weeks (51). At harvest, the cultures were dried in an oven at 55°C for 3 to 4 days and ground to a fine powder using a domestic coffee grinder. For TLC analysis, 200 mg of each powdered sample was extracted in 100% methanol for 2 hours and TLC was performed as previously described (51). The area on the TLC plate with an RF similar to that of authentic zearalenone was collected and resuspended in methanol, and a spectrum analysis was performed according to Mirocha et al. (40).
Although the trichothecene mycotoxins deoxynivalenol (DON) and 3-acetyl-DON are not derived from a polyketide, their critical importance in the wheat disease process prompted us to examine if any of the PKS mutants were affected in DON production. Quantitative combined measurements on both mycotoxins were made on transformants grown in liquid culture under DON-inducing conditions (39) and the results were compared to those of the wild-type, PH-1. DON quantification data were generated using the commercial competitive enzyme-linked immunosorbent assay-based Veratox 5/5 kit (Neogen Corp., Lansing, Michigan) and using a standard curve for DON ranging from 0.25 to 3.00 ppm. Values for optical density at 650 nm were measured after the addition of the stop solution to the multiwells. To ensure accuracy, each sample was quantified twice.
DON analysis of transformants grown in planta was performed differently. Following virulence assays, seed samples for all transformants and for wild-type PH-1 were tested for DON. Harvested seeds were ground to a fine powder and extracted with 4 ml of acetonitrile-water (86:14) per gram with shaking for 2 h. The solvent extract was filtered through Whatman filter paper and stored at 4°C until analyzed. The concentration of DON was determined by liquid chromatography-mass spectrometry as described in Baker and Roberts (4).
For fusarin C analysis, transformants and the wild-type isolate were grown in liquid YES medium (yeast extract 2%, sucrose 6%, pH 5.5). An equal volume of acetonitrile was added to 16-day-old cultures. The resulting solution was mixed briefly and filtered through 0.2-μm nitrocellulose filters. The filtrate was examined for the presence of fusarins by HPLC-UV absorbance-mass spectrometry. The high-pressure liquid chromatography (HPLC) system employed an Intersil ODS3 column (10 cm, 5 mm), a flow rate of 0.3 ml/min, and a gradient solvent system beginning with water-methanol 65:35 (vol/vol) and changed to water-methanol 30:70 (vol/vol) over the course of 5 min. The latter solvent composition was held isocratic for 25 min. The HPLC column was coupled to a UV detector and the API source of a Finnigan LCQ Deca mass spectrometry system (ThermoQuest, San Jose, CA) operated in the electrospray ionization (ESI) mode. The presence of fusarins was determined by their retention time and ultraviolet and mass spectra in comparison with authentic fusarin C. For example, the UV spectrum of a peak with a retention time of 19.2 min showed a single broad peak with an absorption maximum of 365 nm that was identical to the spectrum of fusarin C. The mass spectrum of this component was also identical to that of fusarin C and included signals with masses of 432, 454, and 885, which correspond to the MH+, MNa+, and 2MNa+ ions, respectively.
For aurofusarin analysis, transformants and wild-type PH-1 were grown on potato dextrose agar (Difco Laboratories, Detroit, MI) medium for 10 days. A single 10-cm agar plate per transformant was extracted by first cutting the agar into small pieces and soaking with 30 ml of a 95:5 chloroform-methanol solution overnight. The extract was then filtered through Whatman no. 1 paper. The filtrate was examined by HPLC-UV absorbance-mass spectrometry and found to contain a broad peak at 17.0 min with an associated molecular ion of 571 mU. This mass is consistent with that of aurofusarin, which has been characterized (4).
Expression analysis of PKS genes.
To determine when the PKS genes were expressed in wild-type isolate, we employed reverse transcription PCR (RT-PCR). mRNA was harvested from mycelia grown under a range of conditions designed to represent all stages of the life cycle, including growth in the host plant. Gene expression during sexual development was examined using perithecia harvested from carrot agar 4, 5, and 6 days after induction of sexual development, representing very young, near-mature, and mature perithecia, respectively (54). Conidia and mycelia were harvested 3 days after inoculation of carboxymethylcellulose medium, when conidial production is optimal (13). Gene expression during plant infection and colonization was examined using wheat tissue harvested from infected heads 1, 4, and 5 days postinoculation to represent the early infection and rachis colonization (29). Growth on rice agar (41) and moist cracked corn represented the stages at which mycotoxin (particularly zearalenone) production takes place in the field. DON-inducing medium was used to analyze PKS gene expression during production of this nonpolyketide mycotoxin (39).
Gene expression during growth on a variety of standard laboratory growth media was also examined. These conditions included an undefined medium rich in reduced nitrogen (YES) to simulate saprophytic growth under ideal conditions, a natural plant-based medium (potato dextrose broth [PDB]; Difco Laboratories, Detroit, MI), a synthetic, defined minimal medium with nitrate as the sole nitrogen source [Czapek-Dox broth (0.6 g NaNO3, 0.6 g K2HPO4, 0.2 g MgSO4, 0.2 g KCl, 0.01 g FeSO4 · 7H2O, 30 g sucrose per liter)] and starvation conditions (Czapek-Dox broth lacking either a carbon or a nitrogen source). The last three conditions were selected to simulate the conditions encountered as the fungus survives winter in plant debris. Mycelia grown in YES and PDB were harvested at 48 h, when mycelia are actively growing and nutrients are not yet depleted, and at 60 h days postinoculation, when growth slows and nutrients become limited. The latter represents idiophase, when many secondary metabolites are induced (17). The mycelia grown in minimal medium were harvested after 4 days due to slower growth in these media. Mycelia grown in starvation conditions were initiated in standard Czapek-Dox broth for 4 days before being transferred to starvation media, where they were harvested after 24 h incubation (52, 55).
All samples were harvested from two to eight independent replicate cultures, pooled, frozen at −80°C, and lyophilized overnight. RNA was extracted using TRIzol reagent (Invitrogen Life Technology, Carlsbad, CA) and treated with RNase-free DNase I (Roche Diagnostics Corporation, Indianapolis, IN) as per the manufacturer's instructions. First-strand cDNA synthesis was carried out with SuperScript II RNase H reverse transcriptase (Invitrogen Life Technologies, Carlsbad, CA) using random hexamers.
Gene-specific primers (Table 2) were used to amplify regions of each PKS gene to identify the conditions under which they were expressed. The primers used to generate the disruption vector were used to amplify the genes AUR1, GzFUS1, PGL1, PKS2, PKS11, PKS14, and PKS17. For genes KSA1, PLSP1, ZEA1, PKS1, PKS5, PKS6, PKS7, and PKS9, alternate primers were designed. In all cases, except for PKS11 and PLSP1, primers were designed either to amplify a product that included an intron to distinguish between amplicons generated from contaminating DNA and mRNA or to span an exon-exon junction to ensure that no residual DNA was amplified. Although the primers used to amplify the mRNA of PKS11 and PLSP1 could not distinguish between a DNA and mRNA template, they were not redesigned because these two genes had very specific expression patterns, indicating genomic DNA was not being amplified. In all analyses, amplification was repeated three to six times to confirm results. To thoroughly test expression of all 15 PKS genes under conditions representing those encountered during the life cycle, 18 different treatments were designed. Due to this large number of conditions, quantitative PCR was prohibitively expensive.
TABLE 2.
Primers used to generate disruption fragments of polyketide synthase genes
| Gene designation | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|
| Disruption | ||
| AUR1 | GGATTCACCGTGCCCGACAT | TTCAACGCAGAGCTCCATTACGAC |
| GzFUS1 | CTTGGGCCACGGCTGAGATTC | ATTGGCACTTCCTGTCGCTTCC |
| PGL1 | TCCTAGGCGCGTATACGAACCATT | GAGGCCCAGAGCTTGCTTAGTGTG |
| PKS1 | ATCGCTATCGCTATCTGGGCTGTA | AGGTCTTGATTGGCGTGATGTGTT |
| PKS2 | GATGAACGGGCCAACGGGTATGC | ATGGCGCCTGGAGATGATGC |
| PKS5 | TTTGCGCCGAGACGAATCCA | TGCGCCCACCATCCAGACAG |
| PKS6 | GTCCCAGTGCCGGTGCTTGTGA | GTGGAGAACGTTGGAGGCGATGAG |
| PKS7 | CTGCGCAAAGTCTCCGAACAAAAG | TAGCGCGGAAAACCTCAAAACACA |
| PKS9 | AAGTTCGGTGGTTGGGTCTAA | GTGTCTTTGGATACTCGCTTTTG |
| PKS11 | CCCCTGGAATGACTGTGGCAATGA | AGGCGATGACAACACCCGACTGAA |
| GRS1 | ATCAGAAGCGCCCAAGTCAACCA | TGCAACGAGGCGGAATGTATCAC |
| PLSP1 | CAGTCCGTCATCCTCAGTCACAGA | CGGTAGAGATGGCTTGCGATTTC |
| PKS17 | CTGGCGAGTGTGACGGTGCTGTTA | TTGTCGGCGTTGTTCTCCTCGTG |
| ZEA1 | GCAATGCGTCCAGCTCCAAAAG | TCGGTTCACCTCGGTCAAATCCAG |
| ZEA2 | GGAGGATATGGGCGTGGTGAAGGA | GGCTGGCTGATCTCGGGCAAAGT |
| Expression | ||
| PLSP1 | CTGGCGTATGCTGGACTCGGAATG | AGTGTTAGCGCTTGGCCCTGTTGA |
| PKS6 | GGCAAGCCTGGAAGAACGAGTA | GAGCCGATGGGAAGTGGTGAGTCT |
| ZEA1 | GAAGAGGCCCCGGTAGCGATAAC | TGAAGCCACTCCAGCAGCAGATT |
| PKS9 | TTACAAACGGAACGAACGGACTC | GGGCAGCCAGGGAAATACG |
| PKS7 | CGCTGCCTTTGACGCTTCTT | TGATTGCCCTTTAGTCCACGAG |
| PKS5 | GAGGCACATGGAACTGGAACACAA | GGCCCAACCTCGTGAAAACACTC |
| KSA1 | TGAGCAAAGGCGAATGTCCAAGTT | GCCGAAGCCTGCAAATGTCAAG |
| PKS1 | GTCGCGGCTGGCACAACACTGA | ACAAAAAGCTCGCGCCCCATCC |
RESULTS
Identification of PKS genes.
Fifteen putative PKS genes were identified by analyzing the raw genomic sequence. PKS genes were designated using the previously assigned designations (34) except where function has been determined during this analysis. Examination of the sequence using HMMER yielded eight putative PKS genes: GzFUS1, AUR1, PGL1, ZEA2, PKS6 PKS9, PKS11 and PKS17. MEME/MAST applications recognized these eight genes and an additional seven genes: ZEA1, PKS1, PKS2, PKS5, PKS7, GRS1, and PLSP1. All 15 predicted genes had expect values of E of ≤10−90. In addition to the 15 predicted genes, a 16th gene, designated KSA1 (ketosynthase-acyl carrier protein 1), was predicted and had the next lowest E-value of those genes predicted, 10−3. We included KSA1 in our expression analysis for comparison. Based on the high similarity of KSA1 to β-ketoacyl-acyl carrier protein synthases (E = 3 × 10−125, RPS-BLAST 2.2.9) it is unlikely that this enzyme belongs to the PKS family. β-Ketoacyl-acyl carrier protein synthases are generally involved in the elongation step of fatty acid biosynthesis.
The complete set of domains comprising each PKS gene was identified by BLASTX, PILEUP and PRETTY analysis (Table 1). These domains were verified by aligning the predicted protein sequences with domains identified in previously characterized PKSs (36, 45) using CLUSTAL W (53). Domains having amino acids present that disrupted the conserved protein sequence were predicted to be inactive in the mature protein (Table 1). According to the domain analysis there are five nonreducing PKSs (AUR1, GRS1, PGL1, PLSP1, and ZEA1) and 10 reducing PKSs (GzFUS1, ZEA2, PKS1, PKS2, PKS5, PKS6, PKS7, PKS9, PKS11, and PKS17).
To initially test if the predicted genes were transcribed, TBLASTN searches were performed against the G. zeae EST database. Both sequence identity (>75%) and expect value (<E−10) were used as cutoffs to identify ESTs that were specific to one of the PKS gene. For PGL1, 59 ESTs were identified in the perithecia library (50). For PKS11, one EST from the perithecia library was identified. Each of these EST hits had a stretch of at least 30 amino acids identical to either PGL1 or PKS11. These results were consistent with our expression analysis. EST hits for other PKS genes contained only very short stretches of identical amino acids and showed much higher E values.
Using the chromosomal map of G. zeae provided by the Broad Institute, we found that 13 of the PKS genes were scattered across the four chromosomes. ZEA1 and ZEA2 appeared to be divergently transcribed from a shared 1 kb promoter, while AUR1 was located 15 kb upstream from ZEA1.
Functional analysis of PKS genes.
To identify the function of the PKS genes, all 15 genes were disrupted individually using the single-crossover integration approach (Table 1) to cause insertional mutagenesis of the gene. Up to 16 transformed hygromycin-resistant transformants were examined for each gene to identify two disrupted transformants and one ectopic transformant for further analysis. Southern analysis was performed on genomic DNA isolated from each transformant and from the wild type to confirm the location of the inserted plasmid in the disrupted strains (results not shown).
Growth, development, pathogenicity, and mycotoxin production were analyzed for the set of disrupted transformants of each gene to determine how the functional disruption affected normal fungal growth and development. Mycelial growth, as measured by colony area relative to the wild type, was similar to the wild type for transformants disrupted in AUR1, ZEA1, PKS5, PKS17, PLSP1, and GzFUS1. For transformants disrupted in ZEA2, PKS6, PKS7, PGL1, PKS9, PKS1, and PKS11, significant increases in growth (113% to 117%) were observed. Significant decreases in growth were noted for the GRS1 mutants and the PKS2 mutants (86% and 68%, respectively). Disruption of the PKS genes did not affect either perithecium production or function in vitro and perithecia developed and discharged ascospores normally. Disruption of the PKS genes did not significantly affect pathogenicity on wheat, and wild-type disease symptoms were observed throughout almost every inoculated head of both wild-type and PKS mutants.
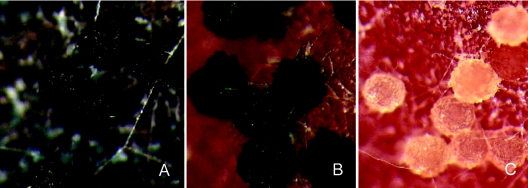
Several of the PKS mutants revealed function based on the loss of a known polyketide. The effects of disruption of ZEA1 and ZEA2 on zearalenone production have been described elsewhere (25). Based on the similarity between GzFUS1 and PKSs from related Fusarium spp. shown to synthesize fusarin C (49), we predicted that GzFUS1 mutants would also be unable to synthesize fusarin C. We found this to be the case as both GzFUS1 mutants failed to synthesize fusarin C, while the ectopic transformant synthesized fusarin C at levels similar to those of the wild type. Similarly, both AUR1 mutants failed to produce aurofusarin, the characteristic red mycelial pigment and mycotoxin, in contrast to ectopic control strains. Examination of the PGL1 mutants indicated that they did not produce the black perithecial pigment characteristic of the perithecia of both the wild-type PH-1 and the ectopic mutant (Fig. 1). The structure of this pigment has not been characterized.
FIG. 1.
PKS mutations affecting perithecium pigmentation. A. AUR1 mutant showing black perithecia and black stroma. B. Wild-type strain showing heavily pigmented, blue-black perithecia on red stroma. C Mutant strain with PGL1 disrupted showing pale perithecia on red stroma. Leica MZ12 stereomicroscope, ×100.
In summary, we disrupted 15 genes and were able to determine the function of five of them: mutants of ZEA1 and ZEA2 revealed two genes responsible for zearalenone production; mutants of AUR1 revealed a gene responsible for aurofusarin biosynthesis; mutants of GzFUS1 revealed a gene responsible for fusarin C biosynthesis and the mutants of PGL1 revealed a gene involved in the biosynthesis of the black perithecial pigment.
Expression analysis of PKS genes.
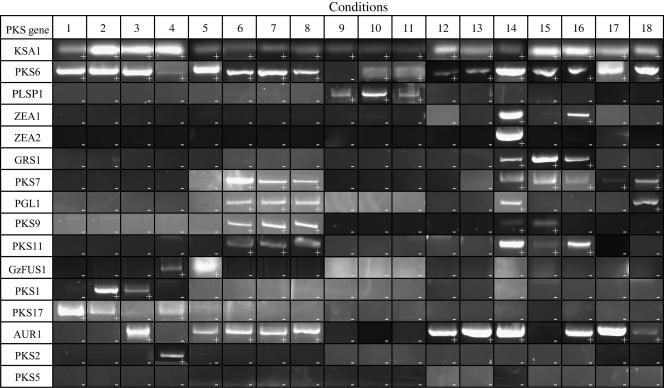
The wild-type PH-1 was grown under 11 different conditions and harvested at one to three time points for each condition. These conditions included three of the conditions (day 6 of perithecial development, carbon and nitrogen starvation) under which the EST libraries were generated. We then harvested total RNA from lyophilized mycelia and used RT-PCR to determine the expression patterns of the 15 PKS genes and, for comparison, KSA1. The results of the expression analysis revealed seven groups of genes with similar expression patterns (Fig. 2). We designated these groups as follows: constitutively expressed (PKS6 and KSA1 also follows this pattern); plant specific (PLSP1); grain specific (GRS1, ZEA1, and ZEA2); sexual development related (PKS7, PGL1, PKS9, and PKS11); mycelial growth related (GzFUS1, PKS1, PKS2, and PKS17); not expressed during infection (AUR1); and expression not detected (PKS5). The sexual development group was also expressed during grain colonization. Interestingly, PKS2, which we relegated to the mycelial growth-related group, was expressed only in YES48, in conditions of active mycelial growth.
FIG. 2.
Expression of putative PKS genes by wild-type G. zeae under various conditions. A representative RT-PCR result is presented. Expression is indicated by the appropriately sized band on an agarose gel (+). Absence of expression is indicated by the lack of an appropriately sized band (−). Culture conditions are indicated as follows: 1) Czapek-Dox, liquid, 10 days; 2) potato dextrose broth, 48 h; 3) potato dextrose broth, 60 h; 4) yeast extract sucrose, liquid, 48 h; 5) yeast extract sucrose, liquid, 60 h; 6 to 8) carrot agar, 4, 5, and 6 days after treatment to induce perithecial development, respectively; 9 to 11) 48, 72, and 96 h postinoculation of wheat head, respectively; 12) carbon starvation conditions; 13) nitrogen starvation conditions; 14) rice agar, 9 days; 15 and 16) corn meal, 48 h and 96 h, respectively; 17) carboxymethylcellulose medium, 4 days; 18) DON-inducing medium.
DISCUSSION
Here we present the first comprehensive functional analysis of all predicted PKS genes from a single filamentous fungus. Our work shows that polyketides affect many aspects of fungal biology, are expressed throughout the life cycle, and are not excluded from actively growing mycelia (Fig. 2). Although the patterns of expression appear to be distinct among the different genes, it is worthwhile to broadly group the genes by expression patterns (Fig. 2), to provide a means for pursuing further functional analyses. For example, GRS1 is expressed during grain colonization, similarly to ZEA1 and ZEA2. All three may function to establish the fungus in the grain and protect that resource. We are currently exploring the functional relationship of polyketides within the expression groups by stacking PKS mutations to determine if the effects of disruption are correspondingly enhanced by the disruption of an entire expression group.
To determine the function of individual PKS genes, each was disrupted independently and the resulting mutants examined for changes in development and pathogenicity. Interestingly, PLSP1 was expressed exclusively during plant infection and PLSP1 mutants remained pathogenic on inoculated wheatheads. PLSP1 appeared to have a nonfunctional acyl carrier protein domain (Table 1) and therefore may not produce an active compound in the wild type. Disruption of GRS1 and PKS2 each resulted in mycelial growth inhibition. Expression of PKS2 was detected only during the early stages of active mycelial growth in YES (Fig. 2). In Aspergillus parasiticus, disruption of the FLUP PKS gene inhibited mycelial growth, eliminated asexual sporulation, and significantly reduced aflatoxin production (62), but the mechanism of these effects has not been reported. The phenotype of the GRS1 and PKS2 mutants leaves open the possibility of their function as regulators of mycelial proliferation. GzFUS1 was also expressed exclusively during mycelial growth. In Magnaporthe grisea, ACE1, a homologue of GzFUS1, was shown to be an avirulence factor recognized specifically by resistant rice (10). Such an interaction has not been observed for GzFUS but supports the hypothesis that a pool of polyketides may be a resource in fungi for adapting to a variety of environments.
Based on the striking phenotype of the disrupted mutants, we identified AUR1 as the PKS responsible for the biosynthesis of the carmine pigment aurofusarin in G. zeae (48). Recent reports were consistent with our findings (31, 37). The two pigment producing genes, AUR1 and PGL1 were both expressed during fruiting body development (Fig. 1 and 2), where they may provide UV and/or desiccation protection for the developing ascospores. The expression of the PGL1 product was affected by the loss of AUR1, as stroma cells produced the black pigment in the AUR1 mutants (Fig. 1). This may be an example of functional redundancy between these pigments, Expression of PKS5 was not observed under the conditions tested and disruption of this gene did not yield a distinctive phenotype. It is possible that PKS5 is expressed either under highly specialized conditions hitherto untested or at very low levels.
The domain analysis of the predicted PKSs revealed the presence of all of the expected domains as well as a few domains not previously described in PKSs. The presence of an acyl-coenzyme A reductase domain in PGL1 and a carnitine O-acyltransferase domain in PLSP1 have not previously been described for type I PKSs. The predicted function of the thioesterase, acyl-coenzyme A reductase, and carnitine O-acyltransferase domains is the same: release of the polyketide from the enzyme. The thioesterase (or cyclase) domain is thought to play a role in cyclizing the nascent carbon chain (24). The acyl-coenzyme A reductase domain may function to release the enzyme and generate a terminal aldehyde (27); while the carnitine O-acyltransferase domain may function to transfer the polyketide to carnitine or a related molecule. PGL1 contains two acyl carrier protein motifs. The presence of a second acyl carrier protein has been observed previously in the fungal PKS wAp (24), but the function of this apparent redundancy is unknown. The subset of reducing PKSs that contained a nonfunctional methyltransferase domain (PKS1, ZEA2, and PKS7) supports previous work suggesting that the ancestral domain structure of these PKSs was ketosynthase-acyltransferase- dehydratase-methyltransferase-enoyl reductase-ketoreductase-acyl carrier protein (34). The pattern of amino acid insertions and deletions scattered throughout the nonfunctional, remnant methyltransferase domains is conserved in the three PKSs and suggests that they share a common ancestor.
Kroken et al. (34) described 16 putative PKS genes in G. zeae in their analysis of several fungal genome sequences, which was based primarily on an analysis of a preliminary 2X coverage of the genomic sequence. Reexamination of their data indicated that PKS14 and PKS16 both corresponded to the GRS1 described here. In addition, PKS1 and PKS8 as described by Kroken et al. encoded a single PKS, which we designated PKS1. Therefore, Kroken et al. (34) described fourteen PKS genes in G. zeae and did not recognize PKS17. The genome analyzed by Kroken et al. (34) and the sequence generated by the Broad Institute each were based on different isolates of G. zeae, and although there is no reason to assume the sequences might vary significantly, it is possible these differences are due to small sequence variations as observed in the trichothecene biosynthetic genes by Ward et al. (58).
There has been speculation regarding the physiological costs of producing extra metabolites that do not contribute directly to the health of the organism (15), but that may serve as auxiliary pools for novel compounds. The results presented here suggest that individual mutations in 7 of 15 PKS genes result in mutants with increased growth on a rich, sterile medium. However, the present data do not preclude the possibility that accumulation of other metabolites, resulting from disruption of a pathway in these mutants, affects growth. In the field, G. zeae is most often associated with plant tissue that it has initially parasitized and must subsequently defend as the plant senesces. In the analysis of PKS genes by Kroken et al. (34), the only fungi that did not have PKS genes in the genome were the yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe. Unicellular yeasts are found in sugary environments, which they colonize when the resource appears, and then become dormant as the sugars become scarce.
Members of the polyketide-rich actinomycetes (15) and the filamentous fungi presented by Kroken and colleagues are soil inhabitants. Fungi, such as G. zeae, that function in several niches, including plant pathogen and soil saprophyte, may require a pool of secondary metabolites from which to develop protective mechanisms. As for functions for these secondary metabolites, the biological activity of the products of 6 of the 15 PKSs are described here. Six others had significant effects on mycelial growth in the PKS mutants, implying a direct or indirect effect on fungal physiology. The latter group should be the object of more intensive efforts to determine their products. As genomic sequences of fungi from a broad group of lifestyles become available, comparisons among studies of PKS genes will further our understanding of the evolution and ecological significance of this diverse group of compounds.
Acknowledgments
We thank our collaborators on the G. zeae sequencing project: Bruce Birren and Li-Jun Ma (Broad Institute, M.I.T.), H. Corby Kistler (USDA, Minnesota), and Jin-Rong Xu (Purdue University). We also thank Lan Xiao of the CANR Biometry Group, Statistical Consulting Center, Michigan State University, for statistical analysis of growth data. Assistance with the DON analyses was generously provided by Benjamin Munn and L. Patrick Hart.
This project was funded by USDA-NRI grant 2001-352-1-10062 to F.T. and the Michigan Agricultural Experiment Station.
REFERENCES
- 1.Bai, G. H., and G. Shaner. 1996. Variation in Fusarium graminearum and cultivar resistance to wheat scab. Plant Dis. 80:975-979. [Google Scholar]
- 2.Bailey, T. L., and C. Elkan. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2:28-36. [PubMed] [Google Scholar]
- 3.Bailey, T. L., and M. Gribskov. 1998. Combining evidence using p-values: application to sequence homology searches. Bioinformatics 14:48-54. [DOI] [PubMed] [Google Scholar]
- 4.Baker, P. M., and J. C. Roberts. 1966. Studies in mycological chemistry. Part XXI. The structure of aurofusarin, a metabolite of some Fusarium species. J. Chem. Soc. 23:2234-2237. [Google Scholar]
- 5.Bennett, J. W. 1983. Differentiation and secondary metabolism in mycelial fungi, p. 1-32. In J. W. Bennett and A. Ciegler (ed.), Secondary metabolism and differentiation in fungi. Marcel Dekker, New York, NY.
- 6.Bennett, J. W. 1995. From molecular genetics and secondary metabolism to molecular metabolites and secondary genetics. Can. J. Botany 73:S917-S924. [Google Scholar]
- 7.Bhatnagar, D., K. C. Ehrlich, and T. E. Cleveland. 2003. Molecular genetic analysis and regulation of aflatoxin biosynthesis. Appl. Microbiol. Biotechnol. 61:83-93. [DOI] [PubMed] [Google Scholar]
- 8.Bingle, L. E. H., T. J. Simpson, and C. M. Lazarus. 1999. Ketosynthase domain probes identify two subclasses of fungal polyketide synthase genes. Fungal Genet. Biol. 26:209-223. [DOI] [PubMed] [Google Scholar]
- 9.Bode, H. B., B. Bethe, R. Höfs, and A. Zeeck. 2002. Big effects from small changes: possible ways to explore nature's chemical diversity. Chembiochem. 3:619-627. [DOI] [PubMed] [Google Scholar]
- 10.Böhnert, H. U., I. Fudal, W. Dioh, D. Tharreau, J. L. Notteghem, and M. H. Lebrun. 2004. A putative polyketide synthase/peptide synthetase from Magnaporthe grisea signals pathogen attack to resistant rice. Plant Cell 16:2499-2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bu'Lock, J. D. 1961. Intermediary metabolism and antibiotic synthesis. Adv. Appl. Microbiol. 3:293-342. [DOI] [PubMed] [Google Scholar]
- 12.Burset, M., I. A. Seledtsov, and V. V. Solovyev. 2001. SpliceDB: database of canonical and non-canonical mammalian splice sites. Nucleic Acids Res. 29:255-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cappellini, R. A., and J. L. Peterson. 1965. Macroconidium formation in submerged cultures by a non-sporulating strain of Gibberella zeae. Mycologia 57:962-966. [Google Scholar]
- 14.Carroll, A. N., J. A. Sweigard, and B. Valent. 1994. Improved vectors for selecting resistance to hygromycin. Fungal Genet. Newsl. 41:22. [Google Scholar]
- 15.Challis, G. L., and D. A. Hopwood. 2003. Synergy and contingency as driving forces for the evolution of multiple secondary metabolite production by Streptomyces species. Proc. Natl. Acad. Sci. USA 100:14555-14561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Čonková, E., A. Laciaková, G. Kováč, and H. Seidel. 2003. Fusarial toxins and their role in animal diseases. Vet. J. 165:214-220. [DOI] [PubMed] [Google Scholar]
- 17.Demain, A. L. 1992. Microbial secondary metabolism: a new theoretical frontier for academia, a new opportunity for industry., p. 3-23. Secondary metbolites: their function and evolution (Ciba Foundation Symposium 171). John Wiley and Sons, Chichester, England. [DOI] [PubMed]
- 18.Desjardins, A. E., D. W. Brown, S. H. Yun, R. H. Proctor, T. Lee, R. D. Plattner, S. W. Lu, and B. G. Turgeon. 2004. Deletion and complementation of the mating type (MAT) locus of the wheat head blight pathogen Gibberella zeae. Appl. Environ. Microbiol. 70:2437-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Devereux, J., P. Haeberli, and O. Smithies. 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Durbin, R., S. Eddy, A. Krogh, and G. Mitchison. 1998. Biological sequence analysis: Probabilistic models of proteins and nucleic acids. Cambridge University Press., Cambridge, England.
- 21.Dvorska, J. E., P. F. Surai, B. K. Speake, and N. H. C. Sparks. 2001. Effect of the mycotoxin aurofusarin on the antioxidant composition and fatty acid profile of quail eggs. Br. Poult. Sci. 42:643-649. [DOI] [PubMed] [Google Scholar]
- 22.Firn, R. D., and C. G. Jones. 2000. The evolution of secondary metabolism—a unifying model. Mol. Microbiol. 37:989-994. [DOI] [PubMed] [Google Scholar]
- 23.Foster, J. W. 1949. Chemical Activities of the fungi. Academic Press, New York, NY.
- 24.Fujii, I., A. Watanabe, U. Sankawa, and Y. Ebizuka. 2001. Identification of claisen cyclase domain in fungal polyketide synthase WA, a naphthopyrone synthase of Aspergillus nidulans. Chem. Biol. 8:189-197. [DOI] [PubMed] [Google Scholar]
- 25.Gaffoor, I., and F. Trail. Characterization of two distinct polyketide synthase genes involved in zearalenone biosynthesis in Gibberella zeae. Submitted. [DOI] [PMC free article] [PubMed]
- 26.Gelderblom, W. C. A., P. G. Thiel, W. F. O. Marasas, and K. J. Vandermerwe. 1984. Natural occurrence of Fusarin C, a mutagen produced by Fusarium moniliforme in corn. J. Agric. Food Chem. 32:1064-1067. [Google Scholar]
- 27.Ishige, T., A. Tani, K. Takabe, K. Kawasaki, Y. Sakai, and N. Kato. 2002. Wax ester production from n-alkanes by Acinetobacter sp. strain M-1: ultrastructure of cellular inclusions and role of acyl coenzyme A reductase. Appl. Environ. Microbiol. 68:1192-1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jurgenson, J. E., R. L. Bowden, K. A. Zeller, J. F. Leslie, N. J. Alexander, and R. D. Plattner. 2002. A genetic map of Gibberella zeae (Fusarium graminearum). Genetics 160:1451-1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kang, Z. S., and H. Buchenauer. 2000. Cytology and ultrastructure of the infection of wheat spikes by Fusarium culmorum. Mycol. Res. 104:1083-1093. [Google Scholar]
- 30.Kerényi, Z., K. Zeller, L. Hornok, and J. F. Leslie. 1999. Molecular standardization of mating type terminology in the Gibberella fujikuroi species complex. Appl. Environ. Microbiol. 65:4071-4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim, J. E., K. H. Han, H. M. Jin, H. Kim, J. C. Kim, S. H. Yun, and Y. W. Lee. 2005. Putative polyketide synthase and laccase genes for biosynthesis of aurofusarin in Gibberella zeae. Appl. Environ. Microbiol. 71:1701-1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klittich, C. J. R., and J. F. Leslie. 1988. Nitrate reduction mutants of Fusarium moniliforme (Gibberella fujikuroi). Genetics 118:417-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kramer, C. Y. 1956. Extension of multiple range tests to group means with unequal numbers of replications. Biometrics 12:307-310. [Google Scholar]
- 34.Kroken, S., N. L. Glass, J. W. Taylor, O. C. Yoder, and B. G. Turgeon. 2003. Phylogenomic analysis of type I polyketide synthase genes in pathogenic and saprobic ascomycetes. Proc. Natl. Acad. Sci. USA 100:15670-15675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuiper-Goodman, T., P. M. Scott, and H. Watanabe. 1987. Risk assessment of the mycotoxin zearalenone. Regul. Toxicol. Pharmacol. 7:253-306. [DOI] [PubMed] [Google Scholar]
- 36.Linnemannstons, P., J. Schulte, M. D. Prado, R. H. Proctor, J. Avalos, and B. Tudzynski. 2002. The polyketide synthase gene pks4 from Gibberella fujikuroi encodes a key enzyme in the biosynthesis of the red pigment bikaverin. Fungal Genet. Biol. 37:134-148. [DOI] [PubMed] [Google Scholar]
- 37.Malz, S., M. N. Grell, C. Thrane, F. J. Maier, P. Rosager, A. Felk, K. S. Albertsen, S. Salomon, L. Bohn, W. Schäfer, and H. Giese. 2005. Identification of a gene cluster responsible for the biosynthesis of aurofusarin in the Fusarium graminearum species complex. Fungal Genet. Biol. 42:420-433. [DOI] [PubMed] [Google Scholar]
- 38.Mayorga, M. E., and W. E. Timberlake. 1992. The developmentally regulated Aspergillus nidulans wA gene encodes a polypeptide homologous to polyketide and fatty acid synthases. Mol. Gen. Genet. 235:205-212. [DOI] [PubMed] [Google Scholar]
- 39.McCormick, S. P., L. J. Harris, N. J. Alexander, T. Ouellet, A. Saparno, S. Allard, and A. E. Desjardins. 2004. Tri1 in Fusarium graminearum encodes a P450 oxygenase. Appl. Environ. Microbiol. 70:2044-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mirocha, C. J. S. V., C. M. Pathre, and Christensen. 1977. Zearalenone, p. 345-36. In J. V. Rodricks, C. W. Hesseltine, and M. A. Mehlman (ed.), Mycotoxins in human and animal health. Pathotox Pulishers, Inc., Park Forest South, IL.
- 41.Nelson, R. R. 1971. Hormonal Involvement in Sexual Reproduction in the fungi with special reference to F-2, a fungal estrogen, p. 181-200. In S. Adai and S. Ouchi (ed.), Morphological and biochemical events in plant-parasite interaction. Phytopathological Society of Japan, Tokyo, Japan.
- 42.Nicholson, T. P., B. A. M. Rudd, M. Dawson, C. M. Lazarus, T. J. Simpson, and R. J. Cox. 2001. Design and utility of oligonucleotide gene probes for fungal polyketide synthases. Chem. Biol. 8:157-178. [DOI] [PubMed] [Google Scholar]
- 43.O'Hagan, D. 1991. The polyketide metabolites. Ellis Horwood Limited, West Sussex, England.
- 44.Pathre, S. V., P. V. Khadikar, and C. J. Mirocha. 1989. Biosynthesis of zearalenone—a simple and efficient method to incorporate [13C]acetate label by using solid cultures. Appl. Environ. Microbiol. 55:1955-1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Proctor, R. H., A. E. Desjardins, R. D. Plattner, and T. M. Hohn. 1999. A polyketide synthase gene required for biosynthesis of fumonisin mycotoxins in Gibberella fujikuroi mating population A. Fungal Genet. Biol. 27:100-112. [DOI] [PubMed] [Google Scholar]
- 46.Proctor, R. H., T. M. Hohn, and S. P. McCormick. 1995. Reduced virulence of Gibberella zeae caused by disruption of a trichothecene toxin biosynthetic gene. Mol. Plant-Microbe Interact. 8:593-601. [DOI] [PubMed] [Google Scholar]
- 47.Sambrook, J., D. W. Russell, and J. Sambrook. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 48.Shibata, S., E. Morishita, T. Takeda, and K. Sakata. 1966. The structure of aurofusarin. Tetrahedron Lett. 7:4855-4860. [Google Scholar]
- 49.Song, Z. S., R. J. Cox, C. M. Lazarus, and T. J. Simpson. 2004. Fusarin C biosynthesis in Fusarium moniliforme and Fusarium venenatum. Chembiochem 5:1196-1203. [DOI] [PubMed] [Google Scholar]
- 50.Steele, J. A., J. R. Lieberman, and C. J. Mirocha. 1974. Biogenesis of zearalenone (F-2) by Fusarium roseum graminearum. Can. J. Microbiol. 20:531-534. [DOI] [PubMed] [Google Scholar]
- 51.Swanson, S. P., R. A. Corley, D. G. White, and W. B. Buck. 1984. Rapid thin-layer chromatographic method for determination of zearalenone and zearalenol in grains and animal feeds. J. AOAC Int. 67:580-582. [PubMed] [Google Scholar]
- 52.Talbot, N. J., H. R. K. McCafferty, M. Ma, K. Moore, and J. E. Hamer. 1997. Nitrogen starvation of the rice blast fungus Magnaporthe grisea may act as an environmental cue for disease symptom expression. Physiol. Mol. Plant Pathol. 50:179-195. [Google Scholar]
- 53.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. Clustal W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Trail, F., and R. Common. 2000. Perithecial development by Gibberella zeae: a light microscopy study. Mycologia 92:130-138. [Google Scholar]
- 55.Trail, F., J. R. Xu, P. San Miguel, R. G. Halgren, and H. C. Kistler. 2003. Analysis of expressed sequence tags from Gibberella zeae (anamorph Fusarium graminearum). Fungal Genet. Biol. 38:187-197. [DOI] [PubMed] [Google Scholar]
- 56.Tuite, J. 1969. Plant pathological methods, fungi and bacteria. Burgess Publishing Co., Minneapolis, Minn.
- 57.Urry, W. H., H. L. Wehrmeister, E. B. Hodge, and P. H. Hidy. 1966. The structure of zearalenone. Tetrahedron Lett. 7:3109-3114. [Google Scholar]
- 58.Ward, T. J., J. P. Bielawski, H. C. Kistler, E. Sullivan, and K. O'Donnell. 2002. Ancestral polymorphism and adaptive evolution in the trichothecene mycotoxin gene cluster of phytopathogenic Fusarium. Proc. Natl. Acad. Sci. USA 99:9278-9283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Watanabe, A., Y. Ono, I. Fujii, U. Sankawa, M. E. Mayorga, W. E. Timber-lake, and Y. Ebizuka. 1998. Product identification of polyketide synthase coded by Aspergillus nidulans wA gene. Tetrahedron Lett. 39:7733-7736. [Google Scholar]
- 60.Weinberg, E. D. 1971. Secondary metabolism—raison d′être. Perspect. Biol. Med. 14:565. [DOI] [PubMed] [Google Scholar]
- 61.Wolf, J. C., and C. J. Mirocha. 1973. Regulation of sexual reproduction in Gibberella zeae (Fusarium roseum graminearum) by F-2 (zearalenone). Can. J. Microbiol. 19:725-734. [DOI] [PubMed] [Google Scholar]
- 62.Zhou, R., R. Rasooly, and J. E. Linz. 2000. Isolation and analysis of fluP, a gene associated with hyphal growth and sporulation in Aspergillus parasiticus. Mol. Gen. Genet. 264:514-520. [DOI] [PubMed] [Google Scholar]