Abstract
Although autophagy is characteristic of type II programmed cell death (PCD), its role in cell death is currently debated. Both cell death-promoting and prosurvival roles of autophagy have been reported depending on the organism and the cell type. In filamentous fungi, a cell death reaction known as an incompatibility reaction occurs when cells of unlike genotype fuse. Cell death by incompatibility is characterized by a dramatic vacuolar enlargement and cell lysis. In Podospora anserina, autophagy is induced early during this cell death reaction. Cell death by incompatibility in Podospora is a model of type II PCD used here to assess the role of autophagy in this type of cell death. We have inactivated PaATG1, the Podospora ortholog of the Saccharomyces cerevisiae ATG1 gene involved in the early steps of autophagy in yeast. The ΔPaATG1 mutant displays developmental defects characteristic of abrogated autophagy in Podospora. Using the green fluorescent protein-PaATG8 autophagosome marker, we show that autophagy is abolished in this mutant. Neither cell death by incompatibility nor vacuolization are suppressed in ΔPaATG1 and ΔPaATG8 autophagy mutants, indicating that a vacuolar cell death reaction without autophagy occurs in Podospora. Our results thus provide a novel example of a type II PCD reaction in which autophagy is not the cause of cell death. In addition, we found that cell death is accelerated in ΔPaATG null mutants, suggesting that autophagy has a protective role in this type II PCD reaction.
Macroautophagy, generally referred to as autophagy, occurs coincidently with cell death in type II programmed cell death (PCD) (17). Autophagy has been first described as a cellular response to nutrient starvation. This intracellular catabolic system allows degradation and recycling of long-lived proteins and organelles in eukaryotic cells (44). During this process, the cytoplasmic material is sequestered in double-membrane vesicles called autophagosomes (44). These vesicles then fuse by their outer membrane to the vacuolar/lysosomal membrane and deliver single-membrane vesicles called autophagic bodies inside the lumen of these organelles for degradation. The autophagic/vacuolar type of cell death (type II) is defined by morphological features that are the presence of autophagosomes or autophagic bodies inside the lysosome/vacuole degradative compartment with or without large vacuoles (8). Thus, type II PCD is defined as autophagic cell death, but this classification does not imply that autophagy directly contributes to cell death. In fact, two opposing views on the role of autophagy in type II PCD have emerged. Autophagy is either viewed as a medium of cellular demise or, on the contrary, as a prosurvival mechanism. Autophagy is also associated with various diseases in humans, including cancer and neurodegenerative disorders (25). Whether autophagy protects from or causes disease is also unclear (45).
The molecular dissection of autophagy has been mostly performed in Saccharomyces cerevisiae and led to the identification of the autophagy-related, or ATG, genes (26). The high degree of conservation of these genes simplified the identification of orthologs in other organisms and the access to their autophagic machinery. The inactivation of these orthologs in higher eukaryotes reveals potential important functions of autophagy in development, stress-induced adaptation, and aging (35).
Cell death by incompatibility in the filamentous fungus Podospora anserina is a cell death reaction associated with autophagy (42). This cell death reaction is ubiquitous in filamentous fungi and occurs in heterokaryotic cells, resulting from the fusion between cells that differ genetically at specific loci called het loci (47). This phenomenon constitutes a mechanism of self/nonself recognition which prevents heterokaryosis between unlike individuals. One beneficial effect of limiting heterokaryosis could be to protect natural populations against the horizontal transfer of mycoviruses (4, 9). The presence of large vacuoles before the cells lyse is the main morphological feature of cell death by incompatibility. At the molecular level, this reaction is associated with the induction of a specific set of genes named idi genes (induced during incompatibility) (6, 13, 41). The idi-6/pspA gene encodes a vacuolar serine protease involved in autophagic body degradation (41, 42). The idi-7/PaATG8 gene (hereafter called PaATG8) is the ortholog of the yeast ATG8 and the human LC-3 autophagy genes (42). The green fluorescent protein (GFP)-PaATG8 fusion protein has been used as a cytological marker of autophagy, as it relocalizes from the cytoplasm to the vacuole when autophagy is induced. Several lines of evidence indicate that autophagy is induced early during cell death by incompatibility (42). This cell death reaction is an autophagic/vacuolar cell death reaction used here as model of type II PCD (17). The aim of the present paper is to determine the role of autophagy in this model of autophagic cell death and, in particular, to establish whether it causes cell death or if, on the contrary, it displays a prosurvival role.
To determine the role of autophagy in this type of death, we chose to genetically inhibit autophagy in Podospora by inactivating autophagy genes. Here we characterized PaATG1, the ortholog of the S. cerevisiae ATG1 gene essential for autophagosome formation (19, 32, 37). We found that autophagy does not occur and that autophagosomes are not formed in a ΔPaATG1 null mutant. This mutant displays the same developmental defects as other Podospora autophagy mutants. Using the ΔPaATG1 and ΔPaATG8 null mutations, we demonstrated that neither cell death nor vacuolization depend on autophagy in Podospora and that cell death is accelerated when autophagy is suppressed, suggesting a protective role for autophagy in this cell death reaction.
MATERIALS AND METHODS
Strains and media.
The s wild-type strain was used as a reference strain. The thermosensitive self-incompatible het-R het-V strain can be isolated in the progeny of a cross between het-R het-V1 and het-r het-V strains, as described by Bourges et al. (6). The preparation and transformation of protoplasts were performed as described previously (3). Synthetic SU medium used to observe pigmentation of the mycelium and differentiation of aerial hyphae was prepared as described elsewhere (42). SA and nitrogen-deprived SA media were as described previously (12). Solid D0 medium, used for observation of fruiting bodies in the fertilization experiment, is a corn meal agar medium (14). In the fertilization experiment, the ΔPaATG mutants were used as females. The strains were grown in the dark for 5 days at 26°C on solid D0 medium. The plates were then transferred to the light to trigger differentiation of sexual organs. Three days later, cultures were overlaid with a suspension containing wild-type male cells of the opposite mating type. The liquid was discarded, and the plates were checked after a few days for the development of fruiting bodies triggered by the fertilization of female organs, termed protoperithecia, by male cells. The male cells were collected after the addition of sterile water to plates containing the wild-type strain grown under the same conditions as the female cells.
Plasmids and plasmid construction.
pGEM-T (Promega) and pCR4-TOPO (Invitrogen) vectors were used for the cloning of amplicons. pCSN43 was used as a template to amplify the hph gene encoding resistance to hygromycin (49). pBluescript II SK+ (Stratagene) plasmid was used for PaATG1 cloning. The pPable plasmid bears the bleomycin resistance gene and was used in cotransformation experiments (11). For the generation of ΔPaATG1 and ΔPaATG8 strains, pBSPaATG1::hph and pBSPaATG8::hph plasmids were constructed. The Escherichia coli GA0AB123BH09 clone belongs to the genomic DNA library constructed by Genoscope (Evry, France) to sequence the Podospora genome. The corresponding plasmid bears about 10 kbp of DNA carrying the PaATG1 locus. This locus was subcloned on an 8-kbp XbaI-SmaI fragment into the pBluescript II SK+ vector and named pBSPaATG1. This plasmid was used in a PCR experiment using divergent oligonucleotides which cover the region upstream of the initiation codon (oli223) and downstream of the stop codon (oli224) of the PaATG1 open reading frame (ORF). The oli223 and oli224 sequences are, respectively, 5′-GGTTAATTAAGGCTACCTGCTGATGTTGGC-3′ and 5′-GGTTAATTAAACGTCTGACGATCCCAAAGC-3′. PacI restriction sites (underlined in the sequences) were introduced in the 5′ half of each oligonucleotide. The amplification was performed with the Expand 20kbPlus PCR system (Roche). The PCR product was restricted with PacI, circularized with T4 DNA ligase, and transformed into DH5α competent cells. A 2.4-kbp fragment containing the hph gene was amplified by PCR using oligonucleotides 107 and 108, introducing PacI sites, and pCSN43 vector as a template. Oligonucleotide 107 and 108 sequences are 5′-GGTTAATTAAGTCGACAGAAGATGATATTG-3′ and 5′-GGTTAATTAATCTAGAAAGAAGGATTACCTC-3′ (PacI restriction sites are underlined in the sequences), respectively. The amplicon was restricted with PacI and cloned into the unique PacI site of the vector obtained previously. The final construct pBSPaATG1::hph contained the hph gene and 3.1 kbp and 2.9 kbp of PaATG1 flanking sequences on the 5′ and 3′ sides, respectively. The pBSPaATG8::hph plasmid was constructed from the pCBidi-7::ura5 plasmid described elsewhere (42). The insert of this plasmid restricted with SalI and SmaI was cloned into pBluescript II SK+ vector. This pBSPaATG8::ura5 plasmid was restricted with PacI and circularized with T4 DNA ligase in the presence of the hph amplicon restricted with PacI to exchange the ura5 gene for the hph gene and construct the pBSPaATG8::hph plasmid. This final construct contained the PaATG8 gene and 2.5 kbp and 2.9 kbp of PaATG8 flanking sequences on the 5′ and 3′ sides, respectively. For complementation of the ΔPaATG1 and ΔPaATG8 strains, the pBSPaATG1 and pGPD-idi7 plasmids described herein and elsewhere were used in cotransformation experiments with the pPable plasmid (42).
DNA and RNA procedures.
Standard molecular techniques followed the recommendation of Sambrook et al. (46). Genomic DNA and RNA from P. anserina were extracted as described previously (6, 33). To map the 3′ end of PaATG1 transcript and the introns, rapid amplification of cDNA end analyses were performed using the GeneRacer kit (Invitrogen). The first intron was mapped using the following oligonucleotides: 5′-ATCGATATGGCCGCCGACAGACAGTTATC-3′ and 5′-GTGCGTGGCCGACTCGACACAGT-3′. The internal oligonucleotide used for 3′-end amplification and mapping of the second intron was 5′-CGGGTGCCGAAGGAGTCATCCTT-3′. Restriction enzymes and other modification enzymes were purchased from Promega. Sequencing was performed by Genome Express (Grenoble, France).
Construction of PaATG1 and PaATG8 mutant strains.
The inactivation of PaATG1 was achieved by gene replacement both in the s reference strain and the self-incompatible het-R het-V strain. Indeed, the construction of the self-incompatible het-R het-V ΔPaATG1 strains by a cross between the het-R het-V strain and the ΔPaATG1 strain could not be performed due to the low fertility of crosses between strains bearing incompatible het-R and het-V genes and to the female sterility conferred by the ΔPaATG mutations (30, 42). Similarly, the het-R het-V ΔPaATG8 strain could not be obtained by a cross with the previously described Δidi-7/ΔPaATG8 strain (42). This strain was thus obtained by repeating the gene replacement strategy in a het-R het-V genetic background.
The pBSPaATG1::hph plasmid was used to transform protoplasts from two hygromycin-sensitive strains: the wild-type s strain and the self-incompatible het-R het-V strain. The pBSPaATG8::hph plasmid was used to transform protoplasts from the het-R het-V strain. The gene replacement event by double crossover will lead to the integration of the hph marker in place of the PaATG1 or PaATG8 ORF, respectively. Homologous recombination is not exclusive in Podospora, and because the double crossover is less frequent than single recombination events, the desired event is minor among the transformed population. From each of the transformation experiments, 250 hygromycin-resistant transformants were subcultured on synthetic SU medium. This medium was used to compare the growth of wild-type and autophagy mutant strains, as some phenotypic traits of these mutants, such as lack of mycelial pigmentation and differentiation of aerial hyphae, appear clearly on this medium. Transformants displaying this phenotype (4 among 250 in each case) were selected, and their DNA was analyzed by Southern blotting (not shown) to identify strains in which gene replacement had occurred. Two of these transformants contained nuclei bearing the corresponding null allele. Homokaryotic strains bearing either the ΔPaATG1 null mutation or the ΔPaATG8 null mutation corresponding, respectively, to the inactivated PaATG1 or PaATG8 locus were obtained, as verified by Southern blot analysis (not shown). Complementation of the phenotypic defects of the ΔPaATG1 strains and the ΔPaATG8 strain were obtained by transformation with the pBSPaATG1 and pBSPaATG8XbaEco plasmids, respectively. Transformants were selected on phleomycin (5 μg/ml)-containing media (CAYLA, France). Strains expressing the GFP-PaATG8 fusion protein were obtained by transformation with the pCBgfp-idi7 plasmid described elsewhere (42).
Light and fluorescence microscopy.
For time course of cell death in wild-type and ΔPaATG self-incompatible (SI) strains, the percentage of dead cells was measured. At different times before and after transfer to 26°C, mycelia were labeled with Evans blue and observed. For each time point, about 1,000 articles (fungal cells) were analyzed. The percentage of cell death corresponds to the percentage of stained cells. For time-lapse video microscopy, Podospora was first cultured on a cellophane sheet. This sheet was transferred onto medium embedded in a plastic core directly on a slide to avoid desiccation.
The mycelia were observed with a Leica DMRXA microscope equipped with a Micromax charge-coupled device (Princeton Instruments). Congo red, Nile red, and Evans blue staining were performed as described previously (12). Images of the GFP-PaATG8 protein were acquired with a Marianas system (Intelligent Imaging Innovations, Inc., Denver, CO). Mycelium was observed in a fully automated Zeiss 200 M inverted microscope (Carl Zeiss, Thornwood, NY) equipped with an MS-2000 stage (Applied Scientific Instrumentation, Eugene, OR), a Lambda LS 175-W xenon light source (Sutter, Novato, CA), and a 100× 1.4NA Plan-Apochromat objective using an Endow GFP longpass emission filter set (Ex HQ470/40, Em HQ500lp, BS Q495lp; Chroma Technology Corp, Rockingham, VT). Images were acquired using a CoolSnap HQ camera (Roper Scientific, Tucson, AZ).
Nucleotide sequence accession number.
The sequence of the PaATG1 ORF was determined and deposited in GenBank under accession no. AY953520.
RESULTS
Molecular characterization of the PaATG1 gene.
To suppress the autophagic process, we chose to inactivate the ortholog of the yeast ATG1 gene in Podospora. The yeast ATG1 gene is involved in the early induction steps of autophagy, and its inactivation leads to the absence of autophagy (22). Mutation of the ATG1 ortholog in Dictyostelium discoideum also abrogates autophagy (39). Atg1p is a serine-threonine kinase composed of an N-terminal kinase domain and a C-terminal domain without any known motif. To identify the Podospora ATG1 ortholog, we analyzed the P. anserina genome sequence database (http://podospora.igmors.u-psud.fr) with the yeast Atg1p sequence using the BLAST algorithm. The identified Podospora anserina ATG1 ortholog has been named PaATG1. Mapping of the introns was performed, and the sequence of the ORF was determined (GenBank accession no. AY953520). The predicted PaATG1 protein is 941 amino acids long and displays a protein kinase domain (amino acids 26 to 331) and a calculated molecular mass of 104 kDa. PaATG1 and Atg1p sequence display strong homology (34% identity and 48% similarity), particularly in the N-terminal part (49% identity and 67% similarity).
The ΔPaATG1 strain has the phenotypic traits of other Podospora autophagy mutants.
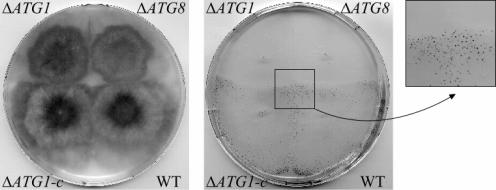
Inactivation of PaATG1 was achieved by gene replacement in the s reference strain (see Materials and Methods). The ΔPaATG1 strain had a slightly slower growth rate (about 10% less) than the wild type. On rich media, this mutant displayed a lower density of aerial hyphae than the wild type and a decreased pigmentation of the mycelium compared to the wild type (Fig. 1). Moreover, the ΔPaATG1 strain produced no protoperithecia (female reproductive organs). This mutant is thus female sterile (Fig. 1). These different phenotypic traits could be suppressed by transforming the ΔPaATG1 strain with a wild-type copy of the PaATG1 gene (Fig. 1). The ΔPaATG1 strain displays the same defects as the ΔpspA and Δidi-7/PaATG8 strains (42). In other words, the ΔPaATG1 strain displays the phenotypic traits of Podospora autophagy mutants. This result confirms that autophagy is required for differentiation of female reproductive organs in P. anserina and thus plays an essential role in fungal development. Differentiation of female organs, occurring during starvation, has to rely on endogenous sources for nutrient supply. The function of autophagy would be to provide amino acid pools from the degradation of preexisting proteins. Thus, as described for other organisms, autophagy is required for differentiation steps occurring under nutrient-limiting conditions (38, 40, 42).
FIG. 1.
Similar differentiation defects between the ΔPaATG1 and the ΔPaATG8 autophagy mutant strains. WT, the s reference strain; ΔATG8, the ΔPaATG8 strain; ΔATG1, the ΔPaATG1 strain; ΔATG1-c, the ΔPaATG1 strain transformed with the wild-type PaATG1 gene. Left panel: phenotype of mycelia. Note that the ΔATG1-c mycelium has a wild-type phenotype. Right panel: differentiation of protoperithecia. Strains are used as female and have been fertilized by wild-type (WT) male cells of the opposite mating type (see Materials and Methods). Fertilization of WT and ΔATG1-c female organs (protoperithecia) by male cells triggers the development of fruiting bodies (black dots, see the enlarged view). Fruiting bodies are observed on the mycelia of strains that support differentiation of female organs. The ΔPaATG8 and ΔPaATG1 mutants do not support differentiation of female organs.
Autophagy is not induced and autophagosomes are not formed in a ΔPaATG1 strain.
In Podospora, autophagy is induced upon starvation or rapamycin treatment, as in other eukaryotes, and is also highly induced during the incompatibility reaction (12, 42). As in yeast and other eukaryotes, when autophagy is induced, the PaATG8 protein redistributes from the cytoplasm to the membranes of autophagosomes and finally reaches the vacuolar lumen after fusion of the autophagosome to the vacuole (23, 24, 42). The corresponding GFP fusion protein thus labels autophagosomes and the vacuolar lytic compartment and constitutes an autophagy marker. To analyze induction of autophagy in the ΔPaATG1 strain, we analyzed the localization of the GFP-PaATG8 autophagy marker during the incompatibility reaction.
Cell death by incompatibility is investigated using the SI thermosensitive het-R het-V strain (29). In this experimental setting, cell death by incompatibility occurs as a generalized process rather than a cell death reaction limited to the fusion cell. This SI strain bears the two incompatible and conditional het-R and het-V genes. At the permissive temperature (32°C), the strain grows normally. The transfer to a nonpermissive temperature (26°C) triggers a generalized cell death reaction. We constructed the self-incompatible het-R het-V ΔPaATG1 gfp-PaATG8 strain (SI ΔPaATG1 gfp-PaATG8) and the control SI gfp-PaATG8 strain and compared the localization of the GFP-PaATG8 fusion protein (Fig. 2A). In both strains grown at permissive temperature, the GFP-PaATG8 protein had mainly a diffuse cytoplasmic distribution and was also occasionally associated with unidentified dot-like structures. These dot-like structures were often larger in the mutant than in the wild type. In the wild type, GFP-PaATG8 localized also to a tubular network corresponding to the tubular vacuoles observed under favorable growth conditions (10, 42). This localization might reflect the basal level of autophagy. Basal activation of autophagy is also observed in yeast and mammalian cells under favorable growth conditions (25). After transfer to the restrictive temperature, in the wild type, GFP-PaATG8 fluorescence localized to the vacuoles and to autophagosomes. Autophagososmes appeared as ring-shaped cytoplasmic structures of 500 nm to 2.1 μm (Fig. 2A). The vacuolar localization of GFP-PaATG8 and the presence of autophagosomes indicated that autophagy was induced in the wild-type strain. In the SI ΔPaATG1 mutant GFP-PaATG8, fluorescence remained cytoplasmic and associated with unidentified dot-like structures that were generally larger and more abundant than at permissive temperature. Importantly, the GFP-PaATG8 protein was excluded from the vacuoles and the ring-shaped fluorescent structures were never observed. These observations indicate that autophagy does not occur and that autophagosomes are not formed in this mutant. A wild-type distribution of GFP-PaATG8 protein was observed in the SI ΔPaATG1 strain transformed with a wild-type copy of the PaATG1 gene (not shown).
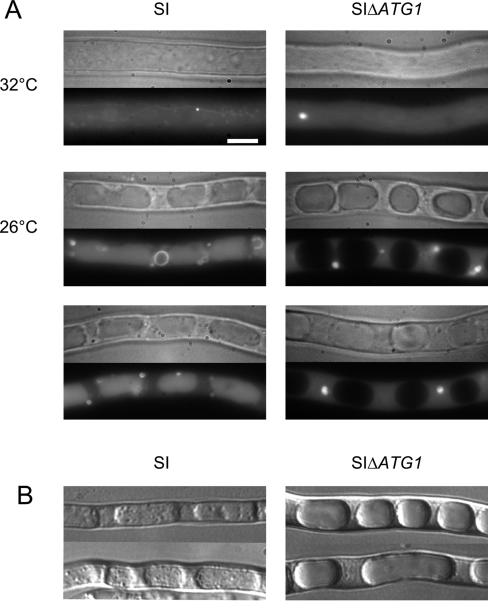
FIG. 2.
Autophagy is abrogated in the ΔPaATG1 mutant strain. (A) The SI gfp-PaATG8 (SI) and SI ΔPaATG1 gfp-PaATG8 (SIΔATG1) strains were grown for 16 h at permissive temperature and transferred to 26°C to trigger the incompatibility reaction. The mycelium was observed by light microscopy (top) and by fluorescence microscopy (bottom) in each panel. Bar 3 μm. Observations were performed before transfer (32°C) and 4 h after transfer to 26°C (26°C). (B) The SI (SI) and SI ΔPaATG1 (SIΔATG1) strains were grown for 16 h at 32°C and transferred to 26°C on fresh medium containing 4 mM PMSF for 6 h before observation by light microscopy.
Induction of autophagy can be evidenced also by the presence of autophagic bodies inside the vacuole when the vacuolar proteases involved in their degradation are inhibited with phenylmethylsulfonyl fluoride (PMSF) (42). The vacuolar content of SI and SI ΔPaATG1 strains was observed after transfer at restrictive temperature on PMSF-containing medium (Fig. 2B). In the presence of PMSF, the vacuoles of the SI strain contained numerous autophagic bodies, while the vacuoles of the SI ΔPaATG1 strain were optically empty. This indicates that autophagic bodies are not formed in the SI ΔPaATG1 mutant. Autophagic bodies were observed in presence of PMSF in the vacuoles of the SI ΔPaATG1 strain transformed with a wild-type copy of the PaATG1 gene (not shown).
Together, the above experiments demonstrate that autophagy does not occur in the SI ΔPaATG1 mutant. We conclude that PaATG1 is required for autophagosome formation and autophagy in Podospora.
Vacuolization is a PaATG-independent process.
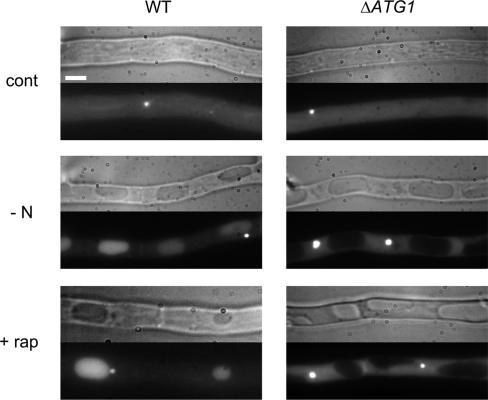
Cellular vacuolization is a main morphological feature of cell death by incompatibility. Vacuolization is also observed upon starvation and rapamycin treatment and is associated in each case with the occurrence of autophagy (12, 42). Rapamycin is a specific inhibitor of the Tor kinase (20). Inhibition of Tor mimics nitrogen starvation and leads to the induction of autophagy in Podospora as in most organisms (12). To determine whether vacuolization is dependent on the autophagic process, we analyzed vacuole formation in the ΔPaATG1 mutant and the wild-type control under conditions of starvation, rapamycin treatment, and incompatibility. Spherical vacuoles were observed in both the wild-type and mutant SI strains during the course of the incompatibility reaction (Fig. 2), while autophagy occurred only in the wild type (Fig. 2). Extensive vacuolization of the cells occurred in both the wild-type and mutant strains upon starvation and rapamycin treatment (Fig. 3). The vacuolar localization of GFP-PaATG8 fluorescence indicated that autophagy was induced upon starvation and rapamycin treatment in the wild type. We could not observe ring-shaped autophagosomes under these conditions, indicating that they were much less abundant than during cell death by incompatibility. The large punctate structures localized in the vicinity of the vacuoles might correspond to autophagosomes or to preautophagosomal structures (50). In the ΔPaATG1 mutant, the GFP-PaATG8 protein did not relocalize upon starvation and rapamycin treatment, indicating that autophagy was not induced (Fig. 3). Again, the dot-like structures seemed larger and more abundant upon starvation and rapamycin treatment than under control conditions and were not perivacuolar. A similar localization of the GFP-ATG8 autophagy marker was observed in the yeast Δatg1 mutant upon rapamycin treatment (51). We conclude that starvation- and rapamycin-induced vacuolization occurred in the ΔPaATG1 mutant without autophagy. Vacuolization took place as efficiently in the autophagy ΔPaATG1 mutant as in the wild type under the three tested conditions (Fig. 2; Fig. 3). This indicates that the morphological modifications of the vacuolar compartment occurring during cell death by incompatibility, starvation, or rapamycin treatment do not require autophagy, thus dissociating autophagy and vacuolization in Podospora.
FIG. 3.
Vacuolization is an ATG-independent mechanism. The gfp-PaATG8 (WT, left) and ΔPaATG1 gfp-PaATG8 (ΔATG1, right) strains were grown on rich medium (SA medium) for 16 h and then transferred for 4 h on fresh SA medium (cont), nitrogen-deprived SA medium (−N), or SA medium containing rapamycin (500 ng/ml) (+ rap). Bar, 3 μm. The Nomarski view is shown on the top of each panel, and the GFP fluorescence of the GFP-PaATG8 protein is shown at the bottom.
Cell death by incompatibility does not require autophagy.
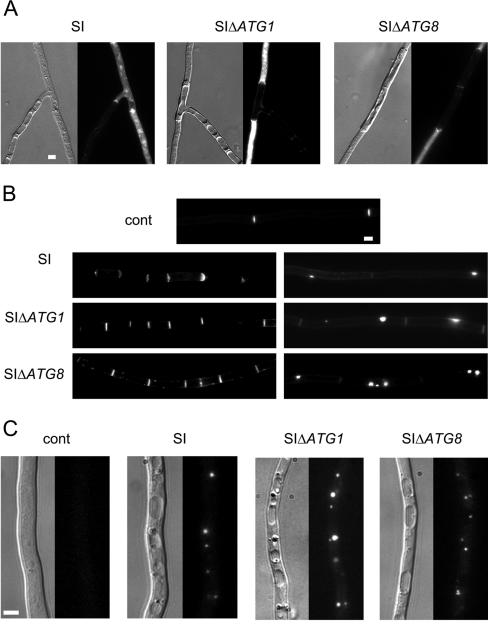
To investigate the role of autophagy in cell death by incompatibility, we constructed the het-R het-V ΔPaATG8 self-incompatible strain (SI ΔPaATG8) in addition to the SI ΔPaATG1 strain (see Materials and Methods). The SI ΔPaATG strains were compared to the wild-type SI strain. For the three strains, the growth stopped rapidly after transfer to restrictive temperature. Cell death was monitored using Evans blue dye, which stains dead cells (Fig. 4A). Cell death occurred in the autophagy mutants. Cell death by incompatibility is characterized by a number of cytological modifications (increased formation of cross walls, abnormal cell wall thickenings, and accumulation of lipid bodies) (12). All of these cytological modifications were also observed in the ΔPaATG SI strains (Fig. 4B and C). Because cell death still occurs in mutants, we conclude that autophagy is not required for this type II PCD reaction.
FIG. 4.
ΔPaATG null mutations do not suppress cell death by incompatibility. Mycelia of SI (SI), SI ΔPaATG1 (SIΔATG1), and SI ΔPaATG8 (SIΔATG8) strains were grown for 16 h at the permissive temperature and then transferred or not (control [cont]) at restrictive temperature. The strains were observed 7 h later. Bar, 3 μm. (A) Vacuolization and cell death occur in all SI strains. For each panel, the Nomarski view is shown on the left and dead cells were revealed using Evans blue dye fluorescence on the right. (B) Congo red labeling allows detection of cross walls on the left and abnormal cell wall deposition on the right. (C) Lipid droplets revealed with Nile red lipophilic dye. Note their accumulation associated with incompatibility. For each panel, the Nomarski view is given on the left and fluorescence of Nile red is shown on the right.
Cell death by incompatibility is accelerated in autophagy mutants.
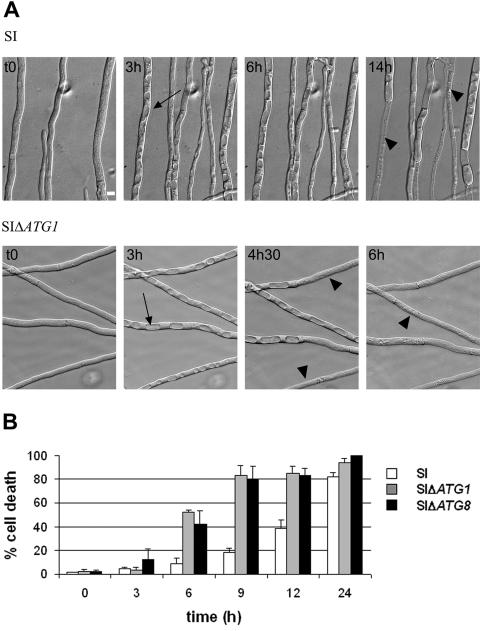
To compare the time course of cell death in the wild type and the ΔPaATG1 mutant, we used video microscopy (Fig. 5A; see Videos SSI and SSI delta PaAtg1 in the supplemental material). The SI (control) and SI ΔPaATG1 strains were cultivated for 1 day on SU medium at 32°C, and then the cultures were examined by microscopy at the restrictive temperature. Important morphological changes to the vacuolar compartment occurred as soon as 45 min in both strains. Spherical vacuoles were observed in place of the classical tubular vacuolar network (10, 42). This vacuolization phenomenon is asynchronous. The spherical vacuoles invaded most of the cell volume and fused, and finally, cell lysis occured. Cell death became apparent after 7 h in the SI strain and earlier (4 h, 30 min) in the ΔPaATG1 strain. All of the ΔPaATG1 cells were dead after 6 h, while noticeable cell survival is still observed after 14 h in the control. The percentage of cell death was then quantified in the wild-type, ΔPaATG1, and ΔPaATG8 SI strains (Fig. 5B). Globally, the rate of cell death was increased in the ΔPaATG mutants. At 6 h after transfer, about 50% of the cells were dead in the ΔPaATG mutants, while the cell death rate only reached 10% in the control. About 80% of cell death occurred 9 h after transfer for the SI ΔPaATG strains, while in the control, the same percentage was only observed 24 h after transfer. No cell death was observed for the three strains maintained at 32°C (data not shown). We conclude that cell death is accelerated in the autophagy mutants and propose that autophagy protects the cells against death in this system.
FIG. 5.
Time course of cell death in autophagy mutants. (A) Nomarski views of mycelium of the self-incompatible strain (SI) or the SI ΔPaATG1 strain (SIΔATG1) grown at 32°C and transferred for the indicated times at restrictive temperature. The most striking event is a rapid increase in the number and size of spherical vacuoles (black arrows). This leads to the invasion of the cellular content by the vacuolar compartment. Finally, cells lyse probably as a result of vacuolar membrane rupture (black arrowheads). Bar, 3 μm. These views were extracted from Videos SSI and SSI delta Atg1 presented in the supplemental material. Images were acquired at 5-min intervals over the course of 14 and 6 h after the temperature drop. The movies are accelerated 4,500-fold. (B) Time course of death in wild-type (SI) and ΔPaATG1 (SIΔATG1) and ΔPaATG8 (SIΔATG8) SI strains. The percentage of cell death corresponds to the percentage of Evans blue-labeled cells measured at different times after transfer to restrictive temperature. Results are mean ± standard deviations of the results from three experiments.
DISCUSSION
Autophagy is a process of protein and organelle degradation by the vacuole (lysosome). This process is conserved in organisms as different as yeast and humans and functions as a cell survival mechanism during nutrient starvation. Autophagy is also associated with type II cell death. Type II PCD is defined by morphological characteristics, namely the presence of autophagosomes (autophagic vacuoles) in the dying cells. The presence of these structures led to the hypothesis that autophagy might have a causative role in autophagic cell death. This idea was in direct contradiction with the proven role of autophagy as a cell survival mechanism. Determining the relationship between autophagy, cell death, and cell survival would allow resolution of this apparent paradox (1). Therefore, the precise role of autophagy in various examples of type II cell death reactions needs to be established. Is autophagy causing the cell death or just accompanying it, or can it even have a prosurvival function?
Here we have used cell death by incompatibility as a model to study autophagic cell death. This cell death reaction is a genetically controlled programmed cell death reaction ubiquitous in filamentous fungi which is accompanied with a strong induction of autophagy in Podospora (42). Podospora is a filamentous fungus, of the ascomycete phylum, capable of differentiation and endowed with a small sequenced haploid genome which does not seem to encode the apoptotic machinery. Studying cell death in this genetically tractable nonanimal eukaryote may reveal mechanisms that are phylogenetically conserved, experimentally more accessible, and less redundant than in animal cells (17).
Role of autophagy in type II cell death mechanisms.
Many of the yeast ATG genes that regulate autophagy have been conserved through evolution. It is now possible to use genetic approaches in model organisms to investigate the role of autophagy during type II cell death. We have identified the Podospora PaATG1 gene involved in autophagosome formation. The ΔPaATG1 mutant is blocked early in the autophagic process and is thus suitable for investigating the role of autophagy. We found that inactivation of autophagy does not suppress cell death by incompatibility, indicating that autophagy does not play a causal role in this type II PCD reaction. The role of autophagy in cell death has also been investigated by ATG1 gene inactivation in another lower eukaryote, namely the protist D. discoideum (27). In this case also, autophagy is not required for cell death. However, the protist atg1 mutant undergoes a cell death reaction without vacuolization (nonvacuolar type of cell death), while the Podospora ΔPaATG1 mutant undergoes a cell death reaction accompanied by vacuolization (vacuolar type of cell death). PaATG gene deletions do not suppress vacuolization in Podospora, while no vacuoles were observed in the Dictyostelium atg1 mutant. This difference might reflect the fact that vacuolization occurs by different mechanisms in these organisms. Our present results demonstrate that cell death is accelerated in the Podospora autophagy mutants. A similar situation is also observed in Dictyostelium (P. Golstein, personal communication). Together, these observations point to autophagy as a protective mechanism during type II cell death in lower eukaryotes. The role of ATG genes has been also investigated in higher eukaryotes by gene inactivation by either gene replacement or RNA interference (RNAi). A prosurvival role of autophagy has been evidenced. ATG genes function in vivo as negative regulators of PCD in the plant hypersensitive response (HR), and RNAi of ATG genes triggers apoptosis in starved mammalian cells (7, 36). This suggests a conservation of this protective effect of autophagy through evolution. These results contrast with findings in three different mammalian models where ATG genes are required to promote cell death (43, 48, 52). Rubinsztein et al. proposed that both roles can be performed depending on cell context (45). Even caspases, normally causal in apoptosis, may be important for cell survival under certain conditions where death by autophagy may prevail (52, 53). Investigation of ATG gene function during cell death revealed that different types of cell death have been classified as type II. This type of cell death has to be further subdivided into cell death with autophagy where autophagy accompanies cell death, as cell death in Podospora or the hypersensitive response in plants, and cell death by autophagy where a causal link exists between cell death and autophagy, as for example, in mouse L929 cells (52). In these cells, caspase-8 inhibition induced autophagic cell death, and this death could be inhibited by RNAi of atg6 and atg7 genes.
Role of vacuoles in cell death by incompatibility.
We observed here that vacuolization of the cells occurs as efficiently in ΔPaATG strains as in the wild type, indicating that autophagy is not required for vacuolization. Thus, the large and numerous autophagosomes revealed by the GFP-PaATG8 marker during cell death by incompatibility in the wild type do not contribute significantly to the vacuole enlargement. The cell death without autophagy that operates in Podopsora PaATG mutants is still associated with a strong vacuolization of the cells, suggesting the contribution of the vacuolar compartment and its lytic activities to cell death. A vacuolar enlargement is described in developmental programmed cell death in plants (28). This enlargement precedes the vacuolar collapse, which causes the release of sequestered hydrolytic enzymes and allows them to attack organelles, leading to cell death. The concept of a similar cell death mechanism where lysosomes and cathepsins function as executioners is emerging in mammals (18, 34). The build-up of various degradative activities, one of which is the vacuolar Podospora serine protease A protease, is a hallmark of cell death by incompatibility, and in video microscopy, cell demise often coincides with vacuolar rupture (2, 5, 41). Vacuolization is a common feature of cell death by incompatibility observed also in Neurospora crassa, where bursting of these vacuoles is apparent (16). It is therefore likely that vacuole membrane permeabilization or rupture, followed by liberation of lytic enzymes and acidification of the cytoplasm, is responsible for cell death by incompatibility in filamentous fungi.
Biological role of the prosurvival function of autophagy.
Autophagy retards cell death by incompatibility in Podospora. Autophagy might promote cell survival in different ways. It could eliminate “prodeath” signals, as for example, a toxic complex formed between incompatible het gene products or damaged organelles, such as mitochondria. The fact that autophagy negatively regulates cell death by incompatibility raises the question of the biological significance of a simultaneous induction of cell death and cell survival mechanisms. Cell death by incompatibility is viewed as a fungal host defense mechanism protecting against the transfer of mycoviruses (4, 9). When PCD occurs as a host defense in the HR in plants or in apoptosis triggered by viral infection, the cell death reaction has to be precisely spatially restricted (15, 31). Liu et al. have shown that autophagy negatively regulates PCD in the plant hypersensitive response and functions to restrict HR PCD to the site of pathogen infection (36). Like the HR, cell death by incompatibility is precisely spatially restricted. Cell death by incompatibility occurs when strains of different het genotype fuse, but this cell death reaction is limited to the heterokaryotic cell resulting from the fusion and never spreads throughout the parental strains, despite their syncytial structure. In Podospora, the hyphae are divided in plurinucleated cells, termed articles, separated by perforated cross walls, ensuring cytoplasmic continuity between articles. To survive an incompatible interaction, fungal strains have to carefully control PCD. Soon after the fusion, the syncytial structure is lost as perforated cross walls bordering the heterokaryotic article are occluded by the Woronin bodies (21). The induction of autophagy in cells surrounding the heterokaryotic article could contribute together with cross wall sealing to the protection of neighboring cells from cell death. Autophagy may be required to eliminate “prodeath” signal(s) or damaged organelles moving out of the heterokaryotic cell into adjacent homokaryotic cells. It will be technically challenging but of great interest to determine whether, in the absence of ATG genes, the PCD reaction spreads beyond the heterokaryotic article to adjacent cells.
We show here that the type II vacuolar/autophagic cell death observed in Podospora is a cell death mechanism with autophagy and not by autophagy. In other words, autophagy accompanies but is not the cause of cell death by incompatibility. We show also that autophagy negatively regulates cell death by incompatibility, as cell death is accelerated in PaATG mutants, suggesting a role for autophagy in cell survival. Protection against PCD could correspond to a conserved function of autophagy shared by fungi, plants, and mammals.
Supplementary Material
Acknowledgments
We thank Robert Debuchy and the Genoscope (Evry, France) for the kind gift of the clone GA0AB123BH09, Pierre Golstein for giving us access to unpublished data and for helpful discussions, Sven J. Saupe for helpful discussions and for critical reading of the manuscript, Jérôme Serand for help in vector construction and gene disruption, and Quentin Fournier for help in strain construction.
This work was supported by the European Commission (Transdeath, contract no. 511983). B.P.-L. was funded by the Ministère de la Recherche.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Baehrecke, E. H. 2005. Autophagy: dual roles in life and death? Nat. Rev. Mol. Cell Biol. 6:505-510. [DOI] [PubMed] [Google Scholar]
- 2.Bégueret, J., and J. Bernet. 1973. Protoplasmic incompatibility: possible involvement of proteolytic enzymes. Nat. New Biol. 243:94-96. [PubMed] [Google Scholar]
- 3.Bergès, T., and C. Barreau. 1989. Heat-shock at elevated temperature improves transformation efficiency of protoplasts from Podospora anserina. J. Gen. Microbiol. 135:601-604. [DOI] [PubMed] [Google Scholar]
- 4.Biella, S., M. L. Smith, J. R. Aist, P. Cortesi, and M. G. Milgroom. 2002. Programmed cell death correlates with virus transmission in a filamentous fungus. Proc. R. Soc. Lond. B 269:2269-2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boucherie, H., and J. Bernet. 1978. Protoplasmic incompatibility and self-lysis in Podospora anserina: enzymes activities associated with cell destruction. Can. J. Bot. 56:2171-2176. [Google Scholar]
- 6.Bourges, N., A. Groppi, C. Barreau, C. Clavé, and J. Bégueret. 1998. Regulation of gene expression during the vegetative incompatibility reaction in Podospora anserina: characterization of three induced genes. Genetics 150:633-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boya, P., R. A. Gonzalez-Polo, N. Casares, J. L. Perfettini, P. Dessen, N. Larochette, D. Metivier, D. Meley, S. Souquere, T. Yoshimori, G. Pierron, P. Codogno, and G. Kroemer. 2005. Inhibition of macroautophagy triggers apoptosis. Mol. Cell. Biol. 25:1025-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bursch, W. 2001. The autophagosomal-lysosomal compartment in programmed cell death. Cell Death Differ. 8:569-581. [DOI] [PubMed] [Google Scholar]
- 9.Caten, C. E. 1972. Vegetative incompatibility and cytoplasmic infection in fungi. J. Gen. Microbiol. 72:221-229. [DOI] [PubMed] [Google Scholar]
- 10.Cole, L., D. A. Orlovich, and A. E. Ashford. 1998. Structure, function, and motility of vacuoles in filamentous fungi. Fungal Genet. Biol. 24:86-100. [DOI] [PubMed] [Google Scholar]
- 11.Coppin, E., and R. Debuchy. 2000. Co-expression of the mating-type genes involved in internuclear recognition is lethal in Podospora anserina. Genetics 155:657-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dementhon, K., M. Paoletti, B. Pinan-Lucarré, N. Loubradou-Bourges, M. Sabourin, S. J. Saupe, and C. Clavé. 2003. Rapamycin mimics the incompatibility reaction in the fungus Podospora anserina. Eukaryot. Cell 2:238-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dementhon, K., S. J. Saupe, and C. Clavé. 2004. Characterization of IDI-4, a bZIP transcription factor inducing autophagy and cell death in the fungus Podospora anserina. Mol. Microbiol. 53:1625-1640. [DOI] [PubMed] [Google Scholar]
- 14.Esser, K. 1974. Podospora anserine, p. 531-551. In R. C. King (ed.), Handbook of genetics. Plenum Press, New York, N.Y.
- 15.Everett, H., and G. McFadden. 1999. Apoptosis: an innate immune response to virus infection. Trends Microbiol. 7:160-165. [DOI] [PubMed] [Google Scholar]
- 16.Glass, N. L., and I. Kaneko. 2003. Fatal attraction: nonself recognition and heterokaryon incompatibility in filamentous fungi. Eukaryot. Cell 2:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Golstein, P., L. Aubry, and J. P. Levraud. 2003. Cell-death alternative model organisms: why and which? Nat. Rev. Mol. Cell Biol. 4:798-807. [DOI] [PubMed] [Google Scholar]
- 18.Guicciardi, M. E., M. Leist, and G. J. Gores. 2004. Lysosomes in cell death. Oncogene 23:2881-2890. [DOI] [PubMed] [Google Scholar]
- 19.Harding, T. M., A. Hefner-Gravink, M. Thumm, and D. J. Klionsky. 1996. Genetic and phenotypic overlap between autophagy and the cytoplasm to vacuole protein targeting pathway. J. Biol. Chem. 271:17621-17624. [DOI] [PubMed] [Google Scholar]
- 20.Heitman, J., N. R. Movva, and M. N. Hall. 1991. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science 253:905-909. [DOI] [PubMed] [Google Scholar]
- 21.Jedd, G., and N. H. Chua. 2000. A new self-assembled peroxisomal vesicle required for efficient resealing of the plasma membrane. Nat. Cell Biol. 2:226-231. [DOI] [PubMed] [Google Scholar]
- 22.Kamada, Y., T. Funakoshi, T. Shintani, K. Nagano, M. Ohsumi, and Y. Ohsumi. 2000. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J. Cell Biol. 150:1507-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim, J., W. P. Huang, and D. J. Klionsky. 2001. Membrane recruitment of Aut7p in the autophagy and cytoplasm to vacuole targeting pathways requires Aut1p, Aut2p, and the autophagy conjugation complex. J. Cell Biol. 152:51-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kirisako, T., M. Baba, N. Ishihara, K. Miyazawa, M. Ohsumi, T. Yoshimori, T. Noda, and Y. Ohsumi. 1999. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J. Cell Biol. 147:435-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klionsky, D. J. 2005. The molecular machinery of autophagy: unanswered questions. J. Cell Sci. 118:7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klionsky, D. J., J. M. Cregg, W. A. Dunn, Jr., S. D. Emr, Y. Sakai, I. V. Sandoval, A. Sibirny, S. Subramani, M. Thumm, M. Veenhuis, and Y. Ohsumi. 2003. A unified nomenclature for yeast autophagy-related genes. Dev. Cell 5:539-545. [DOI] [PubMed] [Google Scholar]
- 27.Kosta, A., C. Roisin-Bouffay, M. F. Luciani, G. P. Otto, R. H. Kessin, and P. Golstein. 2004. Autophagy gene disruption reveals a non-vacuolar cell death pathway in Dictyostelium. J. Biol. Chem. 279:48404-48409. [DOI] [PubMed] [Google Scholar]
- 28.Kuriyama, H., and H. Fukuda. 2002. Developmental programmed cell death in plants. Curr. Opin. Plant Biol. 5:568-573. [DOI] [PubMed] [Google Scholar]
- 29.Labarère, J. 1973. Properties of an incompatibility system in Podospora anserina fungus and value of this system for the study of incompatibility. C. R. Acad. Sci. D 276:1301-1304. [PubMed] [Google Scholar]
- 30.Labarère, J., J. Bégueret, and J. Bernet. 1974. Incompatibility in Podospora anserina: comparative properties of the antagonistic cytoplasmic factors of a nonallelic system. J. Bacteriol. 120:854-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lam, E. 2004. Controlled cell death, plant survival and development. Nat. Rev. Mol. Cell Biol. 5:305-315. [DOI] [PubMed] [Google Scholar]
- 32.Lang, T., E. Schaeffeler, D. Bernreuther, M. Bredschneider, D. H. Wolf, and M. Thumm. 1998. Aut2p and Aut7p, two novel microtubule-associated proteins are essential for delivery of autophagic vesicles to the vacuole. EMBO J. 17:3597-3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lecellier, G., and P. Silar. 1994. Rapid methods for nucleic acids extraction from petri dish-grown mycelia. Curr. Genet. 25:122-123. [DOI] [PubMed] [Google Scholar]
- 34.Leist, M., and M. Jaattela. 2001. Four deaths and a funeral: from caspases to alternative mechanisms. Nat. Rev. Mol. Cell Biol. 2:589-598. [DOI] [PubMed] [Google Scholar]
- 35.Levine, B., and D. J. Klionsky. 2004. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev. Cell 6:463-477. [DOI] [PubMed] [Google Scholar]
- 36.Liu, Y., M. Schiff, K. Czymmek, Z. Talloczy, B. Levine, and S. P. Dinesh-Kumar. 2005. Autophagy regulates programmed cell death during the plant innate immune response. Cell 121:567-577. [DOI] [PubMed] [Google Scholar]
- 37.Matsuura, A., M. Tsukada, Y. Wada, and Y. Ohsumi. 1997. Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae. Gene 192:245-250. [DOI] [PubMed] [Google Scholar]
- 38.Melendez, A., Z. Talloczy, M. Seaman, E. L. Eskelinen, D. H. Hall, and B. Levine. 2003. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science 301:1387-1391. [DOI] [PubMed] [Google Scholar]
- 39.Otto, G. P., M. Y. Wu, N. Kazgan, O. R. Anderson, and R. H. Kessin. 2004. Dictyostelium macroautophagy mutants vary in the severity of their developmental defects. J. Biol. Chem. 279:15621-15629. [DOI] [PubMed] [Google Scholar]
- 40.Otto, G. P., M. Y. Wu, N. Kazgan, O. R. Anderson, and R. H. Kessin. 2003. Macroautophagy is required for multicellular development of the social amoeba Dictyostelium discoideum. J. Biol. Chem. 278:17636-17645. [DOI] [PubMed] [Google Scholar]
- 41.Paoletti, M., M. Castroviejo, J. Bégueret, and C. Clavé. 2001. Identification and characterization of a gene encoding a subtilisin-like serine protease induced during the vegetative incompatibility reaction in Podospora anserina. Curr. Genet. 39:244-252. [DOI] [PubMed] [Google Scholar]
- 42.Pinan-Lucarré, B., M. Paoletti, K. Dementhon, B. Coulary-Salin, and C. Clavé. 2003. Autophagy is induced during cell death by incompatibility and is essential for differentiation in the filamentous fungus Podospora anserina. Mol. Microbiol. 47:321-333. [DOI] [PubMed] [Google Scholar]
- 43.Pyo, J. O., M. H. Jang, Y. K. Kwon, H. J. Lee, J. I. Jun, H. N. Woo, D. H. Cho, B. Choi, H. Lee, J. H. Kim, N. Mizushima, Y. Oshumi, and Y. K. Jung. 2005. Essential roles of Atg5 and FADD in autophagic cell death: dissection of autophagic cell death into vacuole formation and cell death. J. Biol. Chem. 280:20722-20729. [DOI] [PubMed] [Google Scholar]
- 44.Reggiori, F., and D. J. Klionsky. 2002. Autophagy in the eukaryotic cell. Eukaryot. Cell 1:11-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rubinsztein, A. C., M. DiFiglia, N. Heintz, R. A. Nixon, B. Ravikumar, L. Stefanis, and A. Tolkovsky. 2005. Autophagy and its possible roles in nervous system diseases, damage and repair. Autophagy 1:11-22. [DOI] [PubMed] [Google Scholar]
- 46.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning, a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 47.Saupe, S. J. 2000. Molecular genetics of heterokaryon incompatibility in filamentous ascomycetes. Microbiol. Mol. Biol Rev. 64:489-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shimizu, S., T. Kanaseki, N. Mizushima, T. Mizuta, S. Arakawa-Kobayashi, C. B. Thompson, and Y. Tsujimoto. 2004. Role of Bcl-2 family proteins in a non-apoptotic programmed cell death dependent on autophagy genes. Nat. Cell Biol. 6:1221-1228. [DOI] [PubMed] [Google Scholar]
- 49.Staben, C., B. Jensen, M. Singer, J. Pollock, M. Schechtman, et al. 1988. Use of a bacterial hygromycin B resistance gene as a dominant selectable marker in Neurospora crassa transformation. Fungal Genet. Newsl. 35:79-81. [Google Scholar]
- 50.Suzuki, K., T. Kirisako, Y. Kamada, N. Mizushima, T. Noda, and Y. Ohsumi. 2001. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 20:5971-5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Suzuki, K., T. Noda, and Y. Ohsumi. 2004. Interrelationships among Atg proteins during autophagy in Saccharomyces cerevisiae. Yeast 21:1057-1065. [DOI] [PubMed] [Google Scholar]
- 52.Yu, L., A. Alva, H. Su, P. Dutt, E. Freundt, S. Welsh, E. H. Baehrecke, and M. J. Lenardo. 2004. Regulation of an ATG7-beclin 1 program of autophagic cell death by caspase-8. Science 304:1500-1502. [DOI] [PubMed] [Google Scholar]
- 53.Yu, L., M. J. Lenardo, and E. H. Baehrecke. 2004. Autophagy and caspases: a new cell death program. Cell Cycle 3:1124-1126. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.