Abstract
Proteins in the Rho family are small monomeric GTPases primarily involved in polarization, control of cell division, and reorganization of cytoskeletal elements. Phylogenetic analysis of predicted fungal Rho proteins suggests that a new Rho-type GTPase family, whose founding member is Rho4 from the archiascomycete Schizosaccharomyces pombe, is involved in septation. S. pombe rho4Δ mutants have multiple, abnormal septa. In contrast to S. pombe rho4Δ mutants, we show that strains containing rho-4 loss-of-function mutations in the filamentous fungus Neurospora crassa lead to a loss of septation. Epitope-tagged RHO-4 localized to septa and to the plasma membrane. In other fungi, the steps required for septation include formin, septin, and actin localization followed by cell wall synthesis and the completion of septation. rho-4 mutants were unable to form actin rings, showing that RHO-4 is required for actin ring formation. Characterization of strains containing activated alleles of rho-4 showed that RHO-4-GTP is likely to initiate new septum formation in N. crassa.
In many organisms, the purpose of cytokinesis is to couple the process of making a new cell with the replication of the nucleus. In eukaryotes, this can be accomplished by positioning the division plane according to the location of the spindle or by moving mitotic machinery to an already determined division plane (15). Most eukaryotes tightly couple cell division with replication of the nucleus, making cell division essential for viability. For example, Saccharomyces cerevisiae moves the mitotic machinery to the division plane specified by “landmark proteins,” which remain at the bud scar (10). In contrast, the division plane in animal cells is determined by the position of either the spindle midzone (56) or astral microtubules (38). In filamentous fungi, which grow by apical tip extension to form highly polarized multinucleate cells called hyphae, cytokinesis occurs by the formation of crosswalls or septa. These septa serve to define the boundary between hyphal compartments. However, septal pores maintain cytoplasmic continuity in a colony and allow nuclei and organelles to travel between cells (23). Three observations suggest that filamentous fungi have partially uncoupled septation from mitosis: (i) hyphal compartments are multinucleate, (ii) septation is observed in anucleate compartments of Neurospora crassa ropy mutants (29), and (iii) Ashbya gossypii Agcky1 mutants lack septa but grow at a wild-type rate (53). Thus, the partial uncoupling of cell division from mitosis allows isolation of viable aseptate mutants in filamentous fungi. Septation in filamentous fungi is required for certain developmental processes, such as conidiation (asexual spore production) and protoperithecial (female sexual structure) development (20, 37). In addition, septa may serve a structural role in maintaining the tubular shape of hyphae. Septa also act as a scaffold for the Woronin body, a septal plug that stops cytoplasmic leaking after hyphal injury (23, 24, 48).
The process of septation in filamentous fungi has been molecularly characterized by work in Aspergillus nidulans (21, 22, 45, 55), A. gossypii (2, 51, 53), and Penicillium marneffei (7). This work has shown that several regulatory components are required for septum formation in filamentous fungi: actin rings, IQGAPs, PAKs, and formins. In A. gossypii, AgCyk1, an IQGAP-related protein, is required for septation and acts upstream of actin ring formation (53). A second A. gossypii gene, AgCLA4, which encodes a PAK kinase, is also required for septation and the transition from immature to mature mycelia (51). In A. nidulans, actin ring formation and constriction occur before new septa are formed (30). SEPA, the only formin in A. nidulans, controls actin ring formation and is required for both septation and apical tip extension (45). SEPA, like other formins, contains a GTPase binding domain. It is predicted that interactions between a GTP-bound Rho-type GTPase and SEPA are likely to activate the formin and thus actin polarization both at sites of septation and hyphal tips (14, 45).
Rho-type GTPases function as key regulators in many cellular processes, including polarization of the actin cytoskeleton, endocytosis, and chemotaxis. The small monomeric Rho-type GTPase, a conserved family within the Ras superfamily, is defined by domains responsible for GTP and GDP binding, plasma membrane localization, and GTPase activity. Rho-type GTPases act as molecular switches: binding GTP activates interaction with downstream effector proteins; hydrolysis of GTP inhibits interaction with effector proteins (13). These effector proteins, such as formins, IQGAPs, or PAK kinases, often control the reorganization of the actin cytoskeleton; formins have been shown to directly and indirectly nucleate actin filaments (14, 35). Although Rho-type GTPases are molecular switches, a host of proteins in turn regulate the activity or localization of Rho-type GTPases, including guanosine nucleotide exchange factors (GEFs) (43), GTPase-activating proteins (GAPs) (3), and guanosine nucleotide disassociation inhibitors (GDIs) (32).
In this study, we characterize the first cloned gene required for septation in N. crassa, rho-4, which encodes a Rho-type GTPase. rho-4 is not allelic with previously identified aseptate mutations, cwl-1 and cwl-2 (17; D. D. Perkins and N. B. Raju, unpublished results). We show that RHO-4 localizes to septa. The phenotype of strains containing activated alleles of rho-4 and the evaluation of actin localization in rho-4 mutants suggest that RHO-4 acts early to initiate septum formation in N. crassa.
MATERIALS AND METHODS
Strains, media, and growth conditions.
The strains used in this study are listed in Table 1. Strains were maintained on Vogel's medium with required supplements (49). Single conidial isolations were done on BdeS plates (8, 12). Crosses were performed on Westergaard's plates (54). Linear growth of strains was determined by growth in race tubes (40). Heterokaryons of mutant strains and the Fungal Genetics Stock Center (FGSC) (27a) strain FGSC 4564 am1 were used as female partners in most crosses in order to complement protoperithecial development or auxotrophic markers (33). Electroporation was done according to Margolin et al. (26) as modified by Robert L. Metzenberg and Kristin Black (1.5 kV for a 1-mm gap cell; personal communication). Transformation and production of spheroplasts were done according to Royer and Yamashiro (39).
TABLE 1.
Strains used in this study
| Strain | Genotype | Reference or origin |
|---|---|---|
| R2-14 | ad-3B cyh-1 aml | FGSC 4564 |
| FGSC 988 | ORS8-1 a | FGSC 988 |
| I-1-83 | ad-3A his-3 A | Gift from A. J. Griffiths |
| R10-17 | Sad1::hph; flP a | Gift from P. K. T. Shiu |
| R1-19 | cyh-1; pyr-4 A | Gift from R. L. Metzenberg |
| CR5-3 | rho-4 A | I-1-83-(rho-4-RIP) × R1-08 |
| CR5-10 | ad3A his-3; rho-4 A | I-1-83-(rho-4-RIP) × R1-08 |
| CR7-7 | rho-4 a | CR5-6 × R1-19 |
| CR19-46 | Sad1::hph; rho-4 a | CR5-10 × R10-17 |
| FGSC 6103 | his-3 A | FGSC 6103 |
| CR#4 | his-3+::Pgpd-HA-rho-4 A | 6103 (pBM61-HA-rho-4) |
| CR21-12 | his-3+::Pgpd-HA-rho-4; rho-4 A | CR19-46 × CR#4 |
| FGSC 5951 | (cwl-1 A + ad-3b cyh-1 aml) | FGSC 5951 |
| CR17-1 | cwl-1 A | FGSC 5951 × R1-08 |
| FGSC 6875 | (cwl-2 A + ad-3b cyh-1 aml) | FGSC 6875 |
| CR18-1 | cwl-2 A | FGSC 6875 × R1-08 |
| CRD6 | his-3+::Pgpd-HA-D126A-rho-4 A | 6103 (pBM61-HA-D126A-rho-4) |
| CRG3 | his-3+::Pgpd-HA-G18V-rho-4 A | 6103 (pBM61-HA-G18V-rho-4) |
| CRQ2 | his-3+::Pgpd-HA-Q69L-rho-4 A | 6103 (pBM61-HA-Q69L-rho-4) |
| CR28-1 | his-3+::Pgpd-HA-Q69L-rho-4; rho-4 A | CRQ2 × CR19-46 |
| CR29-1 | his-3+::Pgpd-HA-G18V-rho-4; rho-4 A | CRG3 × CR19-46 |
| CR29-9 | his-3+::Pgpd-HA-G18V-rho-4 A | CRG3 × CR19-46 |
| CR31-1 | his-3+::Pgpd-HA-D126A-rho-4; rho-4 A | CRD6 × CR19-46 |
| CR31-2 | his-3+::Pgpd-HA-D126A-rho-4 A | CRD6 × CR19-46 |
DNA techniques and extraction.
For small-scale genomic DNA extraction from N. crassa for PCR, the method outlined in reference 58 was used with one modification: 1% Triton X-100 was added to Tris-EDTA (TE) buffer before boiling conidia. For large-scale DNA extractions, the method of Lee et al. (25) was used. Standard approaches for Southern blotting, cloning, and overlap PCR were used (41). The DNA sequence of all constructs was determined by the DNA Berkeley Sequencing Facility (http://mcb.berkeley.edu/barker/dnaseq). Oligonucleotides were obtained from MWG Biotech (www.mwgbiotech.com) and IDT (www.idtdna.com). Oligonucleotide sequences are provided in Table S1 in the supplemental material.
Phylogenetic analysis.
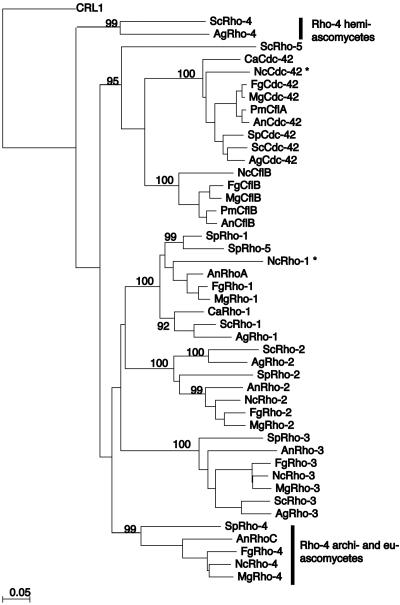
Predicted monomeric GTPases were retrieved as amino acid sequences from GenBank (http://www.ncbi.nih.gov/), the Broad Institute (http://www.broad.mit.edu/annotation/), The Saccharomyces Genome Database (http://genome-www.stanford.edu/Saccharomyces/), The Candida Genome Database (http://www.candidagenome.org/), and The Schizosaccharomyces pombe Genome Database (http://www.sanger.ac.uk/Projects/S_pombe/). Full-length proteins were aligned using ClustalW in MacVector (Oxford Molecular Sciences, Inc.). A neighbor-joining tree was built in MacVector using these parameters: 1,000 bootstrap replicates, gaps ignored, and uncorrected P. Bootstrap support for major family divisions is included on the best tree phylogeny (Fig. 1). The systematic gene names or GenBank accession numbers are listed in Table S2 in the supplemental material.
FIG. 1.
Phylogenetic tree of Rho-type GTPases in fungi. The phylogenetic tree shows the relationship between the Rho-type GTPases in fungi. Rho-type proteins are grouped into the following specific families: Cdc42, CflB, Rho1, Rho2, and Rho3. Predicted Rho4 homologs group into two distinct families: one group is specific to the hemiascomycete species (ScRho4 and AgRho4), while the other is found in the archiascomycete and the euascomycete species. CRL1, a Rho-type GTPase from Candida albicans, was used to root the tree because it has an unusually long C-terminal extension that distinguishes it from the other Rho-type GTPases. Asterisks indicate that the predicted protein may have an error in annotation. Systematic gene names or GenBank accession number are listed in Table S2 in the supplemental material. Ag, Ashbya gossypii; Sc, Saccharomyces cerevisiae; Sp, Schizosaccharomyces pombe; Ca, Candida albicans; Pm, Penicillium marneffei; An, Aspergillus nidulans; Fg, Fusarium graminarium; Nc, Neurospora crassa; Mg, Magnaporthe grisea.
RIP mutagenesis and rho-4 mutant analysis.
Repeat induced point (RIP) mutagenesis is a strategy used in N. crassa to inactivate genes (44). Briefly, N. crassa has evolved the ability to destroy duplicated sequences during a cross by introducing multiple GC-to-AT transitions in both the resident and ectopic copies of a duplicated sequence. An 800-bp internal fragment of the predicted rho-4 open reading frame (ORF; NCU03407) was amplified from genomic DNA using primers RHO4FOR41 and RHO4REV841. The PCR product was ligated into TOPO TA vector (Invitrogen); an XbaI/HindIII fragment was subsequently subcloned into pCB1004, a vector containing the dominant hygromycin resistance cassette (9). pCB1004-Rho-4 was transformed into spheroplasts of strain I-1-83, and three separate hygromycin-resistant transformants were crossed to FGSC 988, a wild-type N. crassa strain. Progeny were screened for sensitivity to hygromycin to ensure that they did not contain the ectopic gene fragment. Restriction fragment length polymorphism (RFLP) differences between wild-type and mutant rho-4 alleles were assessed by PCR amplification using the primers TOTRHO4FOR171 and TOTRHO4REV2374 followed by digestion with the appropriate restriction enzyme. Several mutant rho-4 alleles were cloned, and their DNA sequence was determined.
Construction of rho-4 alleles.
The RHO-4 protein contains a carboxy-terminal prenylation motif that is required for the correct localization of the protein. Thus, a hemagglutinin (HA)-epitope tag was added to the N-terminal region by PCR using the NcoIHARho4FOR and TotRho4REV primers and the polymerase Pfu Turbo (Stratagene). After ligation into the TOPO Blunt vector (Invitrogen), an NcoI/BamHI fragment was cloned into pAN52.1 (36) between the gpd promoter and the trpC terminator. An XbaI/BglI fragment containing the gpd promoter, rho-4, and the trpC terminator was ligated into XbaI/BamHI-digested pBM61 (26) for targeting at the his-3 locus (see below).
Overlap PCR was used to change the amino acid sequences in the rho-4 ORF (41). The amino acid changes predicted to produce dominant active alleles were those used in references 6 and 31. The dominant active alleles are G18V, Q69L, and D126A amino acid substitutions. In each case, the template was the HA-rho-4 ORF in the pAN52.1 vector. The full-length PCR products were ligated into the pAN52.1 vector, and the DNA sequence was determined. A BglII/XbaI fragment was ligated into BamHI/XbaI-digested plasmid pBM61.
Construction of rho-4 strains containing epitope-tagged and mutant rho-4 alleles.
A genetic method was used to introduce both epitope-tagged and mutated rho-4 alleles into the rho-4 mutant strain because the rho-4 mutant could be transformed neither by electroporation nor by spheroplast-based techniques. rho-4 alleles in pBM61 were targeted to the his-3 locus of FGSC 6103 by electroporation followed by selection of His+ prototrophs (16, 26). The resulting transformants contained two copies of rho-4: one ectopic copy at his-3 on LGI (his-3::rho-4) and the resident copy on LGII. Transformants were subsequently crossed to a rho-4− strain, CR19-46, which also contained the dominant mutation Sad-1 to suppress meiotic silencing of unpaired DNA (MSUD) (46). The Sad-1 mutation was included because transformants contained two copies of rho-4+ alleles. During meiosis, the rho-4 allele at the his-3 locus will be unpaired and will trigger the silencing of all copies of rho-4. Such MSUD of rho-4+ in a Sad-1+ strain resulted in sterility; homozygous rho-4 crosses were barren, indicating that rho-4+ is required during meiosis. One-quarter of the progeny from the Sad-1 his-3::rho-4 × rho-4− crosses were his-3::rho-4; rho-4−, allowing complementation and RHO-4 localization to be assessed. Progeny were screened for the his-3::rho-4 allele by PCR using the primers GPDVECT2204FOR and GDPVECT2407REV. The resident rho-4 alleles were amplified by PCR using primers RHO4FOR646 and RHO4REV841, which prohibited amplification of the his-3::rho-4 allele. The PCR product was assessed by RFLP analysis to determine whether or not the strain contained the mutant allele of rho-4. Strains containing both his-3::rho-4 as well as mutations at the rho-4 locus were further analyzed.
Immunofluorescence and fluorescence microscopy.
Conidia or mycelia were inoculated onto slides covered in thin layers of Vogel's minimal medium (MM) or inoculated directly onto petri plates with Vogel's MM solidified with 2% agarose. After growth at room temperature (∼22°C) or 30°C overnight, the samples were prepared for immunofluorescence microscopy following the protocol outlined in reference 22 with the following modifications. (i) Fixation time was reduced to 15 to 30 min. (ii) The cell wall digestion was done with 10 mg/ml Driselase (Sigma), 30 mg/ml lysing enzyme (Sigma), and 50 mg/ml Glucanex (Novo Nordisk Ferment Ltd.) diluted 1:1 with egg white for 20 to 40 min (Steve Harris, personal communication). (iii) Primary antibody (mouse anti-HA; Roche) was diluted 1:500 in 1% bovine serum albumin (BSA)-phosphate-buffered saline (PBS) and incubated overnight at 4°C. Secondary antibody (Alexa 488 goat anti-mouse from Molecular Probes) was added to 1% BSA-PBS (1:500) for 1 h in the dark at room temperature (∼22°C). For actin antibodies, Cy3-conjugated purified mouse immunoglobulin (clone AC-40; Sigma) was used with the protocol described above, except the concentration was changed to 1:200 for primary antibody concentration and samples were then washed three times in 1% BSA-PBS and mounted on slides. Alternatively, clone C4 mouse immunoglobulin (Chemicon) was used with a 1:500 dilution followed by a 1:500 dilution of Alexa 488 goat anti-mouse (Molecular Probes). Photographs of hyphae and germinating conidia were taken using a Hamamatsu digital charge-coupled device camera (Hamamatsu, Japan) and a Zeiss Axioskop II microscope. Samples were measured and recorded using Openlab (Coventry, United Kingdom). The image files were transferred into Adobe Photoshop 7.0 and further processed.
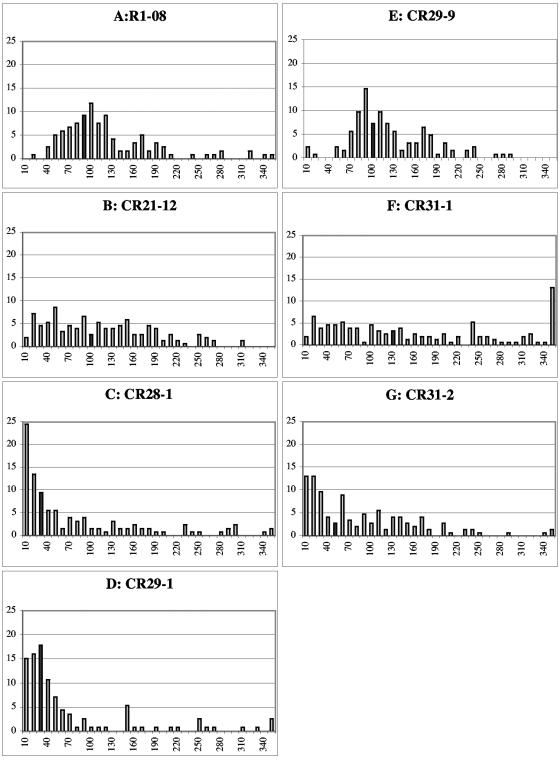
To measure hyphal compartment lengths, conidia were inoculated overnight at 30°C onto Vogel's MM plates. When adequate growth was achieved, the mycelium was stained with calcofluor (1 μg/ml), cut out, placed on a microscope slide, and covered with a coverslip. Fluorescent micrographs were taken of one large hypha starting from the edge and moving towards the interior of the colony. Alternatively, and with similar results, hyphae were observed without the addition of calcofluor by bright-field microscopy. Hyphal compartments in each strain were measured at least three independent times. Measurements were taken using Openlab and transferred into Microsoft Excel for data analysis. A representative histogram of each strain was prepared in Excel with bin lengths of 10 μm (see Fig. 3). For actin ring quantification, unbiased fields of mycelia were photographed first for calcofluor and then for actin immunofluorescence signal until a sufficient sample size (n ≅ 200 septa) was reached. Fisher's exact test (http://www.matforsk.no/ola/fisher.htm) was used to determine if values were statistically significant.
FIG. 3.
Measurement of hyphal compartment lengths. Colonies were grown overnight at 30°C on Vogel's MM and then stained with 1 μg/ml calcofluor. One hypha's compartment lengths were measured from the edge of the colony towards the interior until at least 100 measurements were made. Measurements were binned into 10-μm segments, the frequency was normalized to 100%, and the histogram was plotted. The last bin is 340 μm or greater. The dark gray bar represents the median value for hyphal compartment length. (A) FGSC 988 (wild type [WT]) hyphal compartment lengths. (B) CR21-12 (his-3+::Pgpd-HA-rho-4; rho-4) hyphal compartment lengths. (C) CR28-1 (his-3+::Pgpd-HA-Q69L-rho-4; rho-4) shows a strong bias towards small compartment lengths. (D) CR29-1 (his-3+::Pgpd-HA-G18V-rho-4; rho-4) also shows a strong bias towards small compartment lengths. (E) CR29-9 (his-3+::Pgpd-HA-G18V-rho-4) hyphal compartment lengths are similar to wild type. (F) CR31-1(his-3+::Pgpd-HA-D126A-rho-4; rho-4) hyphal compartment lengths are widely distributed. (G) CR31-2 (his-3+::Pgpd-HA-D126A-rho-4) hyphal compartment lengths are also widely distributed, but show a bias towards smaller compartment lengths.
RESULTS
Comparative analysis of Rho-type GTPases in fungi.
The amino acid sequence of Rho4 was retrieved from the Saccharomyces Genome Database (http://www.yeastgenome.org/). BLASTp (1) was used to determine the closest match in N. crassa genome to Rho4. An ORF was recovered with an e-value of 2e-49 encoding a hypothetical 251-amino-acid protein, b11b23_030 (http://mips.gsf.de/proj/neurospora/) or NCU03407 (http://www.broad.mit.edu/annotation/fungi/neurospora/). Like all Rho-type GTPases, it contained the motifs required for GTP binding (PF00071) and a CAAX box (prenylation motif) for membrane localization (47).
Null mutations in the rho-4 gene in the hemiascomycete species S. cerevisiae and A. gossypii result in strains with either a subtle phenotype or no phenotype (27, 52). In contrast, strains containing null mutations in the rho4 gene in the archiascomycete species Schizosaccharomyces pombe have obvious defects in septation at elevated temperatures (31, 42). To determine the relationship between the Rho-type GTPases in ascomycete species, a neighbor-joining tree was created from the predicted protein products of Rho-type GTPases from the genomes of representative ascomycete species (see Materials and Methods). The protein used as an outgroup is the Candida albicans Rho-type GTPase CRL-1 because it has an unusually long C-terminal region (57). Figure 1 shows that the RHO4 family has more than one branch and therefore potentially more than one function, as suggested by references 4 and 31. The euascomycete and archiascomycete RHO4 family members were more closely related to each other than to other Rho-type GTPases in the hemiascomycete species S. cerevisiae and A. gossypii. Thus, the archiascomycete and euascomycete RHO4 members define a new Rho-type GTPase family found only in fungi, whose founding member is S. pombe RHO4 (31, 42).
rho-4 mutants in N. crassa lack septa.
To generate mutations in the rho-4 locus in N. crassa, we used RIP mutation. RIP is a naturally occurring mutagenic process that occurs during the sexual cycle (see Materials and Methods) (44). Mutations caused by RIP occur stochastically and with a characteristic pattern of GC-to-AT transition mutations in both the resident and ectopic copies of a duplicated sequence. In many cases, this type of mutagenesis leads to loss-of-function mutations caused by introduction of stop codons in the ORF (44). From a cross between transformants containing a duplicated rho-4 sequence and a wild-type strain (FGSC 988), 17% (11/64) of the progeny showed slow growth. The slow-growth phenotype in these progeny was associated with RFLPs within the resident rho-4 allele, suggesting that the slow-growth phenotype resulted from mutations in rho-4. This hypothesis was confirmed by DNA sequence analysis. One mutant (CR5-10) contained a rho-4 allele with amino acid substitutions K36E and Q68STOP. Another rho-4 allele (from CR5-3) had mutations resulting in G17S, T22I, and Q30STOP substitutions. These mutations in rho-4 are likely to be loss-of-function alleles because they have stop codons before the GTP binding or effector binding domains. The sequence of a third rho-4 allele from strain CR7-7 showed amino acid substitutions (M1I, C24Y, S28N, V39I, E43K, C87Y V99I, M100I, C111Y, C131Y, M142I, M167I, and V177I) but no stop codons. The CR5-3, CR5-10, and CR7-7 strains all showed an identical phenotype and were recessive in heterokaryons, indicating that all these strains have rho-4 loss-of-function mutations.
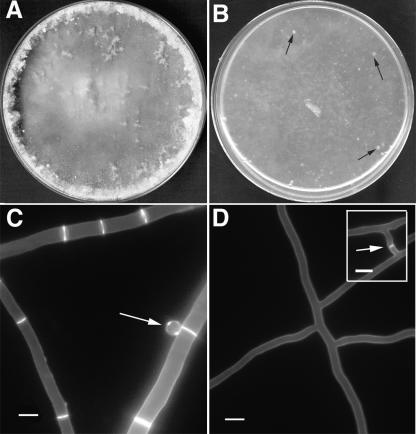
Mutations in the rho-4 locus caused a severe slow-growth phenotype in N. crassa. Strains containing the rho-4 mutations grew at 1 to 2 cm/day, as compared to ∼6 cm/day for wild-type strains, and did not produce any asexual spores (conidia) (Fig. 2B). In addition, the hyphae of rho-4 mutants leaked cytoplasm, producing “lenses” on the agar surface that appeared as white spots in the mycelia (Fig. 2B) (see the movie in the supplemental material). Using osmotic stabilizers such as 1 M sorbitol or 1.2 M NaCl did not remediate cytoplasmic leaking or growth rates in rho-4 mutants. When rho-4 mutants were stained with calcofluor and observed by fluorescence microscopy, it became obvious that their hyphae lacked septa (Fig. 2D). Hyphal tips showed normal polarization (see the movie in the supplemental material), although hyphae of rho-4 mutants were thin and had abnormal branching patterns (Fig. 2D). Despite having thin hyphae, rho-4 mutants were unaffected in ascospore germination. In addition, rho-4 mutants had no detectable defect in cell fusion (Fig. 2D, inset).
FIG. 2.
Phenotype of an N. crassa rho-4 mutant. (A) Wild-type N. crassa hyphae and conidia after 5 days of growth on Vogel's MM plates. (B) A rho-4 mutant on Vogel's MM plates forms only thin mycelia with patches of leaking cytoplasm (black arrows) and no conidia. (C) Wild-type N. crassa hyphae stained with calcofluor (1 μg/ml) show bright staining at septa and cell walls. An arrow points to a conidium. (D) rho-4 mutant hyphae stained with calcofluor (1 μg/ml) show cell wall staining, but no septal staining. (Inset) rho-4 mutants undergo cell fusion. Arrow marks area of recent cell fusion event. Calcofluor staining does not show a septum, but the intervening cell wall of the fusion hyphae, which is degraded upon completion of hyphal fusion. Bar, 10 μm.
rho-4 is not allelic with cwl-1 or cwl-2.
Two mutants have been described in N. crassa that show an aseptate phenotype, cwl-1 and cwl-2 (17, 37; D. D. Perkins, unpublished results). The cwl-1 and cwl-2 loci have been mapped to LGII; the rho-4 locus is also on LGII. A cwl-1 mutant has a similar phenotype to a rho-4 mutant, while a cwl-2 mutant appears to have a denser branching pattern and infrequently produces septa. Both cwl-1 and cwl-2 are recessive mutations (17, 37; D. D. Perkins, unpublished results). A heterokaryon test was used to determine allelism between cw1-1 and rho-4. In this method, hyphal fusion between the cwl mutants and the rho-4 mutant results in the formation of a multinucleate heterokaryon. If mutations in these strains are not allelic, the heterokaryon will show a wild-type phenotype because of complementation. If the mutations are allelic and complementation does not occur, the heterokaryon will have the phenotype of the mutants. A (cwl-1 + rho-4) heterokaryon composed of strains CR17-1 and CR5-10 (Table 1) showed vigorous growth and conidiation, indicating that the cwl-1 and rho-4 mutations were not allelic. Because of potential genetic background differences between cwl-2 and our rho-4 mutants, we assessed allelism between rho-4 and cwl-2 via a cross. A (rho-4 + am1) heterokaryon composed of strains CR5-10 and FGSC 4564 was used as a female in a cross to the cwl-2 strain (FGSC 6875). Although rho-4 homozygous crosses are barren (see below), a cross between rho-4 and cwl-2 strains produced numerous progeny. The DNA sequence of the rho-4 allele from cwl-2 strain CR18-1 was identical to that of the rho-4+ allele (FGSC 2439). These data indicated that cwl-2 and rho-4 were not allelic.
In addition to vegetative growth defects, all rho-4 mutant strains examined (CR5-10, CR7-7, and CR5-3) failed to form female reproductive structures (protoperithecia). When mutants cannot make protoperithecia, a heterokaryon between the mutant and the “helper strain” am1 (FGSC 4564) is commonly used to complement the inability to make female sexual structures (33). Although the (am1 + mutant) heterokaryon forms protoperithecia, the am1 nucleus cannot participate in a cross because of a mutation in the mat a locus. Although the inclusion of the helper strain am1 in a heterokaryon with cw1-1 or cwl-2 complemented the mutants for protoperithecium formation, homozygous crosses [(cwl-1 + am1) × cwl-1 or (cwl-2+ am1) × cwl-2] were still barren and produced only a few asci. Microscopic observation showed that septa were absent in the ascogenous hyphae in the [(cwl-1 + am1) × cwl-1] or [(cwl-2+ am1) × cwl-2)] crosses. Thus, both cwl-1 and cwl-2 are required for the production of septa within the dikaryotic ascogenous hyphae and septa are essential for ascus development (37; and N. B. Raju, unpublished results). To determine if the sexual defect in rho-4 was similar to cwl-1 and cwl-2, heterokaryons between the rho-4 mutants and am1 were made and used as females in crosses. Similar to cwl-1 and cwl-2, homozygous [(rho-4 + am1) × rho-4] crosses were barren, indicating that rho-4 is also required for septum formation in dikaryotic ascogenous hyphae.
RHO-4 localizes to septa.
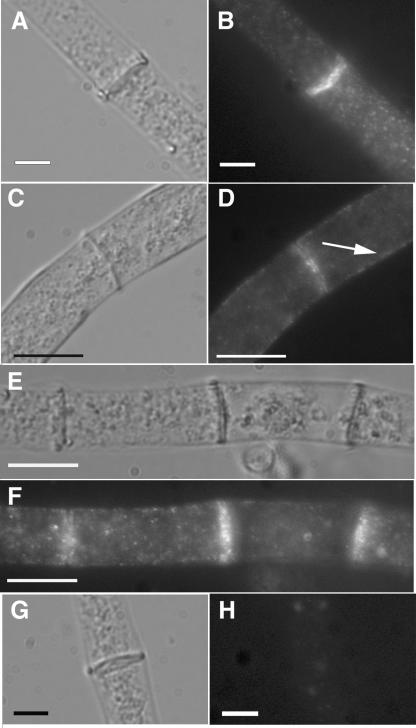
A genetic method was used to introduce all of the rho-4 constructs into a rho-4 mutant background because neither transformation of spheroplasts nor electroporation could be performed with rho-4 mutants (see Materials and Methods). A rho-4 mutant strain, CR21-12, containing an HA-epitope-tagged allele of rho-4 targeted to the his-3 locus and driven by the gpd promoter (see Materials and Methods), grew in a manner similar to wild-type strains at ∼6 cm/day. The introduction of the HA-rho-4 allele into rho-4 mutants also restored septation. The median value of hyphal compartment length in a wild-type strain (FGSC 988) is 87 μm, with a standard deviation of 66 μm (Fig. 3A). The large value for standard deviation in hyphal compartment length reflects the variability of compartment length, and thus placement of septa, in N. crassa. The median value of hyphal compartment length in CR21-12 was 93 μm, with a standard deviation of 71 μm (Fig. 3B). When immunofluorescence microscopy was used to detect the HA-RHO-4 protein in strain CR21-12, it localized to septa (Fig. 4B) and also to the plasma membrane (Fig. 4D). Interestingly, the majority of septa showed a bright fluorescent signal (Fig. 4F), suggesting that RHO-4 localization was not dynamic. In contrast, in A. nidulans, SepA (formin) localization disappears after septum formation (45, 55) and in A. gossypii, septal localization of AgCyk1 (IQGAP) (53) and the landmark protein Bud3 (51) is also transient.
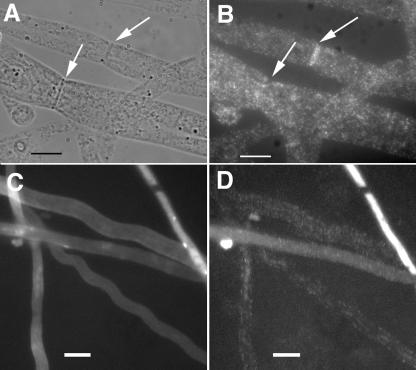
FIG. 4.
Immunofluorescence microscopy of HA-tagged RHO-4. (A) Differential interference contrast (DIC) image of CR21-12 (his-3+::Pgpd-HA-rho-4; rho-4) treated with anti-HA antibodies. (B) Fluorescent micrograph of the same hypha as in panel A. (C) A second DIC micrograph of CR21-12 treated with anti-HA antibodies. (D) The same hypha as that shown in panel C, showing both plasma membrane localization (indicated by an arrow) and septal localization of HA-RHO-4. (E) DIC micrograph of CR21-12 treated with anti-HA antibodies showing multiple septa. (F) Fluorescent micrograph of same hypha as in panel E showing that all septa are labeled. (G) DIC micrograph of FGSC 988 (wild-type strain) treated with anti-HA antibodies. (H) Fluorescent micrograph of the same hypha in panel E shows little nonspecific antibody binding. Bars, 10 μm.
Actin rings are not observed in the rho-4 mutant.
Actin ring formation and constriction have been shown to be required for the formation of septa in fungi (18, 21, 22, 30). To assess actin localization in wild-type and rho-4 mutants, N. crassa hyphae were probed with anti-actin antibodies. In a wild-type strain (FGSC 988), sites of septation and actin colocalized in ∼12% of septa (Table 2; Fig. 5A and B). Septa that show actin rings are presumably in the process of making septa, similar to what has been observed in A. nidulans (22, 30). In the rho-4 mutant strains, actin rings were never observed by immunofluorescence microscopy (Fig. 5C and D). These data indicate that rho-4 mutants are unable to form actin rings and that RHO-4 is required for actin localization during septation.
TABLE 2.
Actin ring formation during septation in wild-type and rho-4− strains containing activated rho-4 alleles
| Strain name (n) | % with actin rings without calcofluor | % with actin rings with calcofluor | % with double ring of actin | % with dot of actin localization | % with septa without actin | % with septa with actin |
|---|---|---|---|---|---|---|
| FGSC 988 (195) | 0.5 | 4.6 | 5.6 | 0.5 | 88.2 | 11.8 |
| CR28-1 (255) | 2.4 | 19.6 | 11 | 3.5 | 65.9 | 34.1b |
| CR29-1 (245) | 0.8 | 9.8 | 2.9 | 0.8 | 86.5 | 13.5 |
| CR31-1 (252) | 0.4 | 4.4 | 0.8 | 0.0 | 94.5 | 5.2+ |
Panels A to E correspond to calcofluor treatment (top panel) and actin localization (bottom panel) in hyphae from wild-type strain FGSC 988 representing the progression of septation. Each bar is 10 μm, except for panel B (5 μm).
Statistically different as determined by Fisher's exact test (http://www.matforsk.no/ola/fisher.htm).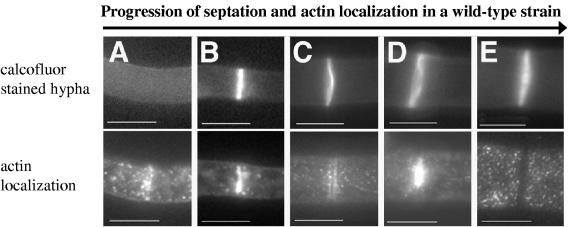
FIG. 5.
Actin localization in wild-type and rho-4 strains. (A) DIC image of wild-type (FGSC 988) hyphae treated with anti-actin antibodies. Arrows show septa. (B) Fluorescent micrograph of hyphae in panel A. Arrows are the same as in panel A. Note absence of actin staining in the lower septa. (C) Calcofluor-stained image of hyphae from rho-4 mutant CR7-7 treated with anti-actin antibodies. Hyphae were stained with calcofluor to show absence of septa in rho-4 mutants. (D) Fluorescent micrograph of hyphae shown in panel C. Bar, 10 μm.
Strains containing activated rho-4 alleles show aberrant septation.
Activated alleles of Rho-type GTPases can help determine downstream effector proteins and elucidate function. Activated alleles hydrolyze GTP more slowly and therefore remain GTP bound for a longer period of time. Based on the high degree of sequence identity within conserved Rho-type GTPase domains, we constructed rho-4 mutant alleles that correspond to activated alleles (5, 11). When these predicted activated rho-4 alleles (Q69L and G18V; see Materials and Methods) were introduced into an N. crassa rho-4 mutant, septation was restored. However, the median hyphal compartment length in CR28-1 (his-3+::Pgpd-HA-Q69L-rho-4; rho-4) was 33 μm, with a standard deviation of 93 μm, while median hyphal compartment length in CR29-1 (his-3+::Pgpd-HA-G18V-rho-4; rho-4) was 31 μm, with a standard deviation of 87 μm (Fig. 3C and D). These data indicate that the introduction of these activated rho-4 alleles into N. crassa results in irregular septation, which is skewed towards decreasing hyphal compartment lengths. In both CR28-1 and CR29-1, hyphae often contained two or more septa in close proximity and which were often not perpendicular to a hypha (Fig. 6A, B, D, and E). Aberrant septa which curved around a hypha occurred in close proximity (<10 μm) to perpendicular septa (Fig. 7A and B). For both CR28-1 and CR29-1, roughly 5% of septa (7/142 and 8/137, respectively) were irregular. In contrast, irregular or aberrant septa were never observed in a wild-type strain (FGSC 988).
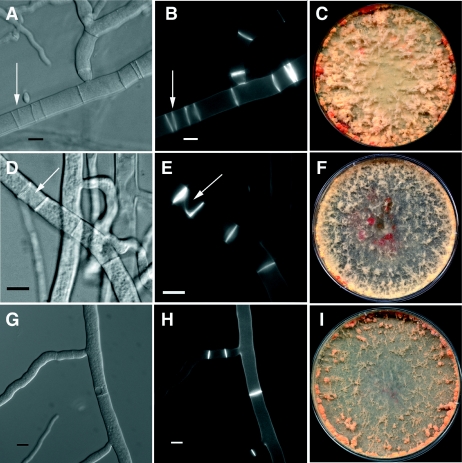
FIG. 6.
Phenotype of strains containing active alleles of rho-4 in N. crassa. (A) Differential interference contrast (DIC) image of CR28-1 (his-3+::Pgpd-HA-Q69L-rho-4; rho-4) showing multiple, closely positioned septa. An arrow points to an irregular septum. (B) Calcofluor-stained fluorescent image of the same hypha as in panel A. (C) Petri dish showing the phenotype of CR28-1 containing the Q69L rho-4 allele. The bright orange dots at the periphery of the plate are due to cytoplasmic bleeding. (D) DIC image of CR29-1 (his3+::Pgpd-HA-G18V-rho-4; rho-4). The arrow points to an incorrectly formed septum. (E) Calcofluor-stained fluorescent image of the same hypha as in panel D. Same arrow as in panel D. (F) Petri dish showing the cytoplasmic bleeding phenotype of CR29-1 (G18V rho-4 allele). (G) DIC image of CR31-1 (his3+::Pgpd-HA-D126A-rho-4; rho-4). (H) Calcofluor-stained hyphae of same hyphae as in panel G. This strain does not show numerous multiple, incorrectly positioned septa as found in CR28-1 or CR29-1, although septation is highly irregular. (I) Petri dish showing the clumpy conidiation phenotype of CR31-1 (D126A rho-4 allele). Bar, 10 μm.
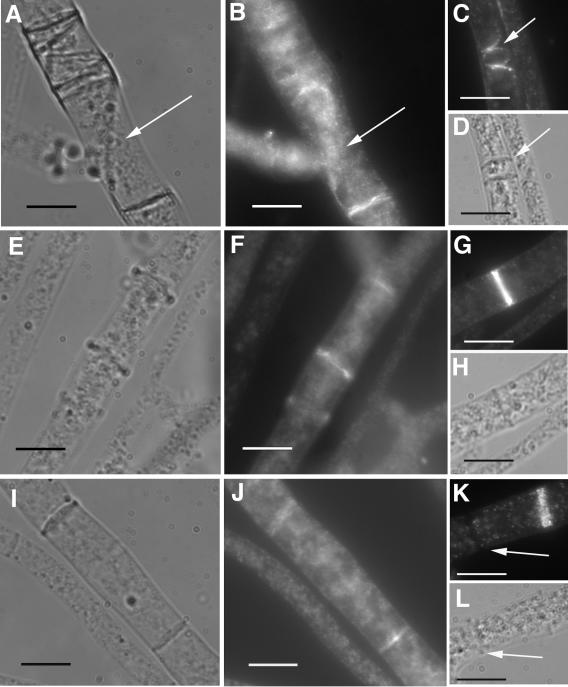
FIG. 7.
Immunolocalization of epitope-tagged activated RHO-4 proteins using anti-HA antibodies. (A) Differential interference contrast (DIC) and (B) fluorescence images of CR28-1 (his3+::Pgpd-HA-Q69L-rho-4; rho-4−) show localization of HA-Q69L-RHO-4 to incorrectly positioned septa (arrow). (C) Actin localization in CR28-1. An arrow points to aberrant actin rings which presumably will lead to aberrant septa. (D) DIC image of panel C showing septa. (E) DIC and (F) fluorescence images of CR29-1 (his3+::Pgpd-HA-G18V-rho-4; rho-4−) show a localization pattern of HA-G18V-RHO-4 at septa that is similar to that of wild-type strains. (G) Actin localization in CR29-1 showing a normal actin ring. (H) DIC image of panel G. (I) DIC and (J) fluorescence images of CR31-1 (his3+::Pgpd-HA-D126A-rho-4; rho-4−) show localization of HA-D126A-RHO-4 to septa. (K) Actin localization in CR31-1. Arrow points to a completed septum without any actin staining. (L) DIC image of K. Bar, 10 μm.
When immunofluorescence microscopy was used to observe HA-Q69L-RHO4 localization in CR28-1 and HA-G18V-RHO-4 localization in CR29-1, signal was apparent at aberrant septa (Fig. 7B and F). Similar to localization of HA-RHO-4 in CR21-12 (Fig. 4), HA-Q69L-RHO-4 localization in CR28-1 and HA-G18V-RHO-4 localization in CR29-1 were not dynamic. Multiple, incorrectly positioned septa and smaller compartment lengths found in CR28-1 and CR29-1, which contain activated rho-4 alleles, suggest that when RHO-4 is bound to GTP, it initiates septum formation. Surprisingly, even though CR28-1 and CR29-1 contained many septa, both strains showed cytoplasm leaking, suggesting that RHO-4 may have an additional role in cell wall integrity (Fig. 6C and F).
Functional RHO-4 is required for localization of actin to sites of septation (Fig. 5). To evaluate actin localization in strains containing activated rho-4 alleles, immunofluorescence was performed on CR28-1 and CR29-1. In CR28-1, 34% of septa (87/255) showed some form of actin localization (Table 2), statistically different from a wild-type strain (P = 1.8e-8). Similar to a wild-type strain, actin rings without concomitant calcofluor staining were also observed. Aberrant actin rings were occasionally observed (Fig. 7C), which presumably will result in the formation of aberrant septa that are observed in this strain (Fig. 6A and B). In CR29-1, 14% of septa showed actin localization, a value statistically equivalent to actin localization in the wild-type strain FGSC 988 (Table 2).
In addition to irregular septation, the growth rate of CR28-1 and CR29-1 was reduced to ∼2 cm/day as compared to ∼6 cm/day for both a wild-type strain (FGSC 988) and a rho-4 strain containing the HA-rho-4 allele (CR21-12). The introduction of the HA-G18V-rho-4 allele into a wild-type (rho-4+) background (CR29-9) mildly affected the position of septum formation, but was semidominant for growth defects. CR29-9 showed reduced growth rates (∼4.5 cm/day) but had hyphal compartments of similar length to those of a wild-type strain, with a median value of 103 μm and a standard deviation of 55 μm (Fig. 3E). Microscopic observation of CR29-9 indicated that aberrant (incomplete or curved) septa occurred in less than 2% of septa (2/122).
A third rho-4 allele was constructed which changed aspartic acid to alanine at amino acid position 126 (D126A). The predicted function of RHO proteins with the D126A mutation is to sequester the GEF; in S. cerevisiae, overexpression of the GEF CDC24 suppresses the phenotype of this mutation in CDC42 (11). Rho-type proteins that contain the D126A substitution show increased GDP disassociation, which should lead to more GTP-bound protein, thereby mimicking an activated allele. Although this mutation in CDC42 causes a dominant-negative phenotype in S. cerevisiae (6, 11), in S. pombe, it shows an activated allele phenotype (28). In N. crassa, the HA-tagged rho-4 allele containing the D126A substitution was introduced into a rho-4 strain (CR31-1). The phenotype of CR31-1 is different from those of strains containing the rho-4 G18V or Q69L mutations. It grew at ∼4 cm/day, multiple rings of septa were infrequent (Fig. 6G and H), and cytoplasmic leaking rarely occurred (Fig. 6I). Hyphal compartment lengths in CR31-1 were very irregular, with a median value of ∼125 μm and a standard deviation of 173 μm, twice that of the wild-type strain (Fig. 3F). Only 1% of septa (1/105) in CR31-1 were aberrant (incomplete or curved). These data suggest that the D126A mutation in rho-4 causes a deregulation in the ability to position septa.
Immunofluorescence microscopy of CR31-1 showed HA-D126A-RHO-4 localization to all septa, similar to the other epitope-tagged alleles of rho-4 (Fig. 7I and J). Actin localization in strain CR31-1 showed significantly fewer actin rings (∼5%) associated with septa (Table 2; P = 8.8e-3).
Similar to rho-4+ strains containing the rho-4 G18V allele (CR29-9), strains containing the D126A substitution in a rho-4+ background (CR31-2) grew at ∼4 cm/day. However, unlike CR29-9 (Fig. 3E), hyphal compartment lengths were variable in CR31-2, which showed a median hyphal compartment length of ∼59 μm with a standard deviation of 84 μm (Fig. 3G). Interestingly, hyphal compartment lengths were skewed towards smaller compartments in CR31-2 as compared to a rho-4 strain containing only the D126A rho-4 allele (CR31-1). Seven percent of septa (10/151) observed in CR31-2 were aberrant (incomplete or curved). These results suggest that, in N. crassa, the rho-4 D126A allele produces a protein that causes a semidominant phenotype with regard to both the regularity of septum deposition and growth.
DISCUSSION
RHO-4 provides a link between small monomeric GTPase signaling and septation.
In this study, we show that mutations in a predicted RHO-type GTPase in N. crassa result in mutants that are aseptate, grow slowly, and show alterations in hyphal morphology. rho-4 is the first N. crassa gene shown to be required for septation, and its product is the first Rho-type GTPase to be implicated in septation in filamentous fungi. Actin rings were not observed in N. crassa rho-4 mutants, indicating that RHO-4 acts early in the septation process and is required for actin ring formation. The frequency of actin ring formation at septa was significantly increased in strain CR28-1 (Q69L rho-4), while strain CR29-1 (G18V rho-4) showed a wild-type frequency of actin rings at septation sites. These observations suggest that although Q69L rho-4 and G18V rho-4 alleles are predicted to encode RHO-4 proteins with a biochemically similar inability to hydrolyze GTP, the G18V RHO-4 protein potentially has more GTPase activity. Strains containing activated rho-4 alleles suggest GTP-bound RHO-4 is involved in the initiation of septation, while cycling to the GDP-bound form may be required for the termination of septation. Thus, our data on rho-4 in N. crassa provide a link between previously known septation mutants in filamentous fungi and small monomeric GTPase signaling. Rho-type GTPases are known to bind to formin homology proteins through the GTPase binding domain (GBD) (50); formins function to nucleate actin (34, 35). The GBD domain of SEPA, the formin in A. nidulans, is sufficient for SEPA localization to septa (45). We speculate that RHO-4 physically interacts with and activates the sole formin (NCU01431) of N. crassa at sites of septation, thus providing a link between a Rho-type GTPase and the formin for construction of the actin ring during septum formation.
When examined by immunofluorescence microscopy, RHO-4 localized to septa. Unlike any of the other proteins known to be required for septation, such as SepA and AspB in A. nidulans (45, 55) or AgCla4 and AgBud3 in A. gossypii (8, 51), RHO-4 in N. crassa does not disappear from the septum once septum formation is complete. Why does RHO-4 stay at the septa? Perhaps RHO-4, besides its role in the early stages of septation, might act later as a landmark protein in fully developed septa. RHO-4 may serve as a signal that informs the hyphal compartment of its boundaries or recruits other proteins for the same purpose.
The phenotype of rho-4 mutants in N. crassa suggests a role for RHO-4 in growth and cell wall integrity.
Similar to an Agcyk1 mutant in A. gossypii, the rho-4 mutant in N. crassa is completely aseptate. However, Agcyk1 mutants grow at a wild-type rate while rho-4 mutants have a growth rate one-sixth that of the wild type. These observations suggest a second role for RHO-4 for optimal growth rate in N. crassa. Two observations support this hypothesis. (i) A spontaneous extragenic suppressor mutation, Drs-1 (for dominant rho-4 suppressor), restores septation and conidiation to rho-4 mutants, but does not restore growth rate (our unpublished results). (ii) The introduction of the G18V-activated rho-4 allele into a rho-4+ strain only mildly perturbed the septation pattern but caused a significant reduction in growth rate. In addition to septation sites, RHO-4 localized to the plasma membrane, suggesting that RHO-4 performs cellular functions required for optimal growth. Other septation mutants, such as a sepA mutant in A nidulans and an A. gossypii Agcla4 mutant, also show impaired growth (or are lethal), suggesting that components involved in septation have multiple functions in filamentous fungi (2, 22).
The phenotype of strains containing activated rho-4 alleles also suggests a role for RHO-4 in cell wall integrity. Lysis at hyphal tips was observed in these strains, even though many septa were formed. The cell lysis defect in strains containing activated rho-4 alleles is not caused by dysfunctional Woronin body localization, as Woronin bodies plug the septa of these strains when wounded (our unpublished results). Perhaps rho-4 mutants in N. crassa, similar to S. pombe rho4Δ (42) and A. nidulans rhoA mutants (19), have alterations in cell wall composition, causing weakening of the cell wall and subsequent lysis of hyphae.
In a wild-type strain of N. crassa, the median hyphal compartment length is ∼75 μm, with a standard deviation of ∼65 μm, indicating that hyphal compartment length in N. crassa is fairly irregular. Wild-type hyphae appear to have a mechanism that prevents the formation of two or more septa within 10 μm. Unlike wild-type strains, mutants containing Q69L and G18V rho-4 alleles showed deregulation of the position of septum formation and often had two or more septa within 10 μm, suggesting that GTP-to-GDP cycling may be required to stop the formation of a new septum. However, even in strains containing these activated rho-4 alleles, there were a significant number of long hyphal compartments, indicating that a signaling factor upstream of RHO-4 is required for initiation of septation.
The Rho-4-type genes in S. pombe and N. crassa define a new family of Rho-type GTPases involved in the formation of septa.
Phylogenetic analysis indicated that S. pombe Rho4 is more similar to RHO-4 in N. crassa than to Rho4 proteins in S. cerevisiae and A. gossypii. In S. pombe, rho4Δ mutants form multiple, aberrant septa at elevated temperature (31, 42) while N. crassa rho-4 mutants lack septa. S. pombe strains containing the dominant active rho4 allele Q61L lack septa, but in N. crassa, the equivalent strain (CR28-1) produces multiple and aberrant septa. Perhaps this difference in phenotype may be due to the different structure of septa in S. pombe versus N. crassa. In S. pombe, the purpose of septation is to divide the cytoplasm and nuclei into two daughter cells (18). The primary septum is formed, followed by the formation of a secondary septum; daughter cells are separated from one another by enzymatic degradation of the primary septum. Santos et al. (42) and Nakano et al. (31) suggest that Rho4 may be responsible for the degradation of the primary septum because an accumulation of septal material as well as clustered vesicles occur near septa in a rho4Δ mutant. In contrast, N. crassa and other filamentous ascomycete fungi have septa that remain incomplete; there is no evidence for enzymatic degradation of septal components. The function of the rho-4 protein may have diverged due to the differentiation of septal formation and structure in S. pombe versus N. crassa. Although the phenotype of rho-4 mutants is different in S. pombe and N. crassa, it is apparent that both proteins are involved in septation and cell wall integrity.
A number of proteins in other systems have been characterized that regulate the activity of Rho-type GTPases. These include GEFs, which remove GDP so that GTP can bind and activate Rho-type GTPases (43); GAPs, which lead to deactivation of Rho-type GTPases by increasing the intrinsic GTPases activity (3); and GDIs, which regulate Rho-type GTPases by preventing nucleotide exchange and inhibiting membrane association (32). The N. crassa genome contains five predicted Rho-type GTPases (Rho1 to -4 and Cdc42) and one predicted Rac homolog (NcCflB). In addition, a large number of predicted GAP, GEF, and GDI homologs are found in the N. crassa genome, which are predicted to regulate Rho-type GTPase activity or localization (4). The relationship between the different Rho-type GTPases, GEFs, GAPs, and GDIs in N. crassa is unclear, as well as the roles of any of them during the septation process. Our studies show that rho-4 is not allelic with previously identified mutations, cw1-1 and cwl-2, which cause an aseptate phenotype (17; D. D. Perkins and N. B. Raju, unpublished results). cw1-1 and cwl-2 may encode effectors that regulate RHO-4 activity or localization and thus septation in N. crassa. In A. gossypii, a component required for septum formation, Bud3, contains a GEF domain, suggesting that it may be involved in recruiting a Rho-type GTPase to sites of septation. Bud3-GFP localizes near newly formed tips as well as to sites of septation (51). However, Rho4 of A. gossypii is more similar to S. cerevisiae Rho4 than it is to N. crassa RHO-4; rho4 mutants in S. cerevisiae are not defective in septum formation. Future experiments in N. crassa will be aimed at determining proteins responsible for targeting RHO-4 to sites of septation and regulating RHO-4 activity.
Supplementary Material
Acknowledgments
We thank Mabel Yang for performing initial studies on hyphal compartment lengths and Emily Chiang for strain maintenance and general lab help. We thank Steve Ruzin and Denise Schichnes (CNR Biological Imaging Facility) for advice on figure preparation. We thank Corinne Clavé for the gift of plasmid pAN52.1. We acknowledge N. B. Raju for sharing unpublished data. We appreciate suggestions from David Drubin, Jeremy Thorner, Andre Fleissner, and Karine Dementhon.
The work in this paper was supported by a grant from the National Science Foundation (MCB-0131355) to N.L.G.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. H. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ayad-Durieux, Y., P. Knechtle, S. Goff, F. Dietrich, and P. Philippsen. 2000. A PAK-like protein kinase is required for maturation of young hyphae and septation in the filamentous ascomycete Ashbya gossypii. J. Cell Sci. 113:4563-4575. [DOI] [PubMed] [Google Scholar]
- 3.Bernards, A. 2003. GAPs galore! A survey of putative Ras superfamily GTPase activating proteins in man and Drosophila. Biochim. Biophys. Acta 1603:47-82. [DOI] [PubMed] [Google Scholar]
- 4.Borkovich, K. A., L. A. Alex, O. Yarden, M. Freitag, G. E. Turner, N. D. Read, S. Seiler, D. Bell-Pedersen, J. Paietta, N. Plesofsky et al. 2004. Lessons from the genome sequence of Neurospora crassa: tracing the path from genomic blueprint to multicellular organism. Microbiol. Mol. Biol. Rev. 68:1-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bourne, H. R., D. A. Sanders, and F. McCormick. 1991. The GTPase superfamily—conserved structure and molecular mechanism. Nature 349:117-127. [DOI] [PubMed] [Google Scholar]
- 6.Boyce, K. J., M. J. Hynes, and A. Andrianopoulos. 2001. The CDC42 homolog of the dimorphic fungus Penicillium marneffei is required for correct cell polarization during growth but not development. J. Bacteriol. 183:3447-3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyce, K. J., M. J. Hynes, and A. Andrianopoulos. 2003. Control of morphogenesis and actin localization by the Penicillium marneffei RAC homolog. J. Cell Sci. 116:1249-1260. [DOI] [PubMed] [Google Scholar]
- 8.Brockman, H. E., and F. J. de Serres. 1963. “Sorbose toxicity” in Neurospora. Am. J. Bot. 50:709-714. [Google Scholar]
- 9.Carroll, A. M., J. A. Sweigard, and B. Valent-Central. 1994. Improved vectors for selecting resistance to hygromycin. Fungal Genet. Newsl. 41:22. [Google Scholar]
- 10.Chant, J., and I. Herskowitz. 1991. Genetic control of bud site selection in yeast by a set of gene products that constitute a morphogenetic pathway. Cell 65:1203-1212. [DOI] [PubMed] [Google Scholar]
- 11.Davis, C. R., T. J. Richman, S. B. Deliduka, J. O. Blaisdell, C. C. Collins, and D. I. Johnson. 1998. Analysis of the mechanisms of action of the Saccharomyces cerevisiae dominant lethal cdc42G12V and dominant negative cdc42D118A mutations. J. Biol. Chem. 273:849-858. [DOI] [PubMed] [Google Scholar]
- 12.Davis, R. H., and F. J. de Serres. 1970. Genetic and microbial research techniques for Neurospora crassa. Methods Enzymol. 17A:79-143. [Google Scholar]
- 13.Etienne-Manneville, S., and A. Hall. 2002. Rho GTPases in cell biology. Nature 420:629-635. [DOI] [PubMed] [Google Scholar]
- 14.Evangelista, M., D. Pruyne, D. C. Amberg, C. Boone, and A. Bretscher. 2002. Formins direct Arp2/3-independent actin filament assembly to polarize cell growth in yeast. Nat. Cell Biol. 4:260-269. [DOI] [PubMed] [Google Scholar]
- 15.Field, C., R. Li, and K. Oegema. 1999. Cytokinesis in eukaryotes: a mechanistic comparison. Curr. Opin. Cell Biol. 11:68-80. [DOI] [PubMed] [Google Scholar]
- 16.Folco, H. D., M. Freitag, A. Ramon, E. D. Temporini, M. E. Alvarez, I. Garcia, C. Scazzocchio, E. U. Selker, and A. L. Rosa. 2003. Histone H1 is required for proper regulation of pyruvate decarboxylase gene expression in Neurospora crassa. Eukaryot. Cell 2:341-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garnjobst, L., and E. L. Tatum.1967. A survey of new morphological mutants in Neurospora crassa. Genetics 57:579-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guertin, D. A., S. Trautmann, and D. McCollum. 2002. Cytokinesis in eukaryotes. Microbiol. Mol. Biol. Rev. 66:155-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guest, G. M., X. Lin, and M. Momany. 2004. Aspergillus nidulans RhoA is involved in polar growth, branching, and cell wall synthesis. Fungal Genet. Biol. 41:13-22. [DOI] [PubMed] [Google Scholar]
- 20.Gull, K. 1978. Form and function of septa in filamentous fungi, p. 78-93. In J. E. Smith and D. R. Berrys (ed.), Developmental mycology. John Wiley and Sons, New York, N.Y.
- 21.Harris, S. D. 2001. Septum formation in Aspergillus nidulans. Curr. Opin. Microbiol. 4:736-739. [DOI] [PubMed] [Google Scholar]
- 22.Harris, S. D., J. L. Morrell, and J. E. Hamer. 1994. Identification and characterization of Aspergillus nidulans mutants defective in cytokinesis. Genetics 136:517-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hunsley, D., and G. W. Gooday.1974. The structure and development of septa in Neurospora crassa. Protoplasma 82:125-146. [DOI] [PubMed] [Google Scholar]
- 24.Jedd, G., and N. H. Chua. 2000. A new self-assembled peroxisomal vesicle required for efficient resealing of the plasma membrane. Nat. Cell Biol. 2:226-231. [DOI] [PubMed] [Google Scholar]
- 25.Lee, S. B., M. G. Milgroom, and J. W. Taylor. 1988. A rapid, high yield mini-prep method for isolation of total genomic DNA from fungi. Fungal Genet. Newsl. 35:23-24. [Google Scholar]
- 26.Margolin, B. S., M. Freitag, and E. U. Selker. 1997. Improved plasmids for gene targeting at the his-3 locus of Neurospora crassa by electroporation. Fungal Genet. Newsl. 44:34-36. [Google Scholar]
- 27.Matsui, Y., and A. Toh-E. 1992. Yeast RHO3 and RHO4 ras superfamily genes are necessary for bud growth, and their defect is suppressed by a high dose of bud formation genes CDC42 and BEM1. Mol. Cell. Biol. 12:5690-5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27a.McCluskey, K. 2003. The Fungal Genetics Stock Center: from molds to molecules. Adv. Appl. Microbiol. 52:245-262. [DOI] [PubMed] [Google Scholar]
- 28.Miller, P. J., and D. I. Johnson. 1994. Cdc42p GTPase is involved in controlling polarized cell growth in Schizosaccharomyces pombe. Mol. Cell. Biol. 14:1075-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Minke, P. F., I. H. Lee, and M. Plamann. 1999. Microscopic analysis of Neurospora ropy mutants defective in nuclear distribution. Fungal Genet. Biol. 28:55-67. [DOI] [PubMed] [Google Scholar]
- 30.Momany, M., and J. E. Hamer. 1997. Relationship of actin, microtubules, and crosswall synthesis during septation in Aspergillus nidulans. Cell Motil. Cytoskel. 38:373-384. [DOI] [PubMed] [Google Scholar]
- 31.Nakano, K., T. Mutoh, R. Arai, and I. Mabuchi. 2003. The small GTPase Rho4 is involved in controlling cell morphology and septation in fission yeast. Genes Cells 8:357-370. [DOI] [PubMed] [Google Scholar]
- 32.Olofsson, B. 1999. Rho guanine dissociation inhibitors: pivotal molecules in cellular signalling. Cell Signal 11:545-554. [DOI] [PubMed] [Google Scholar]
- 33.Perkins, D. D. 1984. Advantage of using the inactive mating type am1 strain as a helper component in heterokaryons. Neurospora Newsl. 31:41-42. [Google Scholar]
- 34.Pring, M., M. Evangelista, C. Boone, C. Yang, and S. H. Zigmond. 2003. Mechanism of formin-induced nucleation of actin filaments. Biochemistry 42:486-496. [DOI] [PubMed] [Google Scholar]
- 35.Pruyne, D., M. Evangelista, C. Yang, E. Bi, S. Zigmond, A. Bretscher, and C. Boone. 2002. Role of formins in actin assembly: nucleation and barbed-end association. Science 297:612-615. [DOI] [PubMed] [Google Scholar]
- 36.Punt, P. J., M. A. Dingemanse, B. J. Jacobs-Meijsing, P. H. Pouwels, and C. A. van den Hondel. 1988. Isolation and characterization of the glyceraldehyde-3-phosphate dehydrogenase gene of Aspergillus nidulans. Gene 69:49-57. [DOI] [PubMed] [Google Scholar]
- 37.Raju, N. B. 1992. Genetic control of the sexual cycle in Neurospora. Mycol. Res. 96:241-262. [Google Scholar]
- 38.Rappaport, R. 1986. Establishment of the mechanism of cytokinesis in animal cells. Int. Rev. Cytol. 105:245-281. [DOI] [PubMed] [Google Scholar]
- 39.Royer, J. C., and C. T. Yamashiro. 1992. Generation of transformable spheroplasts from mycelia, macroconidia and germinating ascospores of Neurospora crassa. Fungal Genet. Newsl. 39:76-79. [Google Scholar]
- 40.Ryan, F. J., G. W. Beadle, and E. L. Tatum. 1943. The tube method of measuring the growth rate of Neurospora. Am. J. Bot. 30:784-799. [Google Scholar]
- 41.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 42.Santos, B., J. Gutierrez, T. M. Calonge, and P. Pérez. 2003. Novel Rho GTPase involved in cytokinesis and cell wall integrity in the fission yeast Schizosaccharomyces pombe. Eukaryot. Cell 2:521-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmidt, A., and A. Hall. 2002. Guanine nucleotide exchange factors for Rho GTPases: turning on the switch. Genes Dev. 16:1587-1609. [DOI] [PubMed] [Google Scholar]
- 44.Selker, E. U. 1990. Premeiotic instability of repeated sequences in Neurospora crassa. Annu. Rev. Genet. 24:579-613. [DOI] [PubMed] [Google Scholar]
- 45.Sharpless, K. E., and S. D. Harris. 2002. Functional characterization and localization of the Aspergillus nidulans formin SEPA. Mol. Biol. Cell 13:469-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shiu, P. K. T., N. B. Raju, D. Zickler, and R. L. Metzenberg. 2001. Meiotic silencing by unpaired DNA. Cell 107:905-916. [DOI] [PubMed] [Google Scholar]
- 47.Sprang, S. R. 1997. G protein mechanisms: insights from structural analysis. Annu. Rev. Biochem. 66:639-678. [DOI] [PubMed] [Google Scholar]
- 48.Tenney, K., I. Hunt, J. Sweigard, J. I. Pounder, C. McClain, E. J. Bowman, and B. J. Bowman. 2000. Hex-1, a gene unique to filamentous fungi, encodes the major protein of the Woronin body and functions as a plug for septal pores. Fungal Genet. Biol. 31:205-217. [DOI] [PubMed] [Google Scholar]
- 49.Vogel, H. J. 1956. A convenient growth medium for Neurospora. Microbiol. Genet. Bull. 13:42-46. [Google Scholar]
- 50.Watanabe, N., P. Madaule, T. Reid, T. Ishizaki, G. Watanabe, A. Kakizuka, Y. Saito, K. Nakao, B. M. Jockusch, and S. Narumiya. 1997. p140mDia, a mammalian homolog of Drosophila diaphanous, is a target protein for Rho small GTPase and is a ligand for profilin. EMBO J. 16:3044-3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wendland, J. 2003. Analysis of the landmark protein Bud3 of Ashbya gossypii reveals a novel role in septum construction. EMBO Rep. 4:200-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wendland, J., Y. Ayad-Durieux, P. Knechtle, C. Rebischung, and P. Philippsen.2000. PCR-based gene targeting in the filamentous fungus Ashbya gossypii. Gene 242:381-391. [DOI] [PubMed] [Google Scholar]
- 53.Wendland, J., and P. Philippsen. 2002. An IQGAP-related protein, encoded by AgCYK1, is required for septation in the filamentous fungus Ashbya gossypii. Fungal Genet. Biol. 37:81-88. [DOI] [PubMed] [Google Scholar]
- 54.Westergaard, M., and H. K. Mitchell. 1947. Neurospora. V. A synthetic medium favoring sexual reproduction. Am. J. Bot. 34:573-577. [Google Scholar]
- 55.Westfall, P. J., and M. Momany. 2002. Aspergillus nidulans septin AspB plays pre- and postmitotic roles in septum, branch, and conidiophore development. Mol. Biol. Cell 13:110-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wheatley, S. P., and Y. Wang. 1996. Midzone microtubule bundles are continuously required for cytokinesis in cultured epithelial cells. J. Cell Biol. 135:981-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Whiteway, M., D. Dignard, and D. Y. Thomas. 1992. Dominant negative selection of heterologous genes: isolation of Candida albicans genes that interfere with Saccharomyces cerevisiae mating factor-induced cell cycle arrest. Proc. Natl. Acad. Sci. USA 89:9410-9414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yeadon, P. J., and D. E. A. Catcheside. 1996. Quick method for producing template for PCR from Neurospora cultures. Fungal Genet. Newsl. 43:71. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.