Abstract
Tandem affinity purification (TAP) allows for rapid and efficient purification of epitope-tagged protein complexes from crude extracts under native conditions. The method was established in yeast and has been successfully applied to other organisms, including mammals and trypanosomes. However, we found that the original method, which is based on the TAP tag, consisting of a duplicate protein A epitope, a tobacco etch virus protease cleavage site, and the calmodulin-binding peptide (CBP), did not yield enough recovery of transcription factor SNAPc (for small nuclear RNA-activating protein complex) from crude trypanosome extracts for protein identification. Specifically, the calmodulin affinity chromatography step proved to be inefficient. To overcome this problem, we replaced CBP by the protein C epitope (ProtC) and termed this new epitope combination PTP tag. ProtC binds with high affinity to the monoclonal antibody HPC4, which has the unique property of requiring calcium for antigen recognition. Thus, analogous to the calcium-dependent CBP-calmodulin interaction, ProtC-tagged proteins can be released from immobilized HPC4 by a chelator of divalent cations. While this property was retained, epitope substitution improved purification in our experiments by eliminating the inefficiency of calmodulin affinity chromatography and by providing an alternative way of elution using the ProtC peptide in cases where EGTA inactivated protein function. Furthermore, HPC4 allowed highly sensitive and specific detection of ProtC-tagged proteins after protease cleavage. Thus far, we have successfully purified and characterized the U1 small nuclear ribonucleoprotein particle, the transcription factor complex TATA-binding protein related factor 4 (TRF4)/SNAPc/transcription factor IIA (TFIIA), and RNA polymerase I of Trypanosoma brucei.
Tandem affinity purification (TAP) has been developed as an efficient tool for protein complex purification under nondenaturing conditions (23, 24). For this method, the protein of interest must be fused to the TAP tag and expressed in the organism or cell line under investigation. The TAP tag contains two immunoglobulin G (IgG)-binding domains of the Staphylococcus aureus protein A (ProtA) and the calmodulin-binding peptide (CBP). Both epitope types are separated by spacer regions and a cleavage site for tobacco etch virus (TEV) protease. In consecutive steps, TAP is achieved by binding of the tagged protein to an IgG column, release of the protein by TEV protease cleavage, binding of the CBP-tagged protein to a calmodulin column, and final elution of the bound protein by a buffer containing a chelating agent of divalent cations such as EGTA. The final elution step is facilitated by calcium cation dependence of CBP binding to calmodulin. The TAP method has been developed in the budding yeast Saccharomyces cerevisiae, and it has been outstandingly successful in this organism, enabling a thorough proteome characterization by systematic analysis of protein complexes (8). TAP has two great advantages over conventional chromatographic methods. First, it is extremely efficient, yielding pure protein in sufficient amounts for mass spectrometric (MS) identification from a few liters of cell culture. For example, purification of yeast RNase P by TAP from 4 liters of cell culture was as efficient as conventional purification of this enzyme from 100 liters of cell culture (3, 23). Second, the whole procedure including the final elution step can be carried out under mild, nondenaturing conditions under which the purified protein complex usually retains functionality. TAP has been successfully applied to other organisms, including mammals (13), insects (7), Escherichia coli (9), plants (25), and trypanosomatids.
The latter are a family of unicellular flagellated eukaryotes harboring the human parasites Trypanosoma brucei, Trypanosoma cruzi, and Leishmania spp. In trypanosomatids, TAP was successfully applied to the exosome (6) and the protein machinery for U insertion/deletion editing of mitochondrial pre-mRNA (1, 2, 22). In contrast to these studies, our own attempts to tandem affinity purify the small nuclear RNA-activating protein complex (SNAPc) from crude trypanosome extracts failed, due to the inefficiency of the calmodulin affinity purification step. Similar unsuccessful results with TAP-tagged proteins were personally communicated to us by other researchers in the field (for example, Elisabetta Ullu and Christian Tschudi, Yale University; Larry Simpson, UCLA). The problem extends to other organisms as well. For example, the original TAP method did not yield enough recovery for protein identification in mammalian cells grown in monolayers, prompting modification of the technique (4).
To overcome the limitation of the calmodulin chromatography step in our purifications, we replaced CBP by the 12-amino-acid short protein C epitope (ProtC). The new epitope combination was designated PTP (for ProtC-TEV-ProtA) tag. ProtC is derived from human protein C, a vitamin K-dependent plasma zymogen specifically expressed in hepatocytes. The monoclonal antibody HPC4 recognizes this epitope with high affinity and, as a unique property, has a calcium-binding site which needs to be occupied for its interaction with ProtC (29). Thus, a ProtC-tagged protein bound to immobilized HPC4 can be released by an EGTA-containing buffer analogously to elution of CBP-tagged proteins from a calmodulin column (26). Thus far, we have PTP tagged, purified, and characterized the six-component transcription factor TATA-binding protein related factor 4 (TRF4)/SNAPc/transcription factor IIA (TFIIA) (28) and the U1 small nuclear ribonucleoprotein particle (21). In this study, we compared TAP versus PTP purification by C-terminal tagging of TbSNAP50, a subunit of SNAPc. We also established N-terminal PTP tagging and purification, and we describe two “in allele” integration vectors in which the coding sequence of a target protein can be fused either N or C terminally to the PTP tag in a single cloning step. To corroborate the general applicability of PTP tagging and purification, we show the purification of two other protein complexes, namely that of RNA polymerase I (Pol I) and TFIIA. Finally, we demonstrate that PTP purification can yield functional proteins.
MATERIALS AND METHODS
Plasmid construction.
pC-PTP-NEO and pN-PURO-PTP are T. brucei genome integration vectors derived from pBluescript SK(+) (Stratagene, La Jolla, Calif.) containing a PTP and a resistance marker cassette each. In pC-PTP-NEO, the PTP cassette contains 745 bp of a TbU1-70K C-terminal coding region (21), a NotI restriction site, the PTP tag coding sequence, the translation stop codon TGA, and 470 bp of 3′ flank from TbRPA1, the gene encoding the largest subunit of RNA polymerase I. The resistance marker cassette contains the neomycin phosphotransferase gene (NEOr) flanked 5′ and 3′ by the intergenic regions of heat shock protein 70 (HSP70) genes 2 and 3 and of β- and α-tubulin genes, respectively. In a further development of the original resistance marker cassette (16), we separated the HSP70 intergenic region from NEOr by an NdeI restriction site and NEOr from the tubulin flank by BamHI, HpaI, and BstBI restrictions sites (Fig. 1). In constructing pN-PURO-PTP, the resistance marker cassette was modified by replacing NEOr by the coding region of the puromycin N-acetyltransferase gene (PUROr). The downstream PTP cassette is composed of 434 bp of 5′ flank from the TbRPA2 gene, encoding the second-largest subunit of RNA polymerase I, a translation initiation codon, the PTP tag coding sequence, and unique restriction sites for fusing target sequences to the tag (Fig. 1).
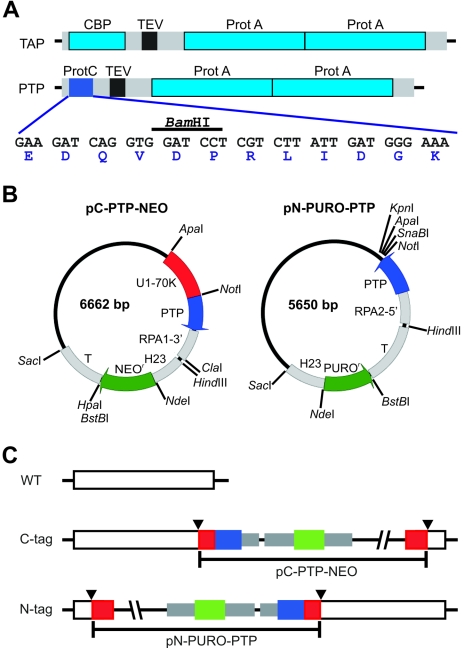
FIG. 1.
PTP tagging. (A) Schematic delineation (to scale) of the TAP and PTP tags. The ProtA and CBP epitopes of the original TAP tag are drawn in light blue, the TEV protease site is black, and spacer sequences are gray. In the PTP tag, the ProtC epitope is depicted in blue; its coding sequence, including a diagnostic BamHI restriction site and the amino acid sequence of the ProtC peptide, is provided below. (B) Circular map (to scale) of the T. brucei genome integration vectors pC-PTP-NEO and pN-PURO-PTP. Constructs are derivatives of pBluescript SK(+) and designed for genome integration. Each of them contains a PTP cassette for fusion of the PTP tag sequence to the target gene and a resistance marker cassette. The PTP sequence, T. brucei gene flanks, and the resistance marker (NEOr and PUROr) coding sequences are drawn in blue, gray, and green, respectively. Arrows indicate the direction of transcription. Most modules of the vectors can be individually excised by unique restriction sites as indicated. Gene flanks providing RNA processing signals for PTP fusion and resistance marker are the 3′ flank of TbRPA1 (RPA1-3′), HSP70 genes 2 and 3 intergenic region (H23), the β-α tubulin intergenic region (T), and the 5′ flank of TbRPA2 (RPA2-5′). While pC-PTP-NEO contains a target sequence (U1-70K, in red) which needs to be replaced, in pN-PURO-PTP a target sequence has to be inserted between the NotI and the following three restriction sites. (C) Illustration of PTP fusion to target genes. For PTP tagging of T. brucei proteins, pC-PTP-NEO and pN-PURO-PTP derivatives must be linearized inside the target sequence, depicted in red. Therefore, a prerequisite for targeted insertion of the constructs by homologous recombination is a unique restriction site within the target sequence (arrowheads). Colors are as above.
pTbSNAP50-PTP-NEO, pTbRPA1-PTP-NEO, and pTbTFIIA-2-PTP-NEO are derivatives of pC-PTP-NEO. In each case, the C-terminal protein-coding region preceding the PTP sequence was exchanged and comprised 462 bp, 410 bp, and 400 bp, respectively. In addition, in pTbSNAP50-PTP-NEO, the BstBI site of the resistance marker cassette was destroyed. pTbSNAP50-TAP-NEO corresponds to its PTP counterpart and has the PTP sequence precisely replaced by the coding region of the original TAP construct. pPURO-PTP-TbSNAP50 was derived from pN-PURO-PTP by insertion of 530 bp of the TbSNAP50 N-terminal coding sequence downstream of the PTP sequence. For genomic integration, pTbSNAP50-PTP-NEO and pTbSNAP50-TAP-NEO were linearized with BstBI, pTbRPA1-PTP-NEO was linearized with SalI, pTFIIA-2-PTP-NEO was linearized with StuI, and pPURO-PTP-TbSNAP50 was linearized with BspMI.
Cell culture and generation of cell lines.
Cultivation of procyclic forms of T. brucei brucei strain 427 was carried out as previously described (15). For generation of cell lines expressing PTP-tagged proteins, 10 μg of linearized PTP vectors was transfected into procyclic 427 cells by electroporation (10). Transfected cells were cloned by limiting dilution and selected with 4 μg of puromycin/ml or 40 μg of G418/ml. Correct integration of constructs was verified by PCR and Southern analysis, and expression of PTP-tagged proteins was analyzed by immunoblotting with HPC4 antibodies.
PTP purification TAP.
For a standard tandem affinity purification, a 2.5-liter procyclic T. brucei culture was grown to a density of 2 × 107 cells per ml, which corresponded to a packed cell volume of approximately 4 ml. Cell extract from this material was prepared as described previously (15), except that the extract was not concentrated. The extract had a volume of 6.5 ml and contained 150 mM sucrose, 150 mM potassium chloride, 20 mM potassium l-glutamate, 3 mM MgCl2, 20 mM HEPES-KOH (pH 7.7), 1 mM dithiothreitol, 10 μg/ml leupeptin, 10 μg/ml aprotinin, 0.1% Tween 20, and half of a Complete Mini, EDTA-free protease inhibitor cocktail tablet (Roche, Indianapolis, IN). For IgG affinity chromatography, the extract was transferred to a 0.8- by 4-cm Poly-Prep chromatography column (Bio-Rad, Hercules, CA) in which a 200-μl settled bead volume of IgG Sepharose 6 Fast Flow beads (Amersham, Piscataway, NJ) had been equilibrated with PA-150 buffer (150 mM potassium chloride, 20 mM Tris-HCl [pH 7.7], 3 mM MgCl2, 0.5 mM dithiothreitol, 0.1% Tween 20). PTP- or TAP-tagged proteins were bound to IgG Sepharose by rotation of the closed column for 2 h at 4°C. After the flowthrough was collected, beads were washed with 35 ml of PA-150 buffer and equilibrated with 15 ml of TEV protease buffer (PA-150 with 0.5 mM EDTA). Tagged proteins were eluted by resuspending the beads in 2 ml of TEV protease buffer containing 300 units of AcTEV protease (Invitrogen, Carlsbad, CA) and rotating the closed column overnight at 4°C. The TEV protease eluate was diluted to 6.5 ml by a wash of the IgG Sepharose beads with PC-150 buffer (PA-150 buffer containing 1 mM calcium chloride) and the contents of the second half of the protease inhibitor tablet. For anti-ProtC affinity purification, calcium chloride was added to the eluate to a final concentration of 2 mM. The eluate was combined in a new column with a 200-μl settled bead volume of either anti-protein C affinity matrix (Roche, Indianapolis, IN) or calmodulin affinity resin (Stratagene, La Jolla, CA) equilibrated in buffer PC-150 and rotated for 2 h at 4°C. After the flowthrough was collected, the matrix was washed with 60 ml of PC-150 and ProtC- or CBP-tagged proteins were eluted with EGTA elution buffer (5 mM Tris-HCl [pH 7.7], 10 mM EGTA, 5 mM EDTA, 10 μg/ml leupeptin). Alternatively, ProtC-tagged proteins were eluted with peptide elution buffer (transcription buffer with 0.5 mg/ml ProtC peptide). The peptide eluate was used without further concentration in functional assays. The volume of the EGTA eluate was reduced from 3 ml to approximately 600 μl by evaporation in a vacuum concentrator. Subsequently, the proteins were bound to 10 μl of hydrophobic StrataClean resin (Stratagene, La Jolla, CA), released into sodium dodecyl sulfate (SDS) loading buffer at 80°C, separated on SDS-polyacrylamide gel electrophoresis (PAGE) gels, and Coomassie stained with the GelCode blue stain reagent (Pierce, Rockford, IL). A detailed step-by-step protocol of PTP purification is available upon request.
Proteins were identified by liquid chromatography-tandem MS and quantified by densitometry against Coomassie-stained bovine serum albumin and IgG standards which were analyzed in parallel. In immunoblot analyses, PTP- or ProtC-tagged proteins were separated on SDS-PAGE gels, electroblotted onto a polyvinylidene difluoride membrane, and detected by either the ProtA-specific PAP (peroxidase-anti-peroxidase soluble complex) reagent (Sigma, St. Louis, MO) or the anti-ProtC antibody HPC4 in combination with the BM Chemiluminescence Blotting substrate according to the manufacturer's protocol (Roche, Indianapolis, IN). Relative signal strengths on one blot were determined by densitometry.
In vitro transcription assay.
In vitro transcription reactions of SLins19 template DNA (12) and GPEET-trm template DNA (15) were conducted as previously described in detail (14, 27) except that only 4 μl instead of 8 μl of extract was used. For SLins19 transcription activation, 12 μl of peptide eluate was added to a transcription reaction mixture. To specifically detect GPEET-trm and SLins19 transcripts, total RNA was prepared from each reaction mixture and analyzed by extension of 32P-end-labeled primers Tag_PE (15) and SLtag (12) which are complementary to unrelated oligonucleotide tags of GPEET-trm and SLins19, respectively. The primer extension products were separated on 6% polyacrylamide-50% urea gels and visualized by autoradiography.
Nucleotide sequence accession numbers.
The complete sequences of pC-PTP-NEO (accession number DQ172900) and pN-PURO-PTP (accession number DQ172901) were submitted to GenBank.
RESULTS
Vectors for PTP tagging of T. brucei proteins.
The PTP tag is a derivative of the original TAP tag and has CBP replaced by ProtC (Fig. 1A), reducing the overall size of the tag from 184 to 169 amino acids and the molecular mass from 20.6 to 18.9 kDa. After TEV protease cleavage, 44 and 29 amino acids accounting for 5.1 and 3.4 kDa, respectively, remain on TAP- and PTP-tagged proteins. To facilitate stable C- and N-terminal PTP tagging of proteins in T. brucei, we generated the genome integration vectors pC-PTP-NEO and pN-PURO-PTP (Fig. 1B). Each of these constructs consists of a PTP and a resistance marker cassette. In the PTP cassettes, the PTP coding sequence is fused to a gene flank, providing essential RNA processing signals on one side, and to a NotI restriction site, which facilitates fusion of a protein coding region to the PTP tag in a single cloning step on the other side.
The constructs are designed for targeted and stable integration of the PTP sequence into an endogenous allele. In this way, PTP-tagged proteins are expressed at normal levels, and potential nonspecific protein-protein interactions due to overexpression (23) from strong class I promoters available for T. brucei are avoided. Since targeted genome integration of linear DNA by homologous recombination is a very efficient and accurate process in T. brucei, typically requiring only 100 to 300 bp of target sequence on either side of the linearization site (17, 30), only the 5′ (N-terminal tagging) or the 3′ (C-terminal tagging) terminal coding sequence needs to be cloned into these vectors, a further advantage in case the protein of interest is large.
For targeted insertion of the construct into the genome, the vectors must be linearized within the target sequence (Fig. 1C). Thus, a prerequisite for successful tagging is the identification of a unique restriction site inside this sequence. To facilitate identification of a unique restriction site, the complete sequences of pC-PTP-NEO and pN-PURO-PTP were submitted to GenBank.
Tandem affinity purification of PTP- and TAP-tagged TbSNAP50.
Our initial failure to tandem affinity purify SNAPc by C-terminal TAP tagging of TbSNAP50 prompted us to develop the PTP method. By employing the latter, we were able to purify and characterize a stable transcription factor complex consisting of TRF4, three subunits of SNAPc (TbSNAP50, TbSNAP2, and TbSNAP3), and two subunits of TFIIA (TFIIA-1 and TFIIA-2) (28). This complex is essential for transcription of the spliced leader (SL) RNA gene encoding the small nuclear RNA that serves as a substrate in SL addition trans splicing of nuclear pre-mRNA (for a review, see reference 19).
To compare TAP and PTP purifications of TbSNAP50 in this study, we tagged this protein in three different ways: N-terminal with the PTP tag (PTP-SNAP50), C-terminal with the PTP tag (SNAP50-PTP), and C-terminal with the TAP tag (SNAP50-TAP). In each case, a clonal cell line was generated which had the construct integrated in one allele and retained the wild type in the other allele (data not shown). Immunodetection of the ProtA epitope revealed that each of these cell lines specifically expressed a tagged protein of approximately 70 kDa in comparable amounts (Fig. 2A). The apparent sizes of the tagged proteins corresponded well with the expected masses, which were 69.1 kDa, 70.1 kDa, and 71.9 kDa for PTP-SNAP50, SNAP50-PTP, and SNAP50-TAP, respectively.
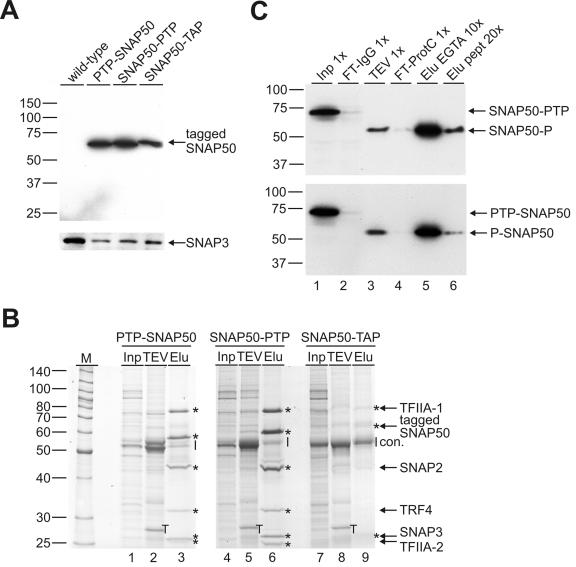
FIG. 2.
PTP and TAP purification of TbSNAP50. (A) Immunoblot analysis of whole-cell lysates from wild-type cells and from cells expressing TbSNAP50 with an N-terminal PTP tag (PTP-SNAP50), with a C-terminal PTP tag (SNAP50-PTP), or with a C-terminal TAP tag (SNAP50-TAP). Tagged proteins were specifically detected with the PAP reagent directed against the ProtA domains. As a protein loading control, the same blot was reprobed with an anti-TbSNAP3 antibody. On the left, masses of protein markers (in kilodaltons) are indicated. (B) Coomassie staining of protein fractions. For each purification, 0.002% of input material (Inp), 5% of TEV protease eluate (TEV), and 100% of the final EGTA eluate (Elu) were separated on a 12.5% SDS-PAGE gel and stained with Coomassie stain. Input material was loaded to visualize the most abundant proteins. Asterisks mark detectable subunits of the TRF4/SNAPc/TFIIA complex as specified on the right, the vertical line identifies a region of contaminating proteins (con.), and T indicates the TEV protease band. SNAP50-PTP and SNAP50-TAP fractions were analyzed on the same gel. Marker sizes are specified on the left. (C) Immunoblot analysis of SNAP50-PTP (top) and PTP-SNAP50 (bottom) purifications. PTP-tagged and, after TEV protease cleavage, P-tagged SNAP50 (indicated on the right) were detected with anti-ProtC antibody in input material (Inp), flowthrough of the IgG Sepharose column (FT-IgG), TEV protease eluate (TEV), flowthrough of the anti-ProtC matrix (FT-ProtC), final EGTA eluate (Elu EGTA), and final peptide eluate (Elu pept). Values of x indicate relative amounts of each fraction analyzed. Marker sizes are indicated on the left.
A standard tandem affinity purification procedure was carried out with each of these cell lines. The starting material was derived from a 2.5-liter procyclic cell culture containing 5 × 1010 cells. Cells were broken, and a crude extract was prepared which contained a mixture of cytoplasmic and extracted nuclear components (15). PTP- and TAP-tagged proteins were treated exactly the same except that the anti-ProtC matrix was replaced by calmodulin affinity resin for purification of SNAP50-TAP (see Materials and Methods). For each purification, input material, TEV protease eluate of the IgG column, and final EGTA eluate were separated on an SDS-PAGE gel and visualized by Coomassie staining (Fig. 2B). While the TEV eluate looked similar in all three cases (Fig. 2B, compare lanes 2, 5, and 8), the final eluates differed considerably. PTP-tagged SNAP50 was purified in the range of 10 to 20 pmol, independently of whether the tag was fused N or C terminally (Fig. 2B, lanes 3 and 6). In both purifications, the three SNAPs copurified in approximately stoichiometric amounts; the quantities of TRF4 and TFIIA subunits were not much lower, indicating that the conditions of the PTP purification were mild enough to quantitatively preserve the integrity of SNAPc and of TRF4/SNAPc/TFIIA. Despite these mild conditions, there was very little contaminating material detectable. A consistent but minor contamination was detected above the 50-kDa marker band (Fig. 2B, con.) which, according to MS analysis, consisted mainly of α- and β-tubulin, as well as IgG heavy chain (data not shown). To assess whether the purified material was suitable for protein identification, the SNAP50-P band (Fig. 2B, lane 6) was excised and analyzed by liquid chromatography-tandem MS. A total of 19 peptides covering 40% of the amino acid sequence were identified which allowed unambiguous identification of SNAP50 by an omniblast search of the GeneDB database (http://www.genedb.org/) (data not shown). In contrast to the PTP purifications, the eluate of the SNAP50-TAP purification contained very little tagged SNAP50 and components of TRF4/SNAPc/TFIIA were hardly detectable, while tubulin contamination was notably stronger (Fig. 2B, lane 9).
To evaluate the efficiency of each step in SNAP50-PTP and PTP-SNAP50 purifications, we employed immunoblotting using the anti-ProtC antibody (Fig. 2C). The results for both purifications were very similar. A 2-h-long incubation at 4°C of crude extract and IgG Sepharose was sufficient to bind >90% of the tagged protein to the matrix; only trace amounts of the protein were detectable in the column flowthrough (Fig. 2C, compare lanes 1 and 2). This result was in accordance with those of the original TAP method (23, 24). As a consequence of TEV protease cleavage, SNAP50-PTP and PTP-SNAP50 were shortened to SNAP50-P and P-SNAP50, respectively, resulting in faster-migrating protein bands (Fig. 2C, compare lane 1 with lanes 3 to 6). TEV protease cleavage recovered 39% of SNAP50-P and 33.5% of P-SNAP50 relative to input material. Although the PTP tag harbored only one ProtC epitope, anti-ProtC immunoaffinity chromatography, the second purification step, was almost as efficient as IgG affinity chromatography. More than 80% of tagged SNAP50 bound to the anti-ProtC matrix in both purifications (Fig. 2C, compare lanes 3 and 4). After elution of bound protein in an EGTA-containing buffer and concentration of purified protein into a small sample volume, approximately half of this material was recovered (lane 5). For the SNAP50-PTP purification, we calculated that 21% of the protein present in the input material was recovered in the final eluate. This corresponded to a 54% recovery in the anti-ProtC affinity purification step alone. The PTP-SNAP50 purification was slightly less efficient, and the corresponding values were 14% and 42%. EGTA elution, however, has the disadvantage that chelating of divalent cations may irreversibly destroy protein stability and function. In such cases, it is useful if matrix-bound protein can be eluted under native conditions in the absence of chelating agents. We therefore synthesized ProtC peptide and eluted ProtC-tagged SNAP50 in transcription buffer containing 0.5 mg of this peptide/ml. While peptide elution efficiency was about an order of magnitude lower than that of EGTA elution (Fig. 2C, lane 6), it provided a functional TRF4/SNAPc/TFIIA complex (see below). Another advantage of the PTP method in our experiments was the highly specific detection of PTP-tagged proteins by the HPC4 antibody throughout the purification (Fig. 2C). In contrast, a commercially available anti-CBP antibody cross-reacted with a 37-kDa large trypanosome protein, very strongly obscuring the immunoblot analysis of the SNAP-TAP purification (data not shown). By detecting the ProtA epitopes before the TEV protease cleavage, we confirmed that IgG affinity chromatography worked as efficiently for SNAP-TAP as for the PTP-tagged proteins (data not shown). This result strongly indicated that the lower efficiency of the SNAP-TAP purification was due to the calmodulin affinity purification.
PTP purification of T. brucei RNA Pol I and TFIIA.
Besides PTP-tagged SNAPc, we have thus far successfully purified PTP-tagged U1 snRNP (21) and TRF4 (28). In each case, MS analysis identified conserved proteins which had not been annotated before and for which their functional role was experimentally confirmed. In addition, we have previously shown the final eluate on a Coomassie-stained gel of a TFIIA-2-PTP purification (28). In this study, we analyzed TFIIA-2-PTP purification in detail in a second purification; to further demonstrate the applicability of the PTP method to other protein complexes, we PTP tagged and purified T. brucei RNA Pol I.
In a recent study, T. brucei RNA Pol I was characterized by TAP tagging and purification (31). Five RNA Pol subunits were identified, namely TbRPA1, TbRPA2, TbRPC40, Tb1RPB5, and TbRPA12. We tagged the largest subunit TbRPA1 C terminally with the PTP tag and conducted a standard PTP purification. Immunoblotting showed that the purification efficiency was comparable to those of the SNAP50 purifications except that peptide elution for this protein complex was reproducibly less efficient (Fig. 3A). Coomassie staining of purified RNA Pol I revealed seven specific bands (Fig. 3B). An MS analysis identified eight subunits because two subunits comigrated in the band with an apparent mass of 16 kDa. In addition to the five subunits found by Walgraffe et al. (31), three bona fide RNA Pol subunits were identified which will be published elsewhere (T. N. Nguyen et al., manuscript in preparation). TbRPA1-PTP purification resulted reproducibly in higher amounts of copurified α- and β-tubulin than other purifications of PTP-tagged proteins (Fig. 3B, lane Elu). Whether the association of tubulins and RNA Pol I is of functional significance or a particularly strong nonspecific interaction remains to be determined. Finally, the remaining doublet band with an apparent mass of 27 kDa did not reproducibly copurify with RNA Pol I, a phenomenon we have not yet analyzed but observed previously (26). Densitometric quantification of the Coomassie-stained band revealed that approximately 3 μg of TbRPA1-P was purified (note that only 33% of the final eluate was loaded in this case), corresponding to 15.2 pmol or to approximately 180 molecules purified from each trypanosome cell. These numbers underscore the effectiveness of PTP purification.
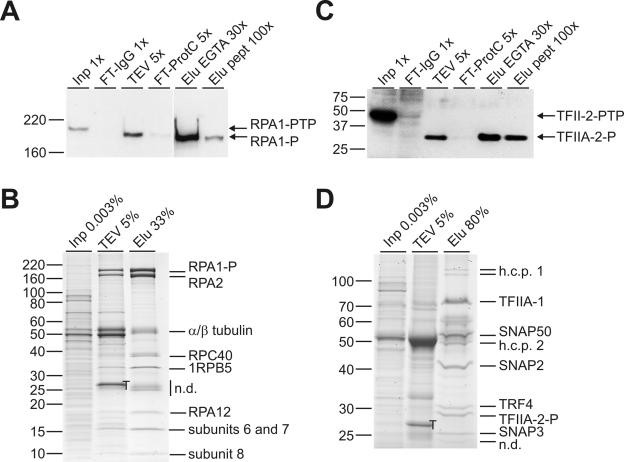
FIG. 3.
PTP purification of T. brucei RNA Pol I and TFIIA. TbRPA1 (RPA1-PTP) and TbTFIIA-2 (TFIIA-2-PTP) were C terminally PTP tagged in individual cell lines and purified according to the standard protocol. (A) Immunoblot analysis of the RPA1-PTP purification. RPA1 fractions, shown as in Fig. 2C, were separated on a 5% SDS-PAGE gel, blotted, and detected with HPC4. Values of x indicate relative amounts of each fraction analyzed. (B) Input material (Inp), TEV eluate, and final EGTA eluate (Elu) of the RPA1-PTP purification were separated on a 10 to 20% SDS-PAGE gradient gel in specified amounts and stained with Coomassie stain. Protein bands below RPA2 were identified by MS. n.d., not determined. (C and D) Corresponding panels for the TFIIA-2-PTP purification except that for immunoblotting and Coomassie staining, proteins were separated on 14% and 12.5% SDS-PAGE gels, respectively. T, TEV protease; h.c.p., hypothetical conserved protein; n.d., not determined. Note that in panels B and D, only part of the final eluate was loaded. On the left of all panels, protein marker masses in kilodaltons are indicated.
Immunoblot analysis of the TFIIA-2-PTP purification uncovered two differences to TbRPA1-PTP purification. While TEV protease eluted tagged TFIIA-2 poorly (Fig. 3A and C, compare lanes Inp with lanes TEV), final elution through the ProtC peptide was highly efficient (compare lanes Elu EGTA with lanes Elu pept). Since these results were obtained in two independent purifications, accessibility of both TEV protease site and ProtC may be influenced sterically by the tagged protein complex. Despite the inefficiency of the TEV protease elution, TFIIA-2-PTP purification resulted in Coomassie-stainable polypeptides. As we have seen before, not only TRF4/SNAPc/TFIIA components copurified with TFIIA-2 but also several minor bands (Fig. 3D, lane Elu). It is unlikely that they were contaminations because they were not detectable in other TRF4/SNAPc/TFIIA purifications (Fig. 2B) (28). Accordingly, MS analysis of the doublet band of approximately 120 kDa (which is the same protein) and of the 49-kDa band identified two conserved trypanosomatid proteins with unknown function, rather than known proteins of high abundance (Fig. 3D, lane Elu, and data not shown).
Taken together, PTP tagging and purification of RNA Pol I and TFIIA-2 in T. brucei revealed highly pure protein complexes in sufficient amounts to identify copurified subunits.
Purified PTP-SNAP50 is functional.
A key concern of protein epitope tagging is the possibility that the epitope interferes with protein function. If the tagged protein is essential for a cell, generation of a cell line which exclusively expresses the tagged protein is a means of showing that tagging is not deleterious. We were able to generate such a cell line expressing TbRPA1 with a C-terminal PTP tag (data not shown). Since this RNA Pol I subunit is essential for T. brucei (11), we concluded that PTP-tagged RNA Pol I was functional in this cell line.
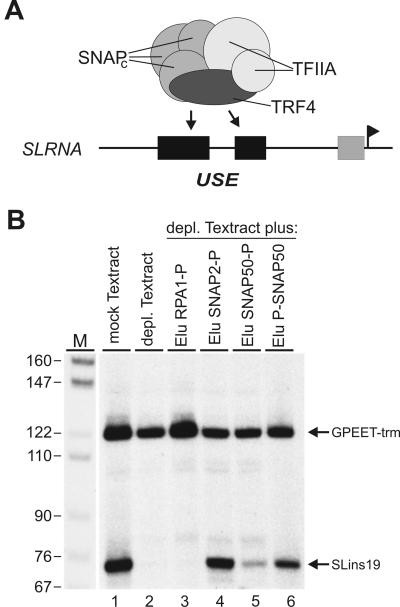
Purification of a functional protein is an alternative way to demonstrate that an epitope tag does not significantly interfere with function. Tandem affinity purification is likely to yield functional proteins because of the low-stringency conditions applied (see, for example, references 2, 5, 18, and 24). Accordingly, we have shown that purified TRF4/SNAPc/TFIIA with C-terminal PTP tags was capable of specifically binding to the upstream sequence element (USE) of the SL RNA gene promoter (Fig. 4A) and of fully reconstituting SL RNA gene transcription in a depleted cell extract (28). Interestingly, the complex was functional when the PTP tag was fused to SNAP2 or TRF4. However, when the tag was C terminally fused to SNAP50, complex functionality was tremendously reduced (reference 28 and see below). To investigate whether N-terminal PTP tagging of SNAP50 could circumvent this defect, we carried out in vitro transcription reactions in which the SL RNA gene template SLins19 was cotranscribed with the control template GPEET-trm. The latter harbors a procyclin gene promoter, which recruits RNA Pol I and does not bind TRF4/SNAPc/TFIIA. As shown in Fig. 4B, SLins19 transcription was abolished in the TRF4/SNAPc/TFIIA-depleted extract, whereas GPEET-trm transcription was not greatly affected (compare lanes 1 and 2). SLins19 transcription was restored when eluate derived from a SNAP2-PTP purification was added to the reaction (lane 4). This transcriptional activation was independent of the tag because PTP-purified RNA Pol I had no effect on SLins19 transcription (lane 3). Although addition of P-SNAP50 eluate resulted only in about 50% of SLins19 transcription compared to the reaction with SNAP2-P eluate (lane 6), it was much more efficient than the SNAP50-P eluate (lanes 5 and 6). Hence, we conclude that N-terminal PTP tagging and purification of SNAP50 results in a partially competent transcription factor complex and that both C- and N-terminal PTP tagging can yield functional proteins.
FIG. 4.
P-SNAP50 eluate restores SL RNA gene transcription in vitro. (A) Diagram of the SL RNA gene promoter and the TRF4/SNAPc/TFIIA complex. The promoter is drawn to scale and consists of the bipartite USE (black boxes) and a second element (gray box) close to the transcription initiation site indicated by the flag. The six-component transcription factor requires both USE elements for optimal DNA binding. (B) In vitro transcription analysis. Templates GPEET-trm and SLins19 were cotranscribed in mock-treated or TRF4/SNAPc/TFIIA-depleted (depl.) transcription extract (Textract). In further reaction mixtures, depleted extract was reconstituted with final peptide eluates of RPA1-PTP (RPA1-P), SNAP2-PTP (SNAP2-P), SNAP50-PTP (SNAP50-P), and PTP-SNAP50 (P-SNAP50) purifications. As confirmed by immunoblot analysis, equal amounts of SNAPs were added back through the SNAP eluates (data not shown). Transcription signals were obtained by primer extension assays, and extension products were separated on 6% polyacrylamide-50% urea gels and visualized by autoradiography. M, MspI-digested pBR322.
DISCUSSION
In this study, we created a new epitope combination for tandem affinity purification, termed PTP. It was generated by fusing two ProtA domains and the TEV protease cleavage site to ProtC, thereby replacing the CBP of the original TAP tag. Introduction of ProtC resulted in several advantages. First, purification of N- or C- terminal PTP-tagged SNAP50 from crude extracts was considerably more efficient than purification of this protein harboring the original TAP tag. The higher efficiency may be crucial if the protein to be purified is expressed at low levels or if it is difficult or expensive to grow a large culture volume of the organism to be investigated. Second, the monoclonal antibody HPC4 was a sensitive and highly specific tool for the detection of ProtC-tagged proteins in our experiments. This is especially important if there is no antibody available for the endogenous protein and the ProtA domains have been removed from the tagged protein by TEV protease cleavage. Third, ProtC offers two options for protein elution. As with CBP, ProtC-tagged proteins can be efficiently eluted from HPC4 beads by EGTA. However, if depletion of divalent cations irreversibly interferes with protein function, elution in the presence of the ProtC peptide is likely to yield functional protein. Since peptide elution can be carried out in any native buffer required for subsequent applications, a dialysis step becomes unnecessary and the eluate can be used directly in a functional assay.
Although we demonstrated successful PTP purification of different protein complexes, the protein yield may still be improved. While in both affinity chromatography steps almost all of the tagged protein bound to the column, only about half of this material was recovered in each case. Elution of tagged protein from IgG Sepharose is mediated by TEV protease. In our standard procedure, TEV protease cleavage was conducted overnight at 4°C to conserve protein function. The recovery rate may be increased if the cleavage reaction is conducted at a higher temperature, e.g., 16°C (23). Furthermore, as a modification to the original TAP protocol we replaced NP-40 by Tween 20 in the TEV protease cleavage buffer, due to our finding that NP-40 interferes with transcriptional activity in our extracts much more strongly than Tween 20 (data not shown). However, if the use of NP-40 is of no concern, this detergent may improve the yield of protein recovery by TEV protease. Protein loss in the second chromatography step cannot be attributed to inefficient elution because only minor amounts of tagged protein remained on the anti-ProtC matrix (data not shown). Therefore, protein loss most likely occurred during concentration of the final eluate. To minimize this loss, we evaluated different concentration procedures and found that a combination of reducing the eluate volume by evaporation and binding of proteins to a hydrophobic resin (32) was most effective (data not shown).
An open question is why the calmodulin affinity step in the original TAP method is efficient in some cases but not in others even when studies are conducted in the same organism. Since endogenous calmodulin can interact with CBP and prevent binding of the epitope to the calmodulin column, it is possible that the efficiency of the calmodulin affinity step is directly dependent on the relative amounts of free endogenous calmodulin and TAP-tagged protein in an extract, e.g., the endogenous calmodulin amount may not suffice to block purification of a more abundant protein, whereas it may efficiently block purification of a protein of low abundance, such as gene expression factors. Successful TAP applications in T. brucei are in accordance with such an explanation because the tagged exosome subunit was overexpressed for purification (6) and RNA editing complexes were purified from mitochondrial extracts, which probably lacked calmodulin (1, 2, 22). At any rate, the PTP method has eliminated this possibility.
Our experiments have been conducted with trypanosomal extracts but except for the extract preparation procedure, the method described here does not contain trypanosome-specific features. Thus, we anticipate that PTP tagging and purification can be applied equally well to other organisms. Protein C is expressed in mammals as a plasma component involved in the regulation of the blood coagulation cascade. The protein is expressed only in hepatocytes, due to transcriptional regulation by three liver-specific elements in its gene promoter (20). Furthermore, the HPC4-binding site of protein C is not well conserved among mammals; accordingly, the antibody does not recognize bovine protein C (29). Thus, specific detection and purification of ProtC-tagged proteins by the HPC4 antibody should be feasible in other organisms, including most mammalian cell lines.
In conclusion, tandem affinity purification of PTP-tagged proteins is a promising alternative to the well-established TAP method, especially in cases where calmodulin affinity chromatography is problematic or where purification efficiency is crucial.
Acknowledgments
This work was supported by a grant from the National Institutes of Health (AI059377) to A.G.
We thank Jens Brandenburg for his advise on keeping purified proteins in solution, the laboratory of Hubert Kalbacher (University of Tübingen, Tübingen, Germany) for synthesizing the ProtC peptide, and Mary Ann Gawinowicz (Protein Core Facility, Columbia University) for excellent MS analyses.
REFERENCES
- 1.Aphasizhev, R., I. Aphasizheva, R. E. Nelson, G. Gao, A. M. Simpson, X. Kang, A. M. Falick, S. Sbicego, and L. Simpson. 2003. Isolation of a U-insertion/deletion editing complex from Leishmania tarentolae mitochondria. EMBO J. 22:913-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aphasizhev, R., I. Aphasizheva, and L. Simpson. 2003. A tale of two TUTases. Proc. Natl. Acad. Sci. USA 100:10617-10622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chamberlain, J. R., Y. Lee, W. S. Lane, and D. R. Engelke. 1998. Purification and characterization of the nuclear RNase P holoenzyme complex reveals extensive subunit overlap with RNase MRP. Genes Dev. 12:1678-1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drakas, R., M. Prisco, and R. Baserga. 2005. A modified tandem affinity purification tag technique for the purification of protein complexes in mammalian cells. Proteomics 5:132-137. [DOI] [PubMed] [Google Scholar]
- 5.Dziembowski, A., A. P. Ventura, B. Rutz, F. Caspary, C. Faux, F. Halgand, O. Laprevote, and B. Seraphin. 2004. Proteomic analysis identifies a new complex required for nuclear pre-mRNA retention and splicing. EMBO J. 23:4847-4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Estevez, A. M., T. Kempf, and C. Clayton. 2001. The exosome of Trypanosoma brucei. EMBO J. 20:3831-3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Forler, D., T. Kocher, M. Rode, M. Gentzel, E. Izaurralde, and M. Wilm. 2003. An efficient protein complex purification method for functional proteomics in higher eukaryotes. Nat. Biotechnol. 21:89-92. [DOI] [PubMed] [Google Scholar]
- 8.Gavin, A. C., M. Bösche, R. Krause, P. Grandi, M. Marzioch, A. Bauer, J. Schultz, J. M. Rick, A. M. Michon, C. M. Cruciat, M. Remor, C. Höfert, M. Schelder, M. Brajenovic, H. Ruffner, A. Merino, K. Klein, M. Hudak, D. Dickson, T. Rudi, V. Gnau, A. Bauch, S. Bastuck, B. Huhse, C. Leutwein, M. A. Heurtier, R. R. Copley, A. Edelmann, E. Querfurth, V. Rybin, G. Drewes, M. Raida, T. Bouwmeester, P. Bork, B. Seraphin, B. Kuster, G. Neubauer, and G. Superti-Furga. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415:141-147. [DOI] [PubMed] [Google Scholar]
- 9.Gully, D., D. Moinier, L. Loiseau, and E. Bouveret. 2003. New partners of acyl carrier protein detected in Escherichia coli by tandem affinity purification. FEBS Lett. 548:90-96. [DOI] [PubMed] [Google Scholar]
- 10.Günzl, A., A. Bindereif, E. Ullu, and C. Tschudi. 2000. Determinants for cap trimethylation of the U2 small nuclear RNA are not conserved between Trypanosoma brucei and higher eukaryotic organisms. Nucleic Acids Res. 28:3702-3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Günzl, A., T. Bruderer, G. Laufer, B. Schimanski, L. C. Tu, H. M. Chung, P. T. Lee, and M. G. Lee. 2003. RNA polymerase I transcribes procyclin genes and variant surface glycoprotein gene expression sites in Trypanosoma brucei. Eukaryot. Cell 2:542-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Günzl, A., E. Ullu, M. Dörner, S. P. Fragoso, K. F. Hoffmann, J. D. Milner, Y. Morita, E. K. Nguu, S. Vanacova, S. Wünsch, A. O. Dare, H. Kwon, and C. Tschudi. 1997. Transcription of the Trypanosoma brucei spliced leader RNA gene is dependent only on the presence of upstream regulatory elements. Mol. Biochem. Parasitol. 85:67-76. [DOI] [PubMed] [Google Scholar]
- 13.Knuesel, M., Y. Wan, Z. Xiao, E. Holinger, N. Lowe, W. Wang, and X. Liu. 2003. Identification of novel protein-protein interactions using a versatile mammalian tandem affinity purification expression system. Mol. Cell. Proteomics 2:1225-1233. [DOI] [PubMed] [Google Scholar]
- 14.Laufer, G., and A. Günzl. 2001. In-vitro competition analysis of procyclin gene and variant surface glycoprotein gene expression site transcription in Trypanosoma brucei. Mol. Biochem. Parasitol. 113:55-65. [DOI] [PubMed] [Google Scholar]
- 15.Laufer, G., G. Schaaf, S. Bollgönn, and A. Günzl. 1999. In vitro analysis of α-amanitin-resistant transcription from the rRNA, procyclic acidic repetitive protein, and variant surface glycoprotein gene promoters in Trypanosoma brucei. Mol. Cell. Biol. 19:5466-5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee, M. G. 1996. An RNA polymerase II promoter in the hsp70 locus of Trypanosoma brucei. Mol. Cell. Biol. 16:1220-1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee, M. G. S., and L. H. T. Van Der Ploeg. 1990. Homologous recombination and stable transfection in the parasitic protozoan Trypanosoma brucei. Science 250:1583-1587. [DOI] [PubMed] [Google Scholar]
- 18.Li, Q., X. Q. Dai, P. Y. Shen, H. F. Cantiello, E. Karpinski, and X. Z. Chen. 2004. A modified mammalian tandem affinity purification procedure to prepare functional polycystin-2 channel. FEBS Lett. 576:231-236. [DOI] [PubMed] [Google Scholar]
- 19.Liang, X. H., A. Haritan, S. Uliel, and S. Michaeli. 2003. trans and cis splicing in trypanosomatids: mechanism, factors, and regulation. Eukaryot. Cell 2:830-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miao, C. H., W. T. Ho, D. L. Greenberg, and E. W. Davie. 1996. Transcriptional regulation of the gene coding for human protein C. J. Biol. Chem. 271:9587-9594. [DOI] [PubMed] [Google Scholar]
- 21.Palfi, Z., B. Schimanski, A. Günzl, S. Lücke, and A. Bindereif. 2005. U1 small nuclear RNP from Trypanosoma brucei: a minimal U1 snRNA with unusual protein components. Nucleic Acids Res. 33:2493-2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Panigrahi, A. K., A. Schnaufer, N. L. Ernst, B. Wang, N. Carmean, R. Salavati, and K. Stuart. 2003. Identification of novel components of Trypanosoma brucei editosomes. RNA 9:484-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Puig, O., F. Caspary, G. Rigaut, B. Rutz, E. Bouveret, E. Bragado-Nilsson, M. Wilm, and B. Seraphin. 2001. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24:218-229. [DOI] [PubMed] [Google Scholar]
- 24.Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Seraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030-1032. [DOI] [PubMed] [Google Scholar]
- 25.Rohila, J. S., M. Chen, R. Cerny, and M. E. Fromm. 2004. Improved tandem affinity purification tag and methods for isolation of protein heterocomplexes from plants. Plant J. 38:172-181. [DOI] [PubMed] [Google Scholar]
- 26.Schimanski, B., B. Klumpp, G. Laufer, R. J. Marhöfer, P. M. Selzer, and A. Günzl. 2003. The second largest subunit of Trypanosoma brucei's multifunctional RNA polymerase I has a unique N-terminal extension domain. Mol. Biochem. Parasitol. 126:193-200. [DOI] [PubMed] [Google Scholar]
- 27.Schimanski, B., G. Laufer, L. Gontcharova, and A. Günzl. 2004. The Trypanosoma brucei spliced leader RNA and rRNA gene promoters have interchangeable TbSNAP50-binding elements. Nucleic Acids Res. 32:700-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schimanski, B., T. N. Nguyen, and A. Günzl. 2005. Characterization of a multi-subunit transcription factor complex essential for SL RNA gene transcription in Trypanosoma brucei. Mol. Cell. Biol. 25:1703-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stearns, D. J., S. Kurosawa, P. J. Sims, N. L. Esmon, and C. T. Esmon. 1988. The interaction of a Ca2+-dependent monoclonal antibody with the protein C activation peptide region. Evidence for obligatory Ca2+ binding to both antigen and antibody. J. Biol. Chem. 263:826-832. [PubMed] [Google Scholar]
- 30.ten Asbroek, A. L., M. Ouellette, and P. Borst. 1990. Targeted insertion of the neomycin phosphotransferase gene into the tubulin gene cluster of Trypanosoma brucei. Nature 348:174-175. [DOI] [PubMed] [Google Scholar]
- 31.Walgraffe, D., S. Devaux, L. Lecordier, J. F. Dierick, M. Dieu, A. J. Van Den, E. Pays, and L. Vanhamme. 2005. Characterization of subunits of the RNA polymerase I complex in Trypanosoma brucei. Mol. Biochem. Parasitol. 139:249-260. [DOI] [PubMed] [Google Scholar]
- 32.Ziegler, J., T. Vogt, O. Miersch, and D. Strack. 1997. Concentration of dilute protein solutions prior to sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Anal. Biochem. 250:257-260. [DOI] [PubMed] [Google Scholar]