Abstract
The CCAAT-binding factor (CBF) is an evolutionarily conserved multimeric transcriptional activator in eukaryotes. In Saccharomyces cerevisiae, the CCAAT-binding factor is composed of four subunits, termed Hap2p, Hap3p, Hap4p, and Hap5p. The Hap2p/Hap3p/Hap5p heterotrimer is the DNA-binding component of the complex that binds to the consensus 5′-CCAAT-3′ sequence in the promoter of target genes. The Hap4p subunit contains the transcriptional activation domain necessary for stimulating transcription after interacting with Hap2p/Hap3p/Hap5p. In this report, we demonstrate that Hap2p, Hap3p, and Hap5p assemble via a one-step pathway requiring all three subunits simultaneously, as opposed to the mammalian CCAAT-binding factor which has been shown to assemble via a two-step pathway with CBF-A (Hap3p homolog) and CBF-C (Hap5p homolog) forming a stable dimer before CBF-B (Hap2p homolog) can interact. We have also found that the interaction of Hap4p with Hap2p/Hap3p/Hap5p requires DNA binding as a prerequisite. To further understand the protein-protein and protein-DNA interactions of this transcription factor, we identified the minimal domain of Hap4p necessary for interaction with the Hap2p/Hap3p/Hap5p-DNA complex, and we demonstrate that this domain is sufficient to complement the respiratory deficiency of a hap4Δ mutant and activate transcription when fused with the VP16 activation domain. These studies provide a further understanding of the assembly of the yeast CCAAT-binding factor at target promoters and raise a number of questions concerning the protein-protein and protein-DNA interactions of this multisubunit transcription factor.
Saccharomyces cerevisiae is a respirofermentative yeast that represses respiratory metabolism when growing in medium containing glucose as the sole carbon source, even in an oxygenated environment (14, 53). Following glucose depletion, cells undergo a major reprogramming of gene expression, known as the diauxic shift, to activate the genes that encode proteins needed for respiration and gluconeogenesis (12, 16, 30, 45). Thus, the organism can utilize the ethanol that was generated during the fermentative metabolism. The CCAAT-binding factor (CBF; the Hap2p/Hap3p/Hap4p/Hap5p complex) is one of the transcriptional activators responsible for the activation of many of the genes involved in respiratory metabolism (12, 16, 45, 57), as well as other genes needed for other metabolic functions, such as ammonia assimilation (10, 11, 41).
The CCAAT-binding factor is a multisubunit transcriptional activator that binds to the 5′-CCAAT-3′ consensus elements within promoters (6, 36). This activator is unique among DNA-binding proteins in that it requires three heterologous subunits, termed Hap2p, Hap3p, and Hap5p, for DNA-binding activity (35, 38). The Hap2p/Hap3p/Hap5p trimer has been shown to be sufficient for CCAAT-specific binding at target promoters (38); however, this complex lacks the ability to activate transcription. A fourth subunit of the complex, termed Hap4p, is necessary for transcriptional activation (15). HAP4 is subject to glucose repression (12, 15), with its expression repressed in the presence of glucose and activated in its absence, while the expression of HAP2, HAP3, and HAP5 is constitutive (12). Thus, the synthesis and interaction of Hap4p with Hap2p/Hap3p/Hap5p modulate the activity of target genes. Mutations that abolish the function of any of the four Hap subunits result in the inability of yeast to grow on nonfermentable carbon sources (15, 24, 38, 40), emphasizing the importance of this protein complex as a global regulator of respiration.
Each of the DNA-binding subunits of the CCAAT-binding factor contains an essential core region that is highly conserved evolutionarily from yeast to humans (35), with the Hap3p and Hap5p core elements displaying amino acid sequence similarities to the histone fold motifs of histones H2B and H2A, respectively (2, 35). In yeast, these core regions have been shown to be essential for the assembly and DNA-binding activity of the heterotrimeric complex (37). Moreover, in vivo expressions of the conserved regions of Hap2p and Hap3p are sufficient for functional complementation of their respective null mutants (39, 55, 56). In addition to the highly conserved core region, Hap5p contains a 32-amino-acid domain that is lacking in the homologous proteins of higher eukaryotes but present in other yeasts and fungi (37). This small domain is required for the association of Hap4p with Hap2p/Hap3p/Hap5p; hence, this region was termed the Hap4p recruitment domain (37). Thus, the Hap4p recruitment domain, together with the histone fold region of Hap5p, is sufficient for functional complementation of a hap5Δ mutant (37).
For several years, Hap4p homologs were not found in other yeast or fungi; however, recent studies have revealed that homologs exist in Kluyveromyces lactis (4) and Hansenula polymorpha (51). These Hap4p homologs were shown to functionally complement an S. cerevisiae hap4Δ mutant in spite of the fact that the homology between these proteins was limited to a 16-amino-acid domain (4, 51). Nevertheless, once this conserved region was identified, it became apparent that Hap4p-like proteins exist in many different yeasts and fungi (51). Moreover, the Hap4 recruitment domain of Hap5p is also conserved in these organisms (unpublished observations), suggesting that these domains may be important for Hap4p-Hap5p interactions. However, biochemical studies that directly assess the domain(s) of Hap4p needed for association with Hap2p/Hap3p/Hap5p have not been performed.
Hap4p is composed of 554 amino acid residues with a C-terminal activation domain that is between amino acid residues 330 and 554 (15). Additional studies of the Hap4p activation domain have revealed two regions capable of stimulating transcription when fused to the LexA DNA-binding domain. One of the activation domains is GCN5 independent and maps between amino acid residues 359 and 476, while the other is GCN5 dependent and lies between amino acid residues 124 and 329 (50).
While the regions of Hap2p, Hap3p, and Hap5p that are important for protein-protein and protein-DNA interactions have been mapped in detail (37, 39, 55, 56), the biochemical pathway by which the Hap2p/Hap3p/Hap5p and Hap2p/Hap3p/Hap4p/Hap5p complexes assemble has not been examined. To date, it has been assumed that the Hap complex assembles in a manner analogous to its mammalian counterparts, termed CBF-A (Hap3p), CBF-B (Hap2p), and CBF-C (Hap5p) in rats or NFY-A (Hap2p), NFY-B (Hap3p), and NFY-C (Hap5p) in mice and humans (35, 48). These proteins follow an ordered two-step assembly pathway whereby CBF-A and CBF-C initially form a stable heterodimer, with CBF-B subsequently binding to form the trimeric DNA-binding complex (48). The stability of the CBF-A/CBF-C heterodimer was such that harsh conditions were required to separate the subunits (34). This raised some interesting contradictions, since we had not observed a stable Hap3p/Hap5p heterodimer in our studies. In fact, we have previously shown that recombinant Hap2p, Hap3p, or Hap5p could efficiently interchange with the endogenous Hap subunits from a yeast cell extract (38), suggesting the Hap3p/Hap5p heterodimer may be less stable than CBF-A/CBF-C. In addition, we have not detected a stable Hap3p/Hap5p heterodimer with purified recombinant Hap3p and Hap5p (unpublished observations), as observed with the mammalian complex (3, 29, 33, 42, 47).
In this study, we examined the assembly of the Hap2p/Hap3p/Hap5p and Hap2p/Hap3p/Hap4p/Hap5p complexes in vitro and found that Hap2p/Hap3p/Hap5p does not follow a two-step assembly pathway analogous to its mammalian counterpart but requires the presence of all three subunits concurrently for stable assembly. In addition, we have determined that the interaction of Hap4p with Hap2p/Hap3p/Hap5p requires the heterotrimer to be bound to its cognate CCAAT site on DNA. To further understand the interaction between Hap4p and Hap2p/Hap3p/Hap5p, we delimited the minimal region of Hap4p necessary for the stable association with Hap2p/Hap3p/Hap5p, and we subsequently demonstrated that this domain, when fused to a heterologous activation domain, can functionally complement a hap4Δ mutant and activate transcription.
MATERIALS AND METHODS
Yeast strains and growth media.
The S. cerevisiae strains used in this study were all isogenic derivatives of FY2 (54) and included FY250 (MATα ura3-52 leu2Δ1 his3Δ200 trp1Δ63), DMY144 (MATα ura3-52 leu2Δ1 his3Δ200 trp1Δ63 hap4Δ::hisG), BY4733 (MATa ura3Δ0 leu2Δ0 his3Δ200 trp1Δ63 met15Δ0), and DMY187 (MATa ura3Δ0 leu2Δ0 his3Δ200 trp1Δ63 met15Δ0 HAP4-3xHA). Yeast strains were grown in yeast extract-peptone (YP), synthetic omission (SC), and 5-fluoroorotic acid (5-FOA) medium prepared as previously described (22) and supplemented with 2% glucose, raffinose, or lactate as needed. The media were solidified with 1.5% agar as appropriate.
Oligonucleotides.
Oligonucleotides used in this study are listed in Table 1.
TABLE 1.
Oligonucleotides used in these studies
| Name | Sequencea |
|---|---|
| oDM0016 | 5′-CCGgaattcAAGCTTCCATTAACTCTCTCATTGTGG-3′ |
| oDM0024 | 5′-CCGgaattcTAGAACCTCCCACCTTCACCAC-3′ |
| oDM0026 | 5′-CCGgaattcTATTGTTGCCTGTATTTAGCC-3′ |
| oDM0054 | 5′-GCCcatatgGAGCAACCATTTTATGTTAATGCC-3′ |
| oDM0056 | 5′-GCCcatatgCTAAGAGAGCAGGACAGATGGCTACCC-3′ |
| oDM0076 | 5′-GCCggatccTGACCGCAAAGACTTTTCTAC-3′ |
| oDM0079 | 5′-GGCGTTTCACACTGGATATTGGTTCCGCCA-3′ |
| oDM0081 | 5′-GCGCTTGGCGCCAGCTCGATTCTGGTACTG-3′ |
| oDM0098 | 5′-GCCcatatgTCAGCAGACGAAACGGATGCG-3′ |
| oDM0099 | 5′-GGCgagctcTTATGTTTTTTTGTCTGCTGCAGCTGC-3′ |
| oDM0100 | 5′-GCCcatatgAATACCAACGAGTCCGAACATGTTAGC-3′ |
| oDM0101 | 5′-GGCgaattcTCAAGGCACCTCTTCGTCGTCCTGCTC-3′ |
| oDM0102 | 5′-GCCcatatgACTGATAGGAATTTCTCACCAC-3′ |
| oDM0115 | 5′-GGCCgcggccgcTTAGGAGGAGGCAGAAGAACG-3′ |
| oDM0128 | 5′-GGGgcggccgcTCAATGGTTTTTTGTTAGCAGACTGTTG-3′ |
| oDM0129 | 5′-GGGggatccTGGAATTCGAAAACGATGGC-3′ |
| oDM0135 | 5′-GGGgcggccgcTCATCTATAACTTGAAAAAGATGGAGATG-3′ |
| oDM0138 | 5′-GGCggatccGAATGGAGCATAATAATATTCCATTGGCGCC-3′ |
| oDM0140 | 5′-GGCCtctagaGCCggatccCTTTTTTTGTTTTTTTTTTGTACTAGAAAATGTAGG-3′ |
| oDM0141 | 5′-GGCCctgcagAATAGGCATGTTGCAATAAAACG-3′ |
| oDM0142 | 5′-GGCCaagcttAGATCTCTCTTGACGTGTTTCACCATACGC-3′ |
| oDM0143 | 5′-GGCCtctagaGCCCCCCCGACCGATGTCAGCCTGGGG-3′ |
| oDM0144 | 5′-GGCCctgcagCTACCCACCGTACTCGTCAATTCC-3′ |
| oDM0169 | 5′-GGCCggatccGAATCATGACCGAGCATAATAATATTCCATTGGCGCC-3′ |
| oDM0170 | 5′-GGCCtctagaCCTATAACTTGAAAAAGATGGAGATG-3′ |
| oDM0187 | 5′-GGCCctgcagTTATCTATAACTTGAAAAAGATGGAGATG-3′ |
| oDM0188 | 5′-GGCCgaattcAGATCTGAATATGAAGTAGGTAGAAGCAGAGG-3′ |
| oDM0197 | 5′-CGACGACCTTGACGAAGATGTCGATTTTTTAAAGGTACAAGTATTTAGGGAACAAAAGCTGG-3′ |
| oDM0198 | 5′-CGTGATTTTTAGTTGTTTTCGTTTTATTGCAACATGCCTATTGTAGGGCGAATTGGG-3′ |
| oDM0230 | 5′-GGGCCCggatccATGTCAGCAGACGAAACGGATGCG-3′ |
| oDM0231 | 5′-GGGCCCgaattcTTATGTTTTTTTGTCTGCTGCAGCTGCG-3′ |
| oDM0232 | 5′-GGGCCCggatccATGAATACCAACGAGTCCGAACATGTTAGC-3′ |
| oDM0233 | 5′-GGGCCCgaattcTCAAGGCACCTCTTCGTCGTCCTGCTC-3′ |
| oDM0234 | 5′-GGGCCCggatccATGACTGATAGGAATTTCTCACCACAACAAGG-3′ |
| oDM0235 | 5′-GGGCCCgaattcTCATTGTGGAAGAGGTCTTCTAGGCAC-3′ |
| oDM0245 | 5′-GGGCAAGATGATAAGGTTGAGCCAGATGAAGAAGAC-3′ |
Relevant restriction enzyme sites are shown in lowercase letters.
Strain construction.
All yeast transformations were performed using the lithium acetate transformation protocol as previously described (17). Strain DMY144 contained a hap4Δ::hisG null allele and it was constructed in FY250 using the plasmid pKS::HAP4ΔhisG as previously described (38). The hap4Δ transformants were initially identified by their inability to grow on YPLactate medium, and Ura− derivatives were subsequently selected on 5-FOA medium. The hap4Δ::hisG disruption was then confirmed by the PCR of purified genomic DNA (26) with the oligonucleotide primers oDM0079 and oDM0081. The 3xHA epitope-tagged allele of HAP4 was generated by the epitope-tagging procedure developed by Schneider et al. (44). The 3xHA-URA3-3xHA cassette was amplified from the template pMPY-3xHA (44) with primers oDM0197 and oDM0198 that contain 40 bp of homology to HAP4 to direct recombination between the last amino acid of Hap4p and the stop codon. The PCR-amplified cassette was transformed into BY4733 (5), and Ura+ transformants were selected and purified. Since the URA3 gene is flanked by duplicated 3xHA sequences, recombination between these duplications yielded Ura− derivatives that retained one copy of the 3xHA epitope tag. Thus, 5-FOA-resistant recombinants were selected, and the HAP4-3xHA allele was confirmed by PCR using genomic DNA and the primers oDM0245 and oDM0142. The resulting strain was designated DMY187.
Plasmid constructions.
The full-length coding regions of HAP2, HAP3, and HAP5 were cloned into pCITE2a (Promega Corp., Madison, WI) for in vitro protein-protein interaction studies. The HAP2 coding sequence was amplified by PCR from the template plasmid pJP103 (40) with primers oDM0098 and oDM0099 that incorporated unique NdeI and SstI restriction sites at the 5′ and 3′ ends of the PCR product, respectively. The DNA was purified, digested with NdeI/SstI, and cloned into the same sites of pCITE2a to generate pDM418. The HAP3 coding region was amplified by PCR from the template pSH94 (24) with primers oDM0100 and oDM0101 that added unique NdeI and EcoRI restriction sites to the 5′ and 3′ ends, respectively; the DNA was purified, digested with NdeI/EcoRI, and cloned into the same sites of pCITE2a to generate pDM410. The coding region of HAP5 was amplified by PCR from the template pDM231 (38) using primers oDM0102 and oDM0016 that added unique NdeI and EcoRI sites, respectively, and the DNA fragment was cloned into pCITE2a to generate pDM413. Plasmid pDM379 has been previously described (37) and contains the coding sequence of Hap4p from amino acid residues 1 through 330 [designated (1-330)] in pCITE2c.
The coding regions of HAP2, HAP3, and HAP5 were expressed in Escherichia coli BL26 (Novagen) as glutathione S-transferase (GST) fusion proteins in the expression plasmid pGEX-2T (Amersham Biosciences). The HAP2, HAP3, and HAP5 coding sequence was amplified by PCR from the template plasmids pJP103 (40), pSH94 (24), and pDM231 (38), respectively, using primer pairs oDM0230/oDM0231, oDM0232/oDM0233, and oDM0234/oDM0235. For each gene, the primers incorporated unique BamHI and EcoRI sites at the 5′ and 3′ ends, respectively. The PCR products and pGEX-2T were digested with BamHI/EcoRI and ligated. The GST-Hap2p-, GST-Hap3p-, and GST-Hap5p-encoding plasmids were designated pDM492, pDM493, and pDM494.
To delimit the region of Hap4p required for interaction with Hap2p/Hap3p/Hap5p, truncated alleles of HAP4 were generated in the plasmid pCITE2a for in vitro transcription/translation. The Hap4p (1-330)-encoding plasmid pDM379 has been previously described (37). It was used to generate Hap4p (1-250) by digestion with HincII and ligation with an XbaI nonsense linker (New England Biolabs, Inc.) to introduce a stop codon after amino acid residue 250, and the plasmid was designated pDM391. The truncated alleles HAP4 (1-210), HAP4 (1-180), and HAP4 (1-165) were generated by PCR amplification with pSLF413 (15) as the template and the primer pairs oDM0076/oDM0128, oDM0076/oDM0135, and oDM0076/oDM0115, which incorporated unique 5′ and 3′ BamHI and NotI restriction sites, respectively, as well as a termination codon after the indicated amino acid residue. The PCR products were digested with BamHI/NotI and cloned into pCITE2c, and the plasmids were designated pDM401, pDM403, and pDM386. To generate HAP4 (46-210), HAP4 was amplified by PCR from the template pSLF413 (15) with primers oDM0128 and oDM0129 that incorporated unique BamHI and NotI restriction sites on the 5′ and 3′ ends, respectively, along with introducing a stop codon after amino acid residue 210. The PCR product and pCITE2c were digested with BamHI/NotI and ligated to generate plasmid pDM400. Plasmid pDM408 contains HAP4 (23-180) and was constructed by PCR amplification from pSLF413 (15) with primers oDM0135 and oDM0138 that incorporated unique BamHI and NotI restriction sites on the 5′ and 3′ ends, respectively, and a termination codon after amino acid residue 180. The PCR product and pCITE2c were digested with BamHI/NotI and ligated.
For the mobility shift studies, the core domains of Hap2p and Hap3p were produced as six-His-tagged fusion proteins in E. coli BL21(DE3). The plasmid encoding the Hap2p core domain was generated by PCR amplification of HAP2 (amino acid residues 158 to 214) from the template plasmid pJP103 (40) with primers oDM00054 and oDM0024 that added unique NdeI and EcoRI sites to the 5′ and 3′ ends of the PCR product, respectively. The DNA was cloned into the NdeI/EcoRI sites of pET-28a (Novagen) to generate pDM341. The plasmid encoding the Hap3p core was generated by PCR amplification of HAP3 (amino acid residues 35 to 127) from the template plasmid pSH94 (24) with primers oDM0056 and oDM0026 that incorporated unique NdeI and EcoRI sites into the 5′ and 3′ ends of the PCR product. The DNA was cloned into pET-28a at the NdeI/EcoRI sites. The GST-Hap5p expression plasmid pDM215 has been previously described and contains amino acid residues 80 to 242 of Hap5p fused to glutathione S-transferase (38). Removal of the affinity tags by thrombin cleavage was carried out as previously described (19).
To determine whether HAP4 (23-180) could functionally complement a hap4Δ mutant, an expression plasmid was generated that contained the native promoter, and the 5′ and 3′ untranslated regions (UTRs) of HAP4, along with the Hap4p (23-180) coding region fused with the VP16 activation domain. The construction of the plasmid required multiple steps. Initially, the HAP4 3′ UTR was amplified by PCR from the template plasmid pSLF413 (15) with primers oDM0141 and oDM0142 that incorporated unique PstI and HindIII sites into the 5′ and 3′ ends, respectively. The DNA was cloned into the PstI/HindIII sites of YCplac33 (18). Next, the HAP4 promoter and 5′ UTR (−1225 to −1 relative to the AUG initiation codon) were amplified by PCR from the same template with primers oDM0188 and oDM0140. Primer oDM0188 added a unique EcoRI site for cloning, while oDM0140 added unique XbaI and BamHI restriction sites. The DNA was digested with EcoRI/XbaI and cloned into the same sites of YCplac33 containing the HAP4 3′ UTR sequence. In the next step, the VP16 activation domain was amplified by PCR from the template plasmid pJR3 (52) using primers oDM0143 and oDM0144 that incorporated unique XbaI and PstI restriction sites into the PCR product at the 5′ and 3′ ends, respectively. The DNA was digested with XbaI/PstI and cloned into YCplac33 containing the HAP4 sequences; this plasmid was designated pDM471. To add the Hap4p (23-180) coding sequence, HAP4 (23-180) was amplified by PCR from the template pSLF413 (15) with primers oDM0169 and oDM0170 that incorporated unique 5′ and 3′ BamHI and XbaI restriction sites, respectively. In addition, oDM0169 added an ATG start codon to HAP4 (23-180). The PCR product and pDM471 were digested with BamHI/XbaI and ligated to create plasmid pDM472. To construct a control plasmid lacking the VP16 activation domain, HAP4 (23-180) was amplified by PCR from the same template using primers oDM0169 and oDM0187 to add a stop codon after amino acid residue 180 and to incorporate a unique PstI site at the 3′ end of the PCR product. The DNA and pDM471 were digested with BamHI/PstI and ligated to create the plasmid pDM473. The BamHI/PstI digestion of pDM471 removed the VP16 activation domain.
Purification of recombinant proteins.
The glutathione S-transferase fusion proteins were expressed in E. coli BL26 and purified from bacterial lysates with glutathione-Sepharose beads (Amersham Biosciences) as previously described (38). For preparation of six-His-Hap2p and six-His--Hap3p, E. coli BL21(DE3) containing the appropriate expression plasmid was inoculated to TB medium (43) containing 40 μg of kanamycin/ml and then grown at 37°C to an optical density at 600 nm (OD600) of ∼0.7. Expression was induced by the addition of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), and the culture was incubated an additional 3 h at 37°C. The cells were harvested by centrifugation at 5,000 × g for 10 min, resuspended in denaturing lysis buffer (20 mM Tris [pH 7.9], 500 mM NaCl, 6 M guanidine hydrochloride, 5 mM imidazole, 1 mM phenylmethylsulfonyl fluoride, and 5 mM benzamidine) and incubated for 1 h at room temperature with agitation. The lysate was sonicated to reduce the viscosity and subsequently centrifuged at 12,000 × g for 15 min. at 4°C. The supernatant was collected and passed over a Ni-nitrilotriacetic acid agarose (QIAGEN) column that had been preequilibrated with lysis buffer. The column was washed with lysis buffer until the absorbance at 280 nm (A280) was <0.01 and then with wash buffer (lysis buffer containing 20 mM imidazole) until A280 was <0.01. The proteins were eluted with lysis buffer containing 300 mM imidazole, and 0.5-ml fractions were collected. Fractions containing the recombinant proteins were identified by A280 values. Small aliquots of each fraction were precipitated with trichloroacetic acid (25), the purity of the fractions was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and the proteins were visualized by Coomassie blue staining. Fractions containing the recombinant proteins were pooled and dialyzed overnight in a renaturation buffer (25 mM Tris [pH 7.9], 100 mM NaCl, 1 mM EDTA, 0.5% Triton X-100, 5% glycerol, and 5 mM β-mercaptoethanol). The proteins were then quantified by the bicinchoninic acid protein assay (Pierce) with bovine serum albumin as the standard, and the activity of the recombinant proteins was verified by mobility shift assays.
In vitro transcription/translation.
To express each of the Hap subunits, in vitro transcription and translation were performed using the Novagen single-tube protein system for coupled transcription/translation according to the manufacturer's protocol. For GST pulldown experiments, the proteins were radiolabeled with [35S]methionine (Dupont-NEN) during translation. For mobility shift studies, the proteins were not radiolabeled, but a [35S]methionine-labeled parallel reaction was performed to confirm the protein size and expression by SDS-12% PAGE, followed by autoradiography.
In vitro protein-protein interaction assays.
For the GST pulldown assays, [35S]methionine-labeled Hap subunits were incubated with GST, GST-Hap2p, GST-Hap3p, GST-Hap5p, or a combination of recombinant proteins in a binding buffer containing 50 mM HEPES (pH 7.9), 100 mM KCl, 1 mM EDTA, 0.1% Tween-20, 5 mM β-mercaptoethanol, and 1 mM phenylmethylsulfonyl fluoride in a final volume of 200 μl. After incubation at room temperature for 30 min, 50 μl of 50% (vol/vol) glutathione-Sepharose resin (Amersham Biosciences), preequilibrated in binding buffer, was added to each reaction mixture, and the incubation was continued for 30 min at room temperature. The mixture was centrifuged, and the resin was washed four times with 200 μl of binding buffer. The resin was then boiled in SDS-PAGE loading buffer (25) and centrifuged, and the supernatant was run on SDS-12% PAGE gels that were subsequently fixed, dried, and exposed to a PhosphorImager screen. The efficiency of the pulldown experiments was quantified on a Molecular Dynamics PhosphorImager. For reactions requiring DNA, the CCAAT box probe was a 37-bp double-stranded oligonucleotide with the sequence derived from CYC1 UAS2UP1 as described previously (23), and the mutated probe was identical to UAS2UP1, except the CCAAT sequence was changed to GGAAG.
DNA-binding assays and gel electrophoresis.
The UAS2UP probe derived from the sequence of the CYC1 gene and containing the CCAAT box was described previously (23). The DNA probe was end labeled with [α-32P]dATP with the Klenow fragment. All DNA-binding reactions were performed in 1× DNA-binding buffer (20 mM HEPES-NaOH [pH 7.9], 100 mM KCl, 1 mM EDTA, 1 mM dithiothreitol, and 20% glycerol) containing 20 ng of each recombinant Hap subunit, 5 μl of the appropriately programmed in vitro transcription/translation extract, and 3 μg of poly(dI-dC) nonspecific competitor DNA. When an in vitro transcription/translation extract was not included in the DNA-binding reaction, the poly(dI-dC) concentration was reduced to 200 ng. Reactions were incubated at room temperature for 30 to 45 min, and the protein-DNA complexes were resolved by gel electrophoresis as previously described (38). The gels were subsequently fixed, and the protein-DNA complexes were visualized by autoradiography and with a PhosphorImager (Molecular Dynamics).
β-Galactosidase assays.
β-Galactosidase assays were performed on cells grown in synthetic omission medium (SC-Ura-Leu) containing 2% glucose, raffinose, or lactate as the sole carbon source. The assays were done by the chloroform-permeabilizing method as previously described (20) with the CYC1-lacZ reporter plasmid pSLFΔ265LEU (15).
Northern blot analysis.
S. cerevisiae strains were grown in rich medium (YP) containing 2% glucose, raffinose, or lactate as the carbon source to an OD600 of ∼1.0. The cells were harvested by centrifugation, and total RNA was prepared by the glass bead-acid phenol method as previously described (1). Approximately 30 μg of each total RNA sample was loaded, separated by formaldehyde-1% agarose gel electrophoresis, and transferred to GeneScreen Plus membranes (Dupont-NEN Research Products) according to the manufacturer's protocol. The membranes were hybridized and washed under standard high-stringency conditions (43). The HAP4, CYC1, and SNR20 probes were obtained by PCR amplification of S. cerevisiae FY250 genomic DNA using oligonucleotide primer pairs oDM0169/oDM187, oDΜ0123/οDΜ124, and oDM206/oDM207, respectively. The PCR products were purified by agarose gel electrophoresis and the GeneClean kit (Qbiogene, Inc.). The probes were radiolabeled with [α-32P]dCTP (Amersham Biosciences) by use of a random primer labeling kit (U.S. Biochemicals) according to the manufacturer's protocol. The transcript levels were quantified on a Molecular Dynamics PhosphorImager.
Western blotting.
S. cerevisiae strains were grown in rich medium (YP) containing 2% glucose, raffinose, or lactate to an OD600 of ∼1.0. The cells were harvested by centrifugation and resuspended in protein lysis buffer (6 mM Tris [pH 6.8], 6 M urea, 2% SDS, 5% β-mercaptoethanol, 10% glycerol, and bromophenol blue) and boiled for 5 min as previously described (27). The cell debris was removed by centrifugation at 12,000 × g for 10 min, and the supernatant was used for SDS-PAGE on 4 to 15% gradient polyacrylamide gels (Bio-Rad) in Tris-glycine buffer. The proteins were transferred to Immobilon-P-polyvinylidene difluoride membranes (Millipore Corp.) (25), and the membranes were blocked in TBST (20 mM Tris [pH 7.6], 137 mM NaCl, and 0.5% Tween 20) containing 5% nonfat dry milk for 2 h at room temperature. The membranes were subsequently probed with anti-HA antibody 12CA5 (BAbCO) or the rabbit anti-TATA-binding protein (TBP) antiserum (a gift from L. Guarente, Massachusetts Institute of Technology) at a 1:5,000 or 1:500 dilution, respectively, for 1 h at room temperature in TBST plus 5% nonfat dry milk. The blots were washed extensively with TBST and probed with either goat anti-mouse or goat anti-rabbit horseradish peroxidase-conjugated secondary antibodies for 1 h at room temperature, and the proteins were detected by the ECL detection kit (Amersham Biosciences). For quantifying the levels of Hap4p and TBP, X-ray films were scanned and analyzed by densitometry after exposure to the ECL reagent.
RESULTS
The assembly pathway of the Hap2p/Hap3p/Hap5p heterotrimer.
The mammalian CCAAT-binding factor has been shown to follow a two-step ordered pathway of assembly, with CBF-A (Hap3p homolog) and CBF-C (Hap5p homolog) initially forming a stable dimer, and CBF-B (Hap2p homolog) subsequently binding to form the trimer (48). Two lines of evidence suggested that this two-step pathway may not exist for the S. cerevisiae CCAAT-binding factor. First, we had not observed a stable Hap3p/Hap5p dimer during our in vitro studies with purified recombinant Hap proteins (data not shown); second, it was previously shown that size variants of Hap3p and Hap5p could be interchanged with their respective counterparts of the CCAAT-binding factor from yeast cell extracts (38). This differed from the stability of the CBF-A/CBF-C dimer, which required denaturing conditions to separate the two subunits (34). Given these contradictions, we investigated the assembly of the Hap2p/Hap3p/Hap5p heterotrimer.
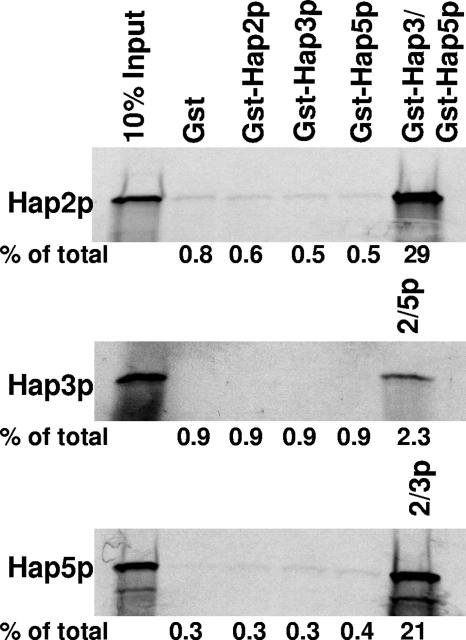
To examine the assembly of Hap2p/Hap3p/Hap5p, full-length [35S]methionine-labeled Hap2p, Hap3p, and Hap5p were synthesized by in vitro transcription/translation and the radiolabeled subunits were subsequently used for in vitro protein-protein interaction studies with purified recombinant GST, GST-Hap2p, GST-Hap3p, and GST-Hap5p. The reactions were precipitated with glutathione-Sepharose resin and the proteins associated with the GST-Hap subunits analyzed by SDS-PAGE (Fig. 1). In this assay, no binary interactions between the Hap subunits were observed, suggesting that Hap3p and Hap5p do not form a stable dimer. In addition, none of the Hap subunits formed homodimers, as indicated by the lack of self-association, consistent with previous studies with the mammalian counterparts (48). The radiolabeled Hap proteins were precipitated only when a combination of the other two GST-Hap subunits were added to the reaction mixture (Fig. 1). From several independent experiments, the efficiency of the precipitation reaction with radiolabeled Hap3p was always lower than those containing radiolabeled Hap2p or Hap5p; however, we do not have an explanation for this observation.
FIG. 1.
The Hap2p/Hap3p/Hap5p heterotrimer assembles via a one-step mechanism. Protein-protein interaction studies were performed as described in Materials and Methods using [35S]methionine-labeled Hap2p, Hap3p, and Hap5p synthesized by in vitro transcription/translation in a rabbit reticulocyte lysate. The GST pulldown reaction mixtures contained the radiolabeled Hap subunit as indicated on the left of each panel along with GST, GST-Hap2p, GST-Hap3, GST-Hap5p, or a combination of GST-Hap subunits as indicated. The first lane of each panel contains 10% of the in vitro-synthesized protein added to each GST pulldown reaction mixture (10% input). The percentage of the total radiolabeled protein precipitated in each reaction is shown below the individual panels.
To evaluate whether the presence of DNA may stabilize a Hap3p/Hap5p intermediate, the same protein-protein interaction studies were repeated in the presence of a 37-bp double-stranded CCAAT site oligonucleotide derived from the sequence of the CYC1 UAS2UP1 (23), a known Hap2p/Hap3p/Hap5p-binding site; again, we observed no stable binary interactions (data not shown). On the basis of these data, we concluded that the yeast CCAAT-binding factor does not follow a two-step assembly pathway like its mammalian homologs but requires the presence of the three Hap subunits for stable complex formation.
Sequence-specific DNA binding by Hap2p/Hap3p/Hap5p is a prerequisite for Hap4p interaction.
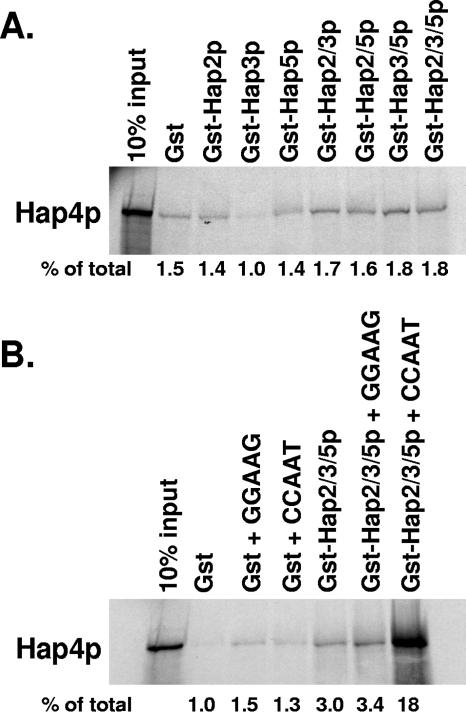
To further dissect the assembly of the yeast CCAAT-binding factor, we examined the requirement for Hap4p interaction. This was particularly important, since the CCAAT-binding factor in higher eukaryotes lacks a Hap4p homolog and the requirements for it to bind with Hap2p/Hap3p/Hap5p have not been investigated. As mentioned previously, the Hap4p recruitment domain of Hap5p is necessary for the Hap4p-Hap2p/Hap3p/Hap5p interaction (37); however, we wanted to determine whether the other subunits of the complex were also necessary. For these studies, we used a truncated allele of HAP4, encoding amino acid residues 1 through 330, since it has been previously shown to interact with Hap2p/Hap3p/Hap5p in mobility shift assays (37). Thus, [35S]methionine-radiolabeled Hap4p (1-330) was synthesized by in vitro transcription/translation and incubated with GST alone or the GST-Hap subunits individually or in various combinations (Fig. 2A). None of the individual or pair wise combinations of GST-Hap subunits formed a stable interaction with Hap4p (Fig. 2A). Unexpectedly, the GST-Hap2p/Hap3p/Hap5p trimer also did not stably associate with Hap4p (Fig. 2A), yet previous studies have shown that Hap4p (1-330) interacted with Hap2p/Hap3p/Hap5p in mobility shift assays (37). This raised a question as to whether DNA, present in the mobility shift studies, was important for the Hap4p-Hap2p/Hap3p/Hap5p complex formation. To address this possibility, protein-protein interaction studies were performed in the presence of a 37-bp double-stranded DNA oligonucleotide containing the sequence of the CCAAT site from CYC1 UAS2UP or an identical control DNA in which the CCAAT site was mutated to GGAAG to abolish binding by Hap2p/Hap3p/Hap5p (Fig. 2B). These data clearly demonstrated that Hap4p (1-330) could interact with Hap2p/Hap3p/Hap5p in the presence of the double-stranded CCAAT-containing oligonucleotide but not when the mutated oligonucleotide was present, suggesting that Hap2p/Hap3p/Hap5p must first bind DNA in a sequence-specific manner as a prerequisite for Hap4p interaction.
FIG. 2.
DNA binding by Hap2p/Hap3p/Hap5p is a prerequisite for Hap4p association. (A) Protein-protein interaction studies were performed as described in Materials and Methods using [35S]methionine-labeled Hap4p (1-330) synthesized by in vitro transcription/translation. Each GST pulldown reaction mixture contained the radiolabeled Hap4p (1-330) incubated with GST, GST-Hap2p, GST-Hap3p, GST-Hap5p, or the indicated combinations. The first lane represents 10% of the radiolabeled Hap4p added to each pulldown reaction mixture (10% input). (B) Protein-protein interaction studies were performed with [35S]methionine-labeled Hap4p (1-330) incubated with GST alone or GST-Hap2p/Hap3p/Hap5p in the presence or absence of a double-stranded DNA oligonucleotide containing a CCAAT box or a mutated oligonucleotide (GGAAG). The first lane represents 10% of the radiolabeled protein in each reaction mixture (10% input). The percentage of the total radiolabeled protein precipitated in each reaction is shown below panels A and B.
The domain of Hap4p required for interaction with Hap2p/Hap3p/Hap5p.
Since the association of Hap4p with Hap2p/Hap3p/Hap5p required the heterotrimer to be bound to DNA, we investigated the region within Hap4p that was essential for the interaction(s). It has been shown that the Hap4p homologs in S. cerevisiae, K. lactis, and H. polymorpha share a 16-amino-acid region that is highly conserved (4, 51); however, it seemed unreasonable to assume that this domain alone would be sufficient to form a stable interaction with Hap2p/Hap3p/Hap5p. To investigate the minimal region of Hap4p sufficient for the stable binding with Hap2p/Hap3p/Hap5p, deletion analysis was performed on HAP4. For these studies, the Hap2p and Hap3p core domains and the Hap5p core plus the Hap4p recruitment domain (termed p92) (37, 38) were expressed in E. coli and purified as outlined in Materials and Methods. Following purification, the affinity tags were removed from each protein by thrombin cleavage. The various HAP4 truncations were synthesized via in vitro transcription/translation using two parallel reactions in which the proteins were synthesized in the presence of [35S]methionine for SDS-PAGE analysis to confirm size and expression (data not shown) or in the presence of nonradioactive methionine for the mobility shift assays.
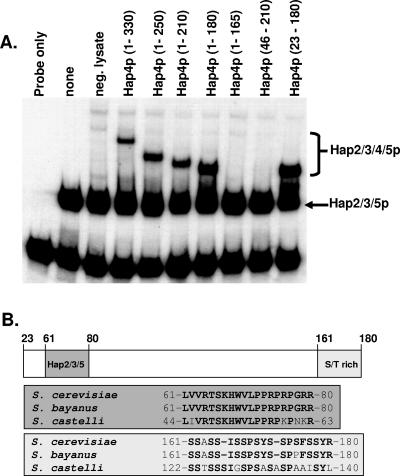
As previously reported (37), Hap4p (1-330) interacted with the Hap2p/Hap3p/Hap5p-DNA complex, as revealed by the slower migration of the complex in the mobility shift assays (Fig. 3A). Further C-terminal deletions of Hap4p yielded proteins competent to interact with the Hap2p/Hap3p/Hap5p-DNA complex until Hap4p (1-165), which failed to interact. Since Hap4p (1-180) could associate with Hap2p/Hap3p/Hap5p, the C-terminal boundary of the Hap2p/Hap3p/Hap5p-DNA interaction domain lies between amino acid residues 165 and 180. To define the N-terminal boundary, we generated a protein that lacked the first 45 amino acids, Hap4p (46-210), and found that it failed to interact. However, Hap4p (23-180) could associate with the Hap2p/Hap3p/Hap5p-DNA complex to alter the mobility of the complex (Fig. 3A). Thus, the N-terminal boundary was mapped to a region between amino acid residues 23 and 46. In summary, the domain of Hap4p required for association with the Hap2p/Hap3p/Hap5p-DNA complex maps between amino acid residues 23 and 180. It should be noted that this region encompasses the evolutionarily conserved domain of Hap4p important for interaction with Hap2p/Hap3p/Hap5p (Fig. 3B) and emphasizes its importance in the assembly of the DNA-bound Hap complex. Furthermore, these data suggest that additional amino acid residues outside the conserved region are also essential for the interaction of Hap4p with the Hap2p/Hap3p/Hap5p-DNA complex.
FIG. 3.
Minimal domain of Hap4p necessary for association with the Hap2p/Hap3p/Hap5p-DNA complex. (A) DNA mobility shift assays were performed with DNA-binding reactions containing a radiolabeled CCAAT box probe incubated with purified recombinant Hap2p core, Hap3p core, and Hap5p (p92) as indicated. Rabbit reticulocyte lysates containing the indicated Hap4p proteins or an unprogrammed lysate (neg. lysate) were added to the binding reaction mixtures as shown. The positions of the Hap2p/Hap3p/Hap5p heterotrimer and the Hap2p/Hap3p/Hap4p/Hap5p heterotetramers are indicated on the right. The free probe is at the bottom of the gel. (B) Schematic diagram depicting Hap4p (23-180) with the numbers above the diagram showing positions of the amino acid residues. The evolutionarily conserved region of Hap4p important for interaction with Hap2p/Hap3p/Hap5p is shown (dark gray box), as is the serine-rich region domain (light shaded box). The amino acid sequences corresponding to each region are shown in the gray and light shaded boxes.
To identify other regions within Hap4p that may be important, we compared the amino acid sequence of Hap4p (23-180) to other Hap4p homologs. Since the amino acid sequence of Hap4p has diverged dramatically in non-Saccharomyces yeasts (4, 51), we compared the sequence of S. cerevisiae Hap4p (23-180) with that of the closely related Saccharomyces species Saccharomyces bayanus (7), and a more evolutionarily diverged species, Saccharomyces castellii (7), to identify any conserved domains (Fig. 3B). Besides the Hap2p/Hap3p/Hap5p interaction domain, which is conserved in all Hap4p homologs (4, 51), the only remarkable conservation was a 20-amino-acid segment at the C terminus of Hap4p (23-180) between amino acid residues 161 and 180. This region was serine rich and conserved in the most divergent species, S. castellii. Furthermore, Hap4p (1-165) removes most of this region, causing the loss of binding to Hap2p/Hap3p/Hap5p (Fig. 3A), indicating that it is functionally important for assembly of the Hap2p/Hap3p/Hap4p/Hap5p complex.
The stoichiometry of Hap4p in the yeast CCAAT-binding factor.
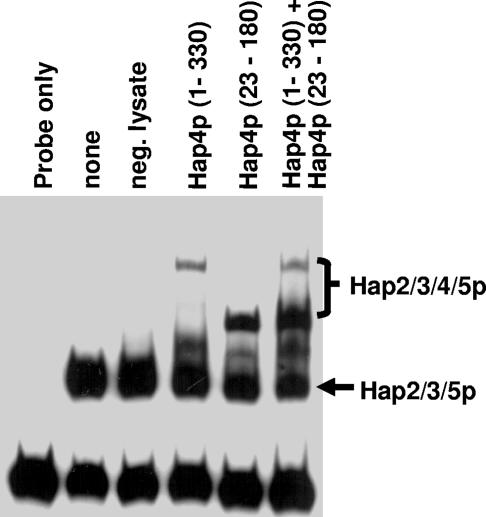
To further explore the interactions between Hap4p and Hap2p/Hap3p/Hap5p, we needed to determine how many copies of Hap4p are associated with each DNA-bound CCAAT-binding complex. Prior studies have demonstrated that each of the DNA-binding subunits of the mammalian CCAAT-binding factor are present in one copy per complex (29, 47), and we have performed mobility shift studies with different size variants of Hap2p, Hap3p, and Hap5p to confirm these data with yeast (data not shown). However, the number of Hap4p molecules per CCAAT-binding complex has not been established. To determine the stoichiometry of Hap4p in the DNA-bound CCAAT complex, mobility shift studies were performed using the in vitro-translated Hap4p (1-330) and Hap4p (23-180) (Fig. 4). Because the protein-DNA complexes contained identical Hap2p, Hap3p, and Hap5p subunits, any difference in mobility must be attributed to the size of the Hap4p protein. When Hap4p (1-330) and Hap4p (23-180) were both present in the DNA-binding assay, two distinct Hap2p/Hap3p/Hap4p/Hap5p complexes were observed, and their mobility corresponded to that of the Hap complexes containing either Hap4p (1-330) or Hap4p (23-180) alone (Fig. 4). If two or more molecules of Hap4p were present in each DNA-bound CCAAT complex, additional DNA-protein complexes of intermediate mobility would have been observed as a result of having a mixture of the two size variants. Thus, these data indicate that Hap4p, like Hap2p, Hap3p, and Hap5p, is present in one copy per DNA-bound CCAAT-binding complex.
FIG. 4.
One Hap4p molecule is present in the Hap2p/Hap3p/Hap4p/Hap5p-DNA complex. DNA mobility shift assays were performed with DNA-binding reaction mixtures containing a radiolabeled CCAAT box probe incubated with purified recombinant Hap2p core, Hap3p core, and Hap5p (p92). Rabbit reticulocyte lysates containing Hap4p (1-330), Hap4p (23-180), or an unprogrammed reticulocyte lysate (neg. lysate) were added to the binding reaction mixtures as indicated. The positions of the Hap2p/Hap3p/Hap5p heterotrimer and the Hap2p/Hap3p/Hap4p/Hap5p heterotetramers are indicated on the right. The free probe is at the bottom of the gel.
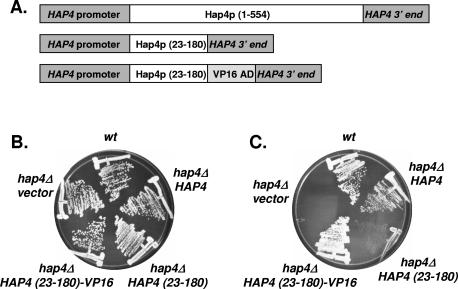
In vivo complementation of a hap4Δ mutant with Hap4p (23-180) fused to the VP16 activation domain.
The studies described above have shown that amino acid residues 23 through 180 are sufficient for Hap4p to associate with the Hap2p/Hap3p/Hap5p-DNA complex in vitro. To investigate whether this region was sufficient in vivo, we determined whether Hap4p (23-180) could functionally complement a hap4Δ mutant of S. cerevisiae. To mimic the normal levels of Hap4p expression in vivo, we generated an autonomously replicating sequence-centromeric (ARS/CEN) plasmid in which Hap4p (23-180) was expressed from the HAP4 promoter with its native 5′ and 3′ untranslated sequences. Two plasmids were generated for these studies. One plasmid contained HAP4 (23-180) lacking an activation domain and the other contained HAP4 (23-180) fused with the VP16 activation domain (Fig. 5A). The VP16 activation domain was chosen to avoid any additional Hap4p sequences within the protein. These plasmids were introduced into strain DMY144 (hap4Δ) and the transformants were tested for their ability to grow on medium containing either glucose (Fig. 5B) or lactate (Fig. 5C) as the sole carbon source. As controls, the wild-type strains FY250 and DMY144 containing an ARS/CEN vector only and DMY144 containing an ARS/CEN plasmid with the full-length HAP4 were included. All of the strains grew normally on glucose-containing medium, demonstrating their viability; however, the strain containing Hap4p (23-180) lacking an activation domain failed to complement the respiratory defect of the hap4Δ mutant. In contrast, Hap4p (23-180) fused to the VP16 activation domain complemented the defect with the strain growing normally on lactate medium compared to the wild-type strain or the hap4Δ strain containing the full-length HAP4.
FIG. 5.
In vivo complementation of a hap4Δ mutant with HAP4 (23-180)-VP16. (A) Schematic diagram of the various HAP4 alleles used to test complementation of the hap4Δ mutant. (B) Yeast strains FY250 (wt) or DMY144 (hap4Δ) were transformed with the indicated plasmids and subsequently grown on SC-Ura medium containing glucose for 3 days at 30°C. (C) Same as in panel B except the strains were grown on SC-Ura medium containing lactate as the sole carbon source. The plates were incubated for 3 days at 30°C.
To ascertain the ability of Hap4p (23-180) to stimulate transcription, the strains containing the different HAP4 alleles were transformed with an LEU2 plasmid containing a CYC1-lacZ reporter (15). The transformants were grown in SC-Ura-Leu synthetic omission medium containing glucose, raffinose, or lactate as the sole carbon source and were subsequently assayed for β-galactosidase activity (Table 2). The Hap4p (23-180) alone did not stimulate β-galactosidase activity beyond that of the hap4Δ control strain; however, addition of the VP16 activation domain to the truncated protein resulted in β-galactosidase activity that was similar to that of the strains containing the wild-type HAP4 gene. Thus, we concluded that Hap4p (23-180) is sufficient for association with Hap2p/Hap3p/Hap5p in vivo and is competent to activate transcription when fused to a heterologous activation domain.
TABLE 2.
Transcriptional activation of the CYC1 promoter by HAP4 (23-180)-VP16
| Strain | Plasmid genea | β-Galactosidase activity (Miller units)b in medium with:
|
||
|---|---|---|---|---|
| Glucose | Raffinose | Lactate | ||
| FY250 (HAP4) | None | 29 (±1) | 257 (±6) | 667 (±62) |
| DMY144 (hap4Δ) | HAP4 | 32 (±4) | 309 (±41) | 695 (±51) |
| DMY144 (hap4Δ) | None | 18 (±2) | 34 (±3) | ND |
| DMY144 (hap4Δ) | HAP4 (23-180) | 19 (±2) | 54 (±3) | ND |
| DMY144 (hap4Δ) | HAP4 (23-180)-VP16 | 38 (±6) | 365 (±62) | 621 (±96) |
Schematic diagram of the HAP4 alleles is shown in Fig. 5A. None, control strains having only the plasmid vector.
Values represent the average of three independent experiments with the standard deviation shown in parentheses. ND, not determined, since the strains would not grow under the indicated conditions.
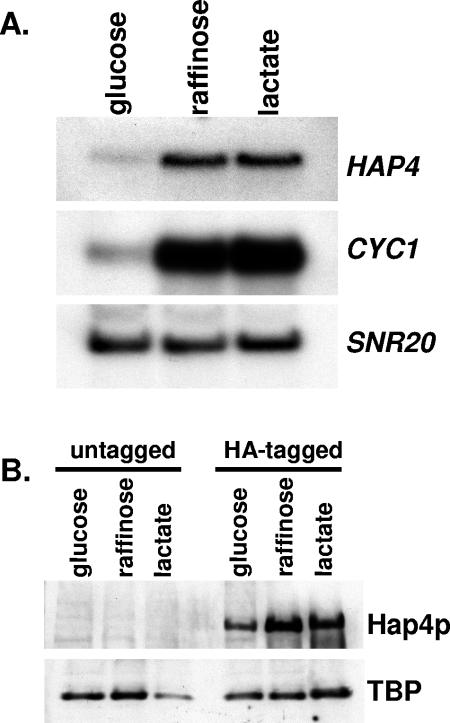
Hap4p protein expression is induced on nonfermentable carbon source.
To understand the association of Hap4p with Hap2p/Hap3p/Hap5p in vivo and how it regulates transcription, it was important to examine the expression of Hap4p protein. It has been shown that HAP4 mRNA levels are repressed when yeasts are grown in glucose-containing medium and induced three- to fourfold in medium containing lactate as the carbon source (15). The HAP4 gene is interesting structurally because it has an unusually long 5′ UTR of 270, 280, and 330 nucleotides (due to three mRNA initiation sites) that contain two short open reading frames, 27 and 9 nucleotides long, before the authentic AUG of the Hap4p open reading frame (15, 28). It has been reported that the HAP4 5′ UTR may serve as an internal ribosome entry site (28); when placed between two reporters in a bicistronic mRNA, it can mediate a carbon source-dependent translational enhancement of the downstream open reading frame (46). This raised a question as to whether the synthesis of the Hap4p protein correlates with the induction of mRNA or whether the upstream open reading frames or the internal ribosome entry site affects protein synthesis in a carbon source-dependent manner. Since Hap4p protein levels have never been examined in vivo, we wanted to investigate whether Hap4p synthesis displayed a pattern of carbon source inducibility similar to the HAP4 mRNA. To examine Hap4p protein levels, we generated an HA epitope-tagged allele of HAP4 in a strain designated DMY187. This strain displayed normal growth kinetics on nonfermentable carbon sources, implying that the HA epitope did not interfere with Hap4p function (data not shown).
To evaluate HAP4 mRNA levels on different carbon sources, strain BY4733 (HAP4) was inoculated to YP-dextrose medium, YPRaffinose, and YPLactate, and the cells were grown at 30°C to an OD of ∼1.0. The cultures were harvested, and total RNA was isolated and analyzed by Northern blotting (Fig. 6A). HAP4 mRNA levels were induced threefold in response to growth in YPRaffinose and YPLactate, consistent with the published data (15). As a control, we examined the induction of a Hap2p/Hap3p/Hap4p/Hap5p-regulated gene, CYC1, whose mRNA levels were induced 10 to 12 fold. To evaluate the induction of Hap4p protein synthesis, the untagged yeast strain BY4733 (HAP4) and epitope-tagged strain DMY187 (HAP4-3xHA) were grown under the conditions described above, and total protein extracts were prepared and separated by SDS-PAGE. The level of Hap4p was evaluated by Western blotting with an anti-HA monoclonal antibody (Fig. 6B). As a control for protein loading, the blot was reprobed with anti-TBP antiserum. The Hap4p protein levels were found to be induced approximately threefold during the growth in YPRaffinose and YPLactate, consistent with the mRNA levels. Interestingly, there was a detectable level of HAP4 mRNA and Hap4p even under glucose-repressing conditions (Fig. 6), which raises a question as to whether other posttranslational regulatory mechanisms are involved in controlling the association of Hap4p with the Hap2p/Hap3p/Hap5p-DNA complex. It seemed surprising that a threefold increase in Hap4p protein levels would yield a 10- to 12-fold activation of the target gene CYC1. Moreover, mobility shift assays performed with extracts prepared from yeast grown on glucose do not show any detectable Hap2p/Hap3p/Hap4p/Hap5p heterotetramer bound to DNA, yet the Hap2p/Hap3p/Hap4p/Hap5p complex is observed in extracts prepared from cells grown in lactate-containing medium (see Fig. 6 of reference 38). In summary, our data show that the inducibility of Hap4p protein levels correlates with the mRNA but suggests that the association of Hap4p with Hap2p/Hap3p/Hap5p may be posttranslationally regulated.
FIG. 6.
Hap4p protein levels correspond to the mRNA induction. (A) Northern blot analysis was performed with total RNA isolated from yeast strain BY4733 (HAP4) that was grown in rich medium containing glucose, raffinose, or lactate as indicated. The membrane was hybridized with radiolabeled probes specific for HAP4, CYC1, or SNR20. SNR20 was used to normalize RNA loading. B. Total protein extracts were prepared from yeast strains BY4733 (untagged HAP4) and DMY187 (3xHA-tagged HAP4) that was grown in rich medium containing glucose, raffinose, or lactate as indicated. The protein extracts were separated by SDS-PAGE using a 4 to 15% gradient gel, transferred to an Immobilon-P membrane, and probed with the anti-HA monoclonal antibody 12CA5 and anti-TBP antiserum. The TBP protein was used as a loading control.
DISCUSSION
In this report, we have made several novel findings related to the assembly of the CCAAT-binding factor in S. cerevisiae. First, the assembly of the Hap2p/Hap3p/Hap5p heterotrimer does not follow a two-step assembly pathway analogous to its mammalian counterpart (48) but assembles via a one-step mechanism requiring the presence of all three subunits for stable complex formation. Second, the interaction of Hap4p with Hap2p/Hap3p/Hap5p requires the heterotrimeric complex be bound to DNA at a CCAAT site. Third, we have identified the minimal region of Hap4p that is necessary for interaction with the Hap2p/Hap3p/Hap5p-DNA complex both in vivo and in vitro and have shown that Hap4p is present in one copy per DNA-bound Hap complex. Fourth, we have shown that the level of Hap4p protein expression under depressing conditions correlates with the induction of HAP4 mRNA, suggesting that protein levels are not likely to be controlled via a translational mechanism that involves the short open reading frames in the 5′ UTR of the mRNA.
We suspected that the assembly of the Hap2p/Hap3p/Hap5p heterotrimer may differ from its mammalian counterparts for the reasons already enumerated. Sinha et al. (47) and Kim et al. (29) have performed detailed mutational studies of CBF-A (Hap3p homolog) and CBF-C (Hap5p homolog), respectively, to delimit the regions of each protein that are necessary for heterodimer formation. A comparison of the amino acid sequences for the yeast and mammalian homologs within these regions revealed several nonconserved amino acid residues that could explain the different stabilities of the heterodimers; however, the functional relevance remains unclear. One possibility may relate to the fact that some organisms, such as plants (13, 21) and fungi (51; our unpublished observations) have multiple genetic loci that encode different variants of each Hap subunit. For example, Candida albicans has two distinct genetic loci encoding Hap3p homologs (unpublished observations), while Arabidopsis thaliana encodes six different nuclear factors YA (NF-YAs), nine NF-YBs, and eight NF-YCs (21), the homologs of Hap2p, Hap3p, and Hap5p, respectively. In contrast, mammalian organisms appear to have a single gene encoding each CCAAT-binding factor subunit (32, 49). Thus, the organisms that encode multiple variants of each subunit may have evolved with a less stable heterodimer to facilitate the interchange of subunits for different roles in gene regulation. Although S. cerevisiae does not have multiple loci encoding each Hap subunit, it may have evolved a similar pattern of stability.
The requirement for Hap2p/Hap3p/Hap5p to be bound to DNA as a prerequisite for Hap4p association raised a number of questions about the protein-protein and/or protein-DNA contacts within the complex. We have previously shown that the Hap4p recruitment domain of Hap5p is necessary for Hap4p to stably associate with Hap2p/Hap3p/Hap5p (37); however, the data in this report show that DNA binding is also important. We have previously reported that the Hap4p recruitment domain of Hap5p fused to the LexA DNA-binding domain is not sufficient to recruit Hap4p to a promoter containing lexA-binding sites (37). Thus, DNA and the Hap4p recruitment domain of Hap5p alone are not sufficient to bring Hap4p to a promoter, implying that Hap2p and Hap3p are also needed. In fact, previous studies have suggested that Hap4p may interact with Hap3p (55); however, the nature of this interaction is not understood. We envision two models for the interaction of Hap4p with the DNA-protein complex which would not be mutually exclusive. The first model involves a conformational change in Hap2p/Hap3p/Hap5p after binding to its cognate CCAAT site that allows the stable association of Hap4p. The alternative model involves Hap4p directly contacting both DNA and Hap2p/Hap3p/Hap5p for stable assembly. By this model, the Hap4p-DNA contact does not have to be sequence specific, since the DNA-bound Hap2p/Hap3p/Hap5p would provide sequence specificity. These hypotheses will be resolved by additional studies using fluorescence spectroscopy and DNA-protein cross-linking experiments.
The minimal functional region of Hap4p required for interaction with the Hap2p/Hap3p/Hap5p-DNA complex lies between amino acid residues 23 and 180. The evolutionarily conserved domain of Hap4p (4, 51) is within this domain, and we presume that it is required for contact with the Hap4p recruitment domain on Hap5p, although this interaction has not yet been established. Interestingly, Stebbins and Triezenberg (50) have shown that Hap4p contains two transcriptional activation domains. By fusing various segments of Hap4p to the LexA DNA-binding domain and assaying their activation potential with a lexA-lacZ reporter, these investigators identified two distinct activation domains. The first activation domain was mapped between amino acid residues 124 and 329, and a second mapped between amino acid residues 359 and 476. They generated point mutations within the first activation domain, namely, a triple point mutant (F148S, L149S, and F151S) that nearly abolished transcriptional activation. Our data raise a question about whether this region is competent to stimulate transcription in the context of the native CCAAT-binding factor, since Hap4p (23-180) failed to activate transcription unless fused to the VP16 activation domain (Table 2). However, we cannot rule out the possibility that additional C-terminal residues between amino acids 180 and 329 may contribute to activated transcription.
Why is the Hap4p subunit absent from the CCAAT-binding complexes of higher eukaryotes? One the basis of studies performed with the mammalian CCAAT-binding factor, this DNA-binding complex functions as a proximal promoter factor that works synergistically with other highly regulated activators to control gene expression (36). The mammalian homologs of Hap2p and Hap5p contain glutamine-rich regions, like that of another proximal promoter factor, Sp1 (8), that function as activation domains (9). Thus, the mammalian CCAAT-binding factor appears to have evolved to serve a more general function in transcription at numerous target genes (36), while the fungal CCAAT-binding factors appear to act as gene-specific transcriptional regulators. Thus, controlling the expression of Hap4p is pivotal in the regulation of downstream target genes.
On the basis of our results, we propose a model for the assembly of Hap2p/Hap3p/Hap4p/Hap5p in S. cerevisiae. Initially, Hap2p, Hap3p, and Hap5p assemble in a single step to form the DNA-binding heterotrimer, which can bind target promoters containing the cognate CCAAT site. Once bound to DNA, the Hap2p/Hap3p/Hap5p complex then recruits Hap4p to facilitate activated transcription under the appropriate environmental conditions. HAP2, HAP3, and HAP5 have been shown to be constitutively expressed in yeast (12). Thus, Hap2p/Hap3p/Hap5p may be constitutively bound at target promoters poised for the interaction with Hap4p and subsequent gene activation; however, constitutive DNA binding by Hap2p/Hap3p/Hap5p has not been demonstrated. If Hap2p/Hap3p/Hap5p were constitutively bound to DNA, then the induction of HAP4 mRNA and protein (Fig. 6) in response to the lack of glucose would permit a rapid reprogramming of gene expression when dictated by changes in nutrient availability. Consistent with the model, previous studies have shown that constitutive overexpression of HAP4 on glucose-containing medium results in the transcriptional induction of many Hap2p/Hap3p/Hap4p/Hap5p-regulated genes (31), implying that Hap2p/Hap3p/Hap5p is competent to bind DNA in the presence of glucose and that Hap4p is limiting for activated transcription. While these studies would imply that posttranslational modification of Hap4p is not necessary for activation on glucose-containing medium, we are cautious in making this assumption, since overexpression may override the normal posttranslational regulatory controls.
Acknowledgments
We thank Fred Winston for generously providing yeast strains and Leonard Guarente for providing plasmids, yeast strains, and the anti-TBP antiserum. D.S.M. expresses his appreciation to Leonard Guarente for his advice and encouragement with this research.
This work was supported by startup funds from the University of Arkansas and NIH grant R01AI51470 to D.S.M.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1994. Current protocols in molecular biology. Greene Publishing Associates and Wiley-Interscience, New York, N.Y.
- 2.Baxevanis, A. D., G. Arents, E. N. Moudrianakis, and D. Landsman. 1995. A variety of DNA-binding and multimeric proteins contain the histone fold motif. Nucleic Acids Res. 23:2685-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bellorini, M., D. K. Lee, J. C. Dantonel, K. Zemzoumi, R. G. Roeder, L. Tora, and R. Mantovani. 1997. CCAAT binding NF-Y-TBP interactions: NF-YB and NF-YC require short domains adjacent to their histone fold motifs for association with TBP basic residues. Nucleic Acids Res. 25:2174-2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bourgarel, D., C. C. Nguyen, and M. Bolotin-Fukuhara. 1999. HAP4, the glucose-repressed regulated subunit of the HAP transcriptional complex involved in the fermentation-respiration shift, has a functional homologue in the respiratory yeast Kluyveromyces lactis. Mol. Microbiol. 31:1205-1215. [DOI] [PubMed] [Google Scholar]
- 5.Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li, P. Hieter, and J. D. Boeke. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115-132. [DOI] [PubMed] [Google Scholar]
- 6.Bucher, P. 1990. Weight matrix descriptions of four eukaryotic RNA polymerase II promoter elements derived from 502 unrelated promoter sequences. J. Mol. Biol. 212:563-578. [DOI] [PubMed] [Google Scholar]
- 7.Cliften, P. F., L. W. Hillier, L. Fulton, T. Graves, T. Miner, W. R. Gish, R. H. Waterston, and M. Johnston. 2001. Surveying Saccharomyces genomes to identify functional elements by comparative DNA sequence analysis. Genome Res. 11:1175-1186. [DOI] [PubMed] [Google Scholar]
- 8.Courey, A., and R. Tjian. 1988. Analysis of SP1 in vivo reveals multiple transcriptional domains, including a novel glutamine rich activation motif. Cell 55:887-898. [DOI] [PubMed] [Google Scholar]
- 9.Coustry, F., S. N. Maity, S. Sinha, and B. de Crombrugghe. 1996. The transcriptional activity of the CCAAT-binding factor CBF is mediated by two distinct activation domains, one in the CBF-B subunit and the other in the CBF-C subunit. J. Biol. Chem. 271:14485-14491. [DOI] [PubMed] [Google Scholar]
- 10.Dang, V. D., C. Bohn, M. Bolotin-Fukuhara, and B. Daignan-Fornier. 1996. The CCAAT box-binding factor stimulates ammonium assimilation in Saccharomyces cerevisiae, defining a new cross-pathway regulation between nitrogen and carbon metabolisms. J. Bacteriol. 178:1842-1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dang, V. D., M. Valens, M. Bolotin-Fukuhara, and B. Daignan-Fornier. 1996. Cloning of the ASN1 and ASN2 genes encoding asparagine synthetases in Saccharomyces cerevisiae: differential regulation by the CCAAT-box-binding factor. Mol. Microbiol. 22:681-692. [DOI] [PubMed] [Google Scholar]
- 12.DeRisi, J. L., V. R. Iyer, and P. O. Brown. 1997. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278:680-686. [DOI] [PubMed] [Google Scholar]
- 13.Edwards, D., J. A. Murray, and A. G. Smith. 1998. Multiple genes encoding the conserved CCAAT-box transcription factor complex are expressed in Arabidopsis. Plant Physiol. 117:1015-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flores, C. L., C. Rodriguez, T. Petit, and C. Gancedo. 2000. Carbohydrate and energy-yielding metabolism in non-conventional yeasts. FEMS Microbiol. Rev. 24:507-529. [DOI] [PubMed] [Google Scholar]
- 15.Forsburg, S. L., and L. Guarente. 1989. Identification and characterization of HAP4: a third component of the CCAAT-bound HAP2/HAP3 heteromer. Genes Dev. 3:1166-1178. [DOI] [PubMed] [Google Scholar]
- 16.Gancedo, J. M. 1998. Yeast carbon catabolite repression. Microbiol. Mol. Biol. Rev. 62:334-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gietz, R. D., R. H. Schiestl, A. R. Willems, and R. A. Woods. 1995. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11:355-360. [DOI] [PubMed] [Google Scholar]
- 18.Gietz, R. D., and A. Sugino. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74:527-534. [DOI] [PubMed] [Google Scholar]
- 19.Guan, K. L., and J. E. Dixon. 1991. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal. Biochem. 192:262-267. [DOI] [PubMed] [Google Scholar]
- 20.Guarente, L., and M. Ptashne. 1981. Fusion of Escherichia coli lacZ to the cytochrome c gene of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 78:2199-2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gusmaroli, G., C. Tonelli, and R. Mantovani. 2001. Regulation of the CCAAT-binding NF-Y subunits in Arabidopsis thaliana. Gene 264:173-185. [DOI] [PubMed] [Google Scholar]
- 22.Guthrie, C., and G. R. Fink. 1991. Guide to yeast genetics and molecular biology. Academic Press, San Diego, CA.
- 23.Hahn, S., and L. Guarente. 1988. Yeast HAP2 and HAP3: transcriptional activators in a heteromeric complex. Science 240:317-321. [DOI] [PubMed] [Google Scholar]
- 24.Hahn, S., J. Pinkham, R. Wei, R. Miller, and L. Guarente. 1988. The HAP3 regulatory locus of Saccharomyces cerevisiae encodes divergent overlapping transcripts. Mol. Cell. Biol. 8:655-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 26.Hoffman, C. S., and F. Winston. 1987. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 57:267-272. [DOI] [PubMed] [Google Scholar]
- 27.Horvath, A., and H. Riezman. 1994. Rapid protein extraction from Saccharomyces cerevisiae. Yeast 10:1305-1310. [DOI] [PubMed] [Google Scholar]
- 28.Iizuka, N., L. Najita, A. Franzusoff, and P. Sarnow. 1994. Cap-dependent and cap-independent translation by internal initiation of mRNAs in cell extracts prepared from Saccharomyces cerevisiae. Mol. Cell. Biol. 14:7322-7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim, I. S., S. Sinha, B. de Crombrugghe, and S. N. Maity. 1996. Determination of functional domains in the C subunit of the CCAAT-binding factor (CBF) necessary for formation of a CBF-DNA complex: CBF-B interacts simultaneously with both the CBF-A and CBF-C subunits to form a heterotrimeric CBF molecule. Mol. Cell. Biol. 16:4003-4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klein, C. J., L. Olsson, and J. Nielsen. 1998. Glucose control in Saccharomyces cerevisiae: the role of Mig1 in metabolic functions. Microbiology 144:13-24. [DOI] [PubMed] [Google Scholar]
- 31.Lascaris, R., H. J. Bussemaker, A. Boorsma, M. Piper, H. van der Spek, L. Grivell, and J. Blom. 2003. Hap4p overexpression in glucose-grown Saccharomyces cerevisiae induces cells to enter a novel metabolic state. Genome Biol. 4:R3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li, X. Y., M. G. Mattei, Z. Zaleska-Rutczynska, R. Hooft van Huijsduijnen, F. Figueroa, J. Nadeau, C. Benoist, and D. Mathis. 1991. One subunit of the transcription factor NF-Y maps close to the major histocompatibility complex in murine and human chromosomes. Genomics 11:630-634. [DOI] [PubMed] [Google Scholar]
- 33.Liang, S. G., and S. N. Maity. 1998. Pathway of complex formation between DNA and three subunits of CBF/NF-Y. Photocross-linking analysis of DNA-protein interaction and characterization of equilibrium steps of subunit interaction and DNA binding. J. Biol. Chem. 273:31590-31598. [DOI] [PubMed] [Google Scholar]
- 34.Maity, S. N., S. Sinha, E. C. Ruteshouser, and B. de Crombrugghe. 1992. Three different polypeptides are necessary for DNA binding of the mammalian heteromeric CCAAT binding factor. J. Biol. Chem. 267:16574-16580. [PubMed] [Google Scholar]
- 35.Mantovani, R. 1999. The molecular biology of the CCAAT-binding factor NF-Y. Gene 239:15-27. [DOI] [PubMed] [Google Scholar]
- 36.Mantovani, R. 1998. A survey of 178 NF-Y binding CCAAT boxes. Nucleic Acids Res. 26:1135-1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McNabb, D. S., K. A. Tseng, and L. Guarente. 1997. The Saccharomyces cerevisiae Hap5p homolog from fission yeast reveals two conserved domains that are essential for assembly of heterotetrameric CCAAT-binding factor. Mol. Cell. Biol. 17:7008-7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McNabb, D. S., Y. Xing, and L. Guarente. 1995. Cloning of yeast HAP5: a novel subunit of a heterotrimeric complex required for CCAAT binding. Genes Dev. 9:47-58. [DOI] [PubMed] [Google Scholar]
- 39.Olesen, J. T., and L. Guarente. 1990. The HAP2 subunit of yeast CCAAT transcriptional activator contains adjacent domains for subunit association and DNA recognition: model for the HAP2/3/4 complex. Genes Dev. 4:1714-1729. [DOI] [PubMed] [Google Scholar]
- 40.Pinkham, J. L., J. T. Olesen, and L. P. Guarente. 1987. Sequence and nuclear localization of the Saccharomyces cerevisiae HAP2 protein, a transcriptional activator. Mol. Cell. Biol. 7:578-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Riego, L., A. Avendano, A. DeLuna, E. Rodriguez, and A. Gonzalez. 2002. GDH1 expression is regulated by GLN3, GCN4, and HAP4 under respiratory growth. Biochem. Biophys. Res. Commun. 293:79-85. [DOI] [PubMed] [Google Scholar]
- 42.Romier, C., F. Cocchiarella, R. Mantovani, and D. Moras. 2003. The NF-YB/NF-YC structure gives insight into DNA binding and transcription regulation by CCAAT factor NF-Y. J. Biol. Chem. 278:1336-1345. [DOI] [PubMed] [Google Scholar]
- 43.Sambrook, J., E. G. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 44.Schneider, B. L., W. Seufert, B. Steiner, Q. H. Yang, and A. B. Futcher. 1995. Use of polymerase chain reaction epitope tagging for protein tagging in Saccharomyces cerevisiae. Yeast 11:1265-1274. [DOI] [PubMed] [Google Scholar]
- 45.Schuller, H. J. 2003. Transcriptional control of nonfermentative metabolism in the yeast Saccharomyces cerevisiae. Curr. Genet. 43:139-160. [DOI] [PubMed] [Google Scholar]
- 46.Seino, A., Y. Yanagida, M. Aizawa, and E. Kobatake. 2005. Translational control by internal ribosome entry site in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1681:166-174. [DOI] [PubMed] [Google Scholar]
- 47.Sinha, S., I. S. Kim, K. Y. Sohn, B. de Crombrugghe, and S. N. Maity. 1996. Three classes of mutations in the A subunit of the CCAAT-binding factor CBF delineate functional domains involved in the three-step assembly of the CBF-DNA complex. Mol. Cell. Biol. 16:328-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sinha, S., S. N. Maity, J. Lu, and B. de Crombrugghe. 1995. Recombinant rat CBF-C, the third subunit of CBF/NFY, allows formation of a protein-DNA complex with CBF-A and CBF-B and with yeast HAP2 and HAP3. Proc. Natl. Acad. Sci. USA 92:1624-1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sinha, S., S. N. Maity, M. F. Seldin, and B. de Crombrugghe. 1996. Chromosomal assignment and tissue expression of CBF-C/NFY-C, the third subunit of the mammalian CCAAT-binding factor. Genomics 37:260-263. [DOI] [PubMed] [Google Scholar]
- 50.Stebbins, J. L., and S. J. Triezenberg. 2004. Identification, mutational analysis, and coactivator requirements of two distinct transcriptional activation domains of the Saccharomyces cerevisiae Hap4 protein. Eukaryot. Cell 3:339-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sybirna, K., B. Guiard, Y. F. Li, W. G. Bao, M. Bolotin-Fukuhara, and A. Delahodde. 2005. A new Hansenula polymorpha HAP4 homologue which contains only the N-terminal conserved domain of the protein is fully functional in Saccharomyces cerevisiae. Curr. Genet. 47:172-181. [DOI] [PubMed] [Google Scholar]
- 52.Triezenberg, S. J., K. L. LaMarco, and S. L. McKnight. 1988. Evidence of DNA:protein interactions that mediate HSV-1 immediate early gene activation by VP16. Genes Dev. 2:730-742. [DOI] [PubMed] [Google Scholar]
- 53.Walker, G. M. 1998. Yeast: physiology and biotechnology. John Wiley & Sons, Ltd., West Sussex, England, United Kingdom.
- 54.Winston, F., C. Dollard, and S. L. Ricupero-Hovasse. 1995. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast 11:53-55. [DOI] [PubMed] [Google Scholar]
- 55.Xing, Y., J. D. Fikes, and L. Guarente. 1993. Mutations in yeast HAP2/HAP3 define a hybrid CCAAT box binding domain. EMBO J. 12:4647-4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xing, Y., S. Zhang, J. T. Olesen, A. Rich, and L. Guarente. 1994. Subunit interaction in the CCAAT-binding heteromeric complex is mediated by a very short alpha-helix in HAP2. Proc. Natl. Acad. Sci. USA 91:3009-3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zitomer, R. S., and C. V. Lowry. 1992. Regulation of gene expression by oxygen in Saccharomyces cerevisiae. Microbiol. Rev. 56:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]