Abstract
Chitin is an essential component of the cell wall of many fungi. Chitin also can be enzymatically deacetylated to chitosan, a more flexible and soluble polymer. Cryptococcus neoformans is a fungal pathogen that causes cryptococcal meningoencephalitis, particularly in immunocompromised patients. In this work, we show that both chitin and chitosan are present in the cell wall of vegetatively growing C. neoformans yeast cells and that the levels of both rise dramatically as cells grow to higher density in liquid culture. C. neoformans has eight putative chitin synthases, and strains with any one chitin synthase deleted are viable at 30°C. In addition, C. neoformans genes encode three putative regulator proteins, which are homologs of Saccharomyces cerevisiae Skt5p. None of these three is essential for viability. However, one of the chitin synthases (Chs3) and one of the regulators (Csr2) are important for growth. Cells with deletions in either CHS3 or CSR2 have several shared phenotypes, including sensitivity to growth at 37°C. The similarity of their phenotypes also suggests that Csr2 specifically regulates chitin synthesis by Chs3. Lastly, both chs3Δ and the csr2Δ mutants are defective in chitosan production, predicting that Chs3-Csr2 complex with chitin deacetylases for conversion of chitin to chitosan. These data suggest that chitin synthesis could be an excellent antifungal target.
Chitin, a β-(1,4)-linked polymer of N-acetylglucosamine (GlcNAc), is one of the most abundant biopolymers found in nature. It is found in a diverse group of organisms, ranging from fungal species to insects to crustaceans (12). Chitin is vital to the integrity of the fungal cell wall and septum, as it provides strength through the hydrogen bonding of multiple chitin chains arranged into microfibrils (32). Interestingly, chitin, with its substantial importance for the well-being of the fungal cell wall, accounts for only 1 to 2% of dry cell weight of yeasts (23), although in filamentous fungi, the proportion of chitin can be as great as 40% (3).
Chitin synthase (EC 2.4.1.16) polymerizes GlcNAc into chitin from cytoplasmic pools of UDP-GlcNAc. Each chitin synthase (CHS) is integrated into the plasma membrane by multiple trans-membrane helices. Through an unknown process, individual chitin chains are extruded across the plasma membrane to become an extracellular component of the cell wall or septum (44). Attachment of some of the chitin occurs via covalent linkages to β-(1,3)-glucan (24, 25). Chitin synthases were initially grouped into three classes (class I, II, III) based on conserved regions of protein sequence of the catalytic domain (5). Three classes have been added (class IV, V, VI) as additional CHSs were sequenced (35, 50). Classes I to III comprise one family. These synthases are generally smaller proteins (average of 900 amino acids) with the catalytic domain, the signature sequence of all chitin synthases, found more towards the N terminus. In comparison, the second family contains classes IV to VI with proteins that have an average size of nearly 1,200 amino acids with the catalytic domain located more towards the C terminus (45). Orthologs of CHSs of the same class appear to have similar function in related fungi as observed by Munro and Gow in their review on chitin synthesis (35). Recently, a group of the class V CHSs was identified that includes a myosin motor-like domain at the N terminus and localizes with the actin cytoskeleton (27, 51).
Chitosan, the deacetylated derivative of chitin, also is an important constituent of the cell wall at various times during the life cycle of some fungal species (11). However, it is also not clear how prevalent chitosan is among fungi. Chitosan is not directly synthesized. Instead, secreted enzymes, the chitin deacetylases (EC 3.5.1.41) (2, 22), are thought to be in close proximity to the regions where chitin traverses the plasma membrane. As chitin is synthesized, the deacetylase enzyme can act efficiently to produce chitosan. Chitosan is a polycation, which is much more soluble than neutrally charged chitin. Of the few fungal species shown to synthesize chitosan, those belonging to the Mucorales have been shown to generate chitosan during vegetative growth (40), whereas Saccharomyces cerevisiae only produces chitosan during sporulation (11).
Various single deletions of CHS and their regulator genes have been shown to have some importance for the virulence of human fungal pathogens, including Candida albicans (CHS1; 35, 36) (CHS7; 47) and Exophiala (Wangiella) dermatitidis (CHSV) (27). Chitin synthases are also important for full virulence of plant pathogens including Fusarium oxysporum (CHSV; 29), Ustilago maydis (CHS6; 18), and Botrytis cinerea (CHS1; 49).
Chitin synthesis is best characterized in S. cerevisiae, which has genes that encode three CHSs. Chs1p, a class I synthase, is suggested to be involved in repair of the cell wall of the daughter cell that is hydrolyzed by chitinase during cell division and separation (7, 8). Chs2p, a class II synthase, is required for formation of the primary septum during cytokinesis (48). CHS2 expression appears to be tightly regulated throughout the cell cycle (10, 21). Finally, Chs3p, a class IV synthase, generates 90% of the chitin of S. cerevisiae (6, 54). Chs3p synthesizes the chitin ring that appears at the bud neck, chitin of the lateral wall, and the chitin of the spore wall that is deacetylated to chitosan (48). Chs3p as well as Chs1p levels remain constant throughout the cell cycle (61). For synthesis of chitin, Chs3p binds with a regulator protein, Skt5 (also known as Chs4p), which is required for enzymatic activity and localization to the septin ring of the bud neck (14, 39, 52). Several proteins are important for the proper trafficking of Chs3p through secretory pathways to the plasma membrane (61).
Cryptococcus neoformans is a dimorphic basidiomycetous human fungal pathogen, found world-wide, affecting mainly the immunocompromised. C. neoformans grows as a yeast but can also form hyphae during its sexual cycle. It is estimated that up to 13% of AIDS patients will develop a life-threatening case of cryptococcal meningitis (9). The cell wall of C. neoformans is essential to its viability and is associated with several virulence factors. These include the large polysaccharide capsule, which is attached to the wall (43), and melanin (57). Acapsular strains of Cryptococcus neoformans are essentially avirulent (16). Melanin, a scavenger of reactive oxygen species, provides protection to Cryptococcus neoformans against the host's defenses. There are two laccase proteins encoded by C. neoformans genes which contribute to the production of melanin. The major laccase, Lac1, is responsible for the majority of melanin production, is localized to the cell wall (33, 58), and is important for virulence (42).
Current antifungal therapies are still inadequate to completely control this disease, posing toxicity, efficacy, and resistance issues (9). Interestingly, a new class of antifungals, the echinocandins, which target β-(1,3)-glucan synthase, an essential enzyme responsible for the synthesis of a major component of the fungal cell wall, is effective for safe treatment of a broad range of fungal infections but is ineffective in treating cryptococcosis (26), even though the cryptococcal β-(1,3)-glucan synthase is sensitive to the drug (30). With its importance for fungal cell integrity and the absence of chitin production in humans, biosynthesis of chitin and the chitin-derived polymer chitosan make excellent targets for future antifungals.
Here, we begin to describe the dynamics of chitin and chitosan biosynthesis by C. neoformans during growth in culture as budding yeast and the importance of specific genes for synthesis of these critical polymers. It was found that C. neoformans encodes eight chitin synthases and three homologues of the S. cerevisiae chitin synthase regulator, Skt5p. Expression analysis of the chitin synthases and gene deletions of each of the above show that CHS3 is the most important chitin synthase for vegetative growth and that one of the Skt5p homologs appears to specifically regulate Chs3.
MATERIALS AND METHODS
Fungal strains and media.
H99, a well-characterized virulent clinical isolate of C. neoformans serotype A, was used as the wild-type strain, and all deletions were made in H99. Strains of S. cerevisiae and C. albicans were BY4741 and CAI4, respectively. Strains were grown on rich medium, YPD (1% yeast extract, 2% Bacto-peptone, and 2% dextrose). Solid medium contained 2% Bacto-agar. Selective YPD medium contained 100 μg/ml nourseothricin (Werner BioAgents, Jena-Cospeda, Germany) or 200 U/ml hygromycin (Calbiochem, La Jolla, CA).
Generation of deletion constructs.
An overlap PCR gene deletion technology (supplemental material) (13, 19) was used to generate gene-specific deletion cassettes of CHS1, CHS3, CHS4, CHS5, CHS6, CHS7, CHS8, and CSR2 that included a nourseothricin cassette (31) and a hygromycin cassette (20) for CHS2, CSR1, and CSR3. This resulted in the deletion of greater than 95% of CHS1, greater than 81% of CHS2, greater than 99% of CHS3, greater than 97% of CHS4, greater than 98% of CHS5, greater than 96% of CHS6, greater than 89% of CHS7, greater than 95% of CHS8, 54% of CSR1, greater than 90% of CSR2, and 19% of CSR3. The GenBank accession numbers of the proteins from the closely related strain, JEC21, are shown in Table 1, and the primers used to disrupt the genes in H99 are shown in supplementary Table S1.
TABLE 1.
Chitin synthases and chitin synthase regulators of C. neoformans strain JEC21
| Gene | Chromosome | GenBank accession no. (gDNA) | No. of introns | GenBank accession no. (cDNA) | cDNA 5′ UTSb (bp) | cDNA 3′ UTS (bp) | Predicted protein (aa) | GenBank accession no. (protein) |
|---|---|---|---|---|---|---|---|---|
| CHS1 | 7 | AE017347 | 5 | XM_572145 | Yes (234) | No | 1236 | AAW44838 |
| CHS2 | 7 | AE017347 | 11 | XM_571995 | Yes (331) | Yes (135) | 947 | AAW44688 |
| CHS3 | 8 | AE017348 | 4 | XM_572399 | Yes (363) | Yes (178) | 1423 | AAW45092 |
| CHS4 | 1 | AE017341 | 10 | XM_566840 | Yes (20) | Yes (122) | 1271 | AAW41021 |
| CHS5 | 6 | AE017346 | 12 | XM_571494 | No | Yes (102) | 1895 | AAW44187 |
| CHS6 | 14 | AE017356 | 25 | XM_568689 | Yes (44) | Yes (149) | 996 | AAW47172 |
| CHS7 | 5 | AE017345 | 24 | XM_570882a | No | No | 931 | AAW43575 |
| CHS8 | 3 | AE017343 | 23 | XM_569357 | No | Yes (110) | 1024 | AAW42050 |
| CSR1 | 6 | AE017346 | 4 | XM_571612 | No | Yes (210) | 754 | AAW44305 |
| CSR2 | 5 | AE017345 | 4 | XM_570973 | Yes (208) | Yes (342) | 490 | AAW43666 |
| CSR3 | 2 | AE017342 | 4 | XM_569085a | No | No | 838 | AAW41778 |
Predicted cDNA does not carry a complete open reading frame as presently reported in GenBank, and there are no EST sequences reported that correspond to these genes.
UTS, untranslated sequence.
Transformation of C. neoformans.
H99 was transformed using biolistic techniques (20, 53). Cells were grown in YPD to late-log phase, concentrated, and plated onto YPD agar for transformation. The cells were bombarded with 0.6-μm gold beads (Bio-Rad, Richmond, CA) that were coated with DNA of the target construct according to the manufacturer's recommendations. Following the transformation, the cells were incubated at 30°C for 4 h on nonselective media to allow for recovery and then transferred with 0.8 ml sterile phosphate-buffered saline (PBS) to the appropriate selective media. Transformants were observed in 3 to 5 days.
Analysis of transformants.
To isolate stable transformants, all transformants were passaged five times on nonselective YPD medium and then tested for resistance to the appropriate selective marker. Only those transformants that grew equally well on selective and nonselective media were considered to be stable transformants. A three-primer PCR screen was used to verify homologous integration at both the 5′ and 3′ ends of the deletion cassette (supplemental material) (38). In this manner, homologous recombinants can be distinguished from the wild type. A PCR screen using primers outside the deletion construct was used to amplify the entire integration region, demonstrating that a single copy of the transforming DNA had been inserted at the desired locus. Southern blots were performed to screen for single integration in the genome. Single bands were observed on all Southern blots when hybridized with a selectable marker-specific probe. All deletion strains generated for this work had a single deletion construct homologously integrated at the appropriate locus and had no other insertions in the genome (data not shown).
Genomic DNA preparation.
Genomic DNA was prepared by a modification of the glass bead DNA extraction protocol described by Fujimura and Sakuma (17). C. neoformans cells were suspended in a microcentrifuge tube in 500 μl lysis buffer (50 mM Tris-HCl, pH 7.5, 20 mM EDTA, 1% sodium dodecyl sulfate [SDS]), with 400 mg glass beads (425 to 600 μm; Sigma G-9268). Cells were disrupted by vortexing for 10 min, followed by a 10-min incubation at 70°C. After brief vortexing, 200 μl 5 M potassium acetate and 150 μl 5 M NaCl were added. The tubes were placed on ice for 20 min and centrifuged at 14,000 rpm for 20 min. The supernatant was mixed with 500 μl phenol-chloroform and spun for 5 min at 14,000 rpm. The aqueous phase was then mixed with 450 μl chloroform and spun for 5 min at 14,000 rpm. The DNA was then precipitated by addition of 200 μl ethanol, washed with 70% ethanol, dried, and resuspended in 50 μl deionized water.
Southern hybridizations.
Approximately 10 μg of genomic DNA from each strain was digested with various restriction endonucleases according to the manufacturer's recommendations. Restriction fragments were separated on a 1% agarose gel and transferred to nylon membranes using a Turbo-Blot apparatus (Schleicher & Schuell) and 10× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) as transfer buffer. Probes for Southern analysis were prepared by random priming (random priming kit; Roche) using 50 μCi [α-32P]dCTP (Amersham AA0005) according to the manufacturer's instructions. The blots were incubated in 10 ml of a 6× SSC, 0.1% SDS, and 5% nonfat dry milk (Carnation) solution for 1 h at 65°C, and then probe was added to this solution and the blots were hybridized at 65°C overnight. The blots were washed twice in 2× SSC, 0.1% SDS at room temperature for 10 min and once for 10 min in 0.2× SSC, 0.1% SDS that had been prewarmed to 65°C.
RNA extraction, cDNA synthesis, and poly(A) RNA purification.
Each C. neoformans strain was initially grown for 16 h at 30°C in liquid YPD medium. All cultures were then diluted to an OD650 of 0.5 in 50 ml of fresh YPD medium and allowed to grow for an additional 2.5 h at 30°C. Cells were collected by centrifugation at 1,800 × g for 5 min, washed once with distilled water, and lyophilized overnight. The lyophilized pellet was then vortexed with 3 ml glass beads (1 mm; Biospec, Inc.) and resuspended in 4 ml TRIzol Reagent (Invitrogen). After sitting at room temperature for 5 min, 800 μl of chloroform was added and the mixture was shaken for 30 s. This cell lysate was then centrifuged at 4,000 rpm for 10 min, and the supernatant was transferred to a new tube. Two milliliters of isopropanol was added, incubated for 10 min at room temperature, and centrifuged at 4,000 rpm for 10 min. After washing the pellet with 75% ethanol, it was resuspended in water and incubated with DNase I at 37°C for 1 h. The RNA was extracted again with TRIzol and chloroform and precipitated with isopropanol as described above. The dried pellet was resuspended in 300 μl RNase-free water (Gibco). Poly(A) RNA was purified from the total RNA sample using an oligoTex RNA purification kit (QIAGEN) following the manufacturer's specifications. First-strand cDNA was made using the First Strand cDNA synthesis kit for reverse transcription-PCR (Roche).
Northern hybridizations.
The procedure we used for Northern hybridizations was adapted from Sambrook and Russell (46). Approximately 1 μg of poly(A) RNA from each strain was mixed with 2 μl of diethyl pyrocarbonate (DEPC) (Sigma)-treated 5× formaldehyde gel running buffer (0.1 mM morpholinepropanesulfonic acid [MOPS] [Fisher], pH 7.0, 40 mM sodium acetate [Sigma], 5 mM EDTA, pH 8.0), 3.5 μl of 37% formaldehyde (Sigma), and 10 μl of formamide (Sigma). Samples were then incubated at 65°C for 15 min and placed directly on ice. Two microliters of DEPC-treated gel loading buffer (50% glycerol [Fisher], 1 mM EDTA [Fisher], 0.25% bromophenol blue [Sigma]) was added, and samples were loaded on a 1.5% agarose (Roche) gel containing formaldehyde (DEPC treated, 1× formaldehyde gel running buffer, 6.6% formaldehyde) submersed in 1× formaldehyde gel running buffer. An RNA size marker (3191A; Promega) sample was prepared in the same way; however, ∼1 μg ethidium bromide (Sigma) was added before the sample was loaded onto the agarose gel. Importantly, a positive binding control ladder, containing 7.5 × 10−19 mol of each gene-specific DNA of interest, was also loaded on the agarose gel. The primers used to amplify the gene-specific DNA fragments for the positive binding control ladder are shown in supplemental Table S1. Gels were prerun in 1× formaldehyde gel running buffer for 5 min at approximately 5 V/cm before samples were loaded and run at approximately 2 V/cm overnight (∼14 h) with constant recirculation of running buffer until the bromophenol blue had traveled 8 to 9 cm. Gels were rinsed in several volumes DEPC-treated water, soaked in several volumes of DEPC-treated 50 mM NaOH (Fisher) for 20 min, and soaked in several volumes 20× SSC for 45 min. Fragments were then transferred to charged nylon membranes using a Turbo-Blot apparatus with 20× SSC as transfer buffer. Membranes were then UV cross-linked. Probes for Northern analysis were prepared by random priming (random priming kit; Roche) using 50 μCi [α-32P]dCTP according to the manufacturer’s instructions. The membranes were incubated in 20 ml of hybridization buffer (0.5 M sodium phosphate [Fisher], pH 7.2, 7% [wt/vol] SDS, 1 mM EDTA, pH 7.0) for 1 h at 65°C, and then probe was added to this solution and the membranes were hybridized at 65°C overnight. The membranes were washed one time in ∼150 ml of a solution of 1× SSC, 0.1% SDS at 25°C for 10 min. Membranes were then washed three times in ∼150 ml of a solution of 0.5× SSC, 0.1% SDS at 68°C for 10 min each wash. When necessary, membranes were stripped of probe by incubating in 1 liter of 10 mM Tris-Cl, pH 7.4, 0.2% SDS, prewarmed to 72°C, for 1.5 h, and subsequently reprobed following instructions above.
Cell wall stress plates.
Solid YPD media were made with designated amounts of SDS, caffeine, or Congo Red (Congo Red stock made in 50% ethanol). C. neoformans strains were grown to mid-log phase in YPD, and 10-fold dilutions were made. Five microliters of each of the 10-fold dilutions of cultures for each strain were spotted onto the solid media and grown at 30°C, 37°C, or 40°C.
Calcofluor white staining.
Cells were grown in 50 ml YPD, diluted in YPD to an OD600 of 0.1 and allowed to grow for 8, 16, 24, and 48 h. Aliquots were taken at each time point, fixed, stained with calcofluor white (Sigma), and mounted following the protocol of Pringle et al. (41). Cells were examined with an Olympus Vanox AHBT3 microscope using a 4,6-diamidino-2-phenylindole filter.
Cellular chitin and chitosan content assay.
To measure the chitin and chitosan content of cells, samples were divided into two aliquots. One aliquot was treated with acetic anhydride to measure chitin plus chitosan, and the second aliquot remained untreated to measure chitin. The difference between the two measurements estimated the amount of chitosan. Cultures were initially grown for 20 h in liquid YPD medium and then diluted to an OD650 of 0.05 in fresh medium and incubated at 30°C with shaking at 225 rpm. The two 0.5- to 1.0-ml aliquots of each culture were transferred to tared 2-ml microcentrifuge tubes. Cells were collected by centrifugation at 14,000 rpm for 2 min, the media were removed, and tubes were spun again at 14,000 rpm for 1 min so as to remove residual media. The weight of the cell pellet of each sample was determined and defined as wet weight, typically 20 to 30 mg. Dry weights were measured following 2 to 3 days of evaporation at 37°C. One aliquot of pelleted cells was resuspended in 1.0 ml 1 M sodium bicarbonate followed by addition of 50 μl acetic anhydride. The acetylation reaction proceeded for 20 min at room temperature with occasional mixing followed by 5 min at 100°C. Cells were pelleted as above. Both aliquots of cells were subsequently extracted with 1 ml 6% KOH at 80°C for 90 min. Samples were centrifuged at 14,000 rpm for 20 min, and the supernatants were discarded. Each pellet was suspended in 1 ml PBS and spun again, and the buffer was discarded. Finally, each pellet was suspended in 0.2 ml of McIlvaine's Buffer (0.2 M Na2HPO4, 0.1 M citric acid, pH 6.0) and frozen at −20°C. Upon thawing, 5 μl of purified Streptomyces plicatus chitinase-63 (5 mg/ml in PBS) was added to hydrolyze chitin to GlcNAc; samples were incubated for 2 to 3 days at 37°C and then stored at −20°C. For colorimetric determination of N-acetylglucosamine (GlcNAc) the Morgan-Elson method was adapted for microplate readers essentially as previously described (6a). Chitinase-treated samples were spun at 14,000 rpm for 1 min, and each 10 μl of sample supernatant was combined with 10 μl 0.27 M sodium borate, pH 9.0, in 0.2-ml PCR strip tubes. Samples were heated in a thermocycler (Techne Incorporation, Princeton, NJ) to 99.9°C for about 60 s, mixed gently, and incubated further at 99.9°C for 10 min. Immediately upon cooling to room temperature, 100 μl of freshly diluted DMAB solution (Ehrlich's reagent, 10 g p-dimethylaminobenzaldehyde in 12.5 ml concentrated HCl, and 87.5 ml glacial acetic acid, diluted 1:10 with glacial acetic acid) was added, followed by incubation at 37°C for 20 min. Ninety microliters of each sample was transferred to 96-well low-evaporation microtiter plates, and absorbance at 585 nm was recorded. Standard curves were prepared from stocks of 0.2 to 2.0 mM of GlcNAc (Sigma).
Analysis of melanin production.
Strains were streaked out from freshly growing YPD medium onto birdseed agar medium (Remel, Lenexa, KS) and incubated at 25°C for 3 days. Cells were also grown overnight in YPD medium, pelleted, washed twice in PBS, and resuspended in PBS to an OD650 of 1.0. Tenfold serial dilutions were made in PBS and 5 μl of each dilution plated on l-3,4-dihydroxyphenylalanine (L-DOPA) medium (13 mM glycine, 15 mM glucose, 29.4 mM KH2PO4, 10 mM MgSO4 · 7H2O, 3 μM thiamine, 5 μM d-biotin, 2% agar, and 1 mM L-DOPA, pH 5.5). The plates were incubated at 25°C for 4 days in the dark. Cells of each strain were also taken from solid YPD medium and resuspended in 2 ml glucose-free asparagine medium (1 g/liter l-asparagine, 0.5 g/liter MgSO4 · 7H20, 3 g/liter KH2PO4, 1 mg/liter thiamine) plus 1 mM L-DOPA at a concentration of 5 × 107 cells/ml. Cells were shaken at 30°C for 24 h, spun down at 652 × g for 10 min, and photographed. The OD400 was measured for the supernatants from three independent cultures.
Analysis of capsule formation.
Strains were streaked out on DME plates (13.4 g/liter Dulbecco's modified Eagle's medium [Sigma, St. Louis, MO], 25 mM MOPS, pH 7.0, 1.8% agar) and incubated for 3 days at 30°C. Individual isolates were resuspended in 1:4 India Ink:H2O solution. Cells were observed through an Olympus AHBT3 microscope at 400× and 1,000× magnification, and capsule diameter was measured.
RESULTS
Cell wall composition changes with increasing time of growth in culture.
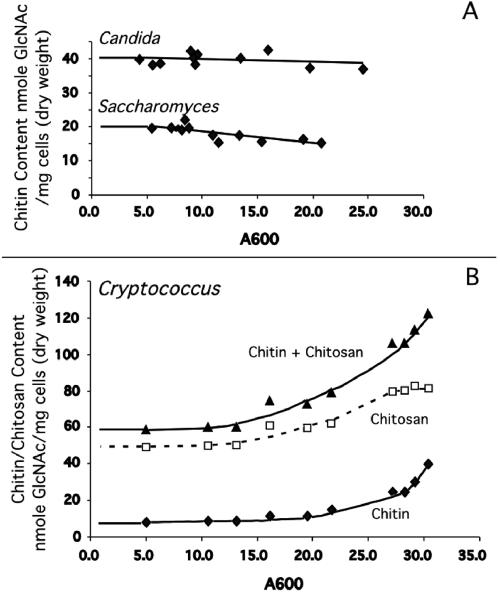
The cellular chitin and chitosan content of wild-type H99 was assessed at different times during growth in YPD liquid medium (Fig. 1). Surprisingly, as the culture grew to increasing density, the amount of both chitin and chitosan of the cell increased. Another unexpected observation was that the amount of chitosan was found to be three to five times greater than the amount of chitin. These results are in direct contrast to the results from S. cerevisiae and C. albicans that have a constant amount of chitin as culture growth time increases and contain no chitosan during vegetative growth (Fig. 1). Chitosan is found in S. cerevisiae (an ascomycete) only during spore formation (11), but it has been found in the cell wall of several zygomycetes (4). Our finding of chitosan in the cell wall of C. neoformans during vegetative growth is the first report for a basidiomycete.
FIG. 1.
Chitin and chitosan measurements of vegetatively growing wild-type C. neoformans strain H99, S. cerevisiae strain BY4741, and C. albicans strain CAI4. YPD cultures were grown and assayed for chitin and chitosan content when cultures reached various absorbance values. Absorbance measurement at 600 nm is indicated at the bottom of the panel. Chitin and chitosan content are indicated at the left of the panel. A. Chitin content of both S. cerevisiae and C. albicans remains virtually constant throughout growth, with no chitosan found for either species during vegetative growth (data not shown). B. Both chitin and chitosan are synthesized by C. neoformans during vegetative growth, the levels of each increasing as culture density increases.
Eight putative chitin synthase genes identified in Cryptococcus neoformans.
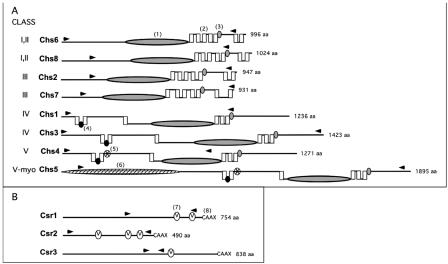
The genome of C. neoformans var. neoformans strain JEC21 has been recently sequenced and annotated (28). Analysis of the genome revealed eight potential chitin synthase genes (Fig. 2, Table 1). Each of these genes was also found in the genome of H99, a var. gattii strain that has been jointly sequenced by Duke University (http://cgt.genetics.duke.edu) and the Broad Institute (http://www.broad.mit.edu/annotation/fungi/cryptococcus_neoformans). National Center for Biotechnology Information (NCBI) BLAST search analysis of each of the C. neoformans putative chitin synthase proteins reveals that all eight contain a chitin synthase domain. Pairwise BLAST alignment of the eight proposed chitin synthase amino acid sequences of C. neoformans indicates 22% identity among the synthases. Each of the eight putative proteins include the motifs QxFEY, A/GEDR, and QRRRW, which are thought to be located at the carboxy terminal of the catalytic domain. Based on enzyme function of point mutants in S. cerevisiae, these sequences are likely important for catalytic activity (37).
FIG. 2.
C. neoformans chitin synthase (Chs) and chitin synthase regulator (Csr) predicted protein structure comparison. The name of each protein is indicated to the immediate left of the protein structure. The predicted amino acid size (from the JEC21 genome sequence) is indicated to the right of each protein structure. A. Chitin synthases. Class designations are indicated by Roman numerals. Chs6 and Chs8 have protein characteristics of both class I and II. Chs5 (class V-myo) has class V protein sequence characteristics as well as a myosin motor domain. Domains were identified by rpsblast searches of the Conserved Domain Database at NCBI (www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). Domains are identified as follows: (1) large shaded ellipse, catalytic domain contains homologous sequence of chitin synthase domains 1 and 2 (pfam01644 and pfam03142); (2) vertical box, trans-membrane span; (3) small, shaded oval, conserved sequence “S/T-W-G-X-T/K-R/G”; (4) small black oval, conserved sequence “AWREK”; (5) small starred oval, cytochrome b5-like heme/steroid binding domain (pfam00173); (6) large hatched ellipse, myosin motor domain (cd 00124). B. Chitin synthase regulators. Domains are identified as (7) small “V” oval, SEL1 domain (smart 00671), which may be sites for protein-protein interaction; (8) C-terminal sequence “CAAX” as potential site for farensylation. The area between the arrow heads for each protein in both A and B indicates the region deleted in each deletion strain.
The phylogenetic relationship of C. neoformans chitin synthases to those of other fungal species was deduced using the conserved catalytic domain found in all chitin synthases. This analysis indicated that C. neoformans contains two putative synthases with homology to both class I and class II synthases (Chs6, Chs8), two class III (Chs2, Chs7), two class IV (Chs1, Chs3), and two class V (Chs4, Chs5) chitin synthases (5, 35, 50). Transmembrane domain analysis of the C. neoformans chitin synthase protein sequences was performed using TMHMM (www.cbs.dtu.dk/services). The catalytic domain of each of the synthases is proposed to be on the cytoplasmic face of the plasma membrane (Fig. 2). Each of the proposed class I, II, and III synthases contains seven transmembrane domains. Five of the domains are found directly after the chitin synthase catalytic domain, with the peptide S/T-W-G-X-T/K-R/G located immediately after the fifth transmembrane stretch, putatively on the outside of the plasma membrane. This peptide is conserved in all known chitin synthase proteins, and based on mutation analysis of ScChs2p, it is important for catalytic activity (59). The last two transmembrane domains are found downstream of this region.
Each of the proposed class IV and V synthases contain six transmembrane domains. Again, these are found in the same relative regions of each synthase. Two closely spaced transmembrane domains precede the catalytic domain separated by the conserved peptide sequence AWREK that putatively faces the cytoplasm. A third transmembrane domain is found several hundred amino acids downstream, before the catalytic domain. Immediately following the catalytic domain are three more transmembrane domains instead of five, as found in the classes I, II, and III mentioned above. However, these three transmembrane stretches do have the characteristic S/T-W-G-X-T/K-R/G peptide region following immediately after, as in class I, II, and III. Although the importance is not understood, the putative class V synthases contain a heme and/or steroid binding domain directly downstream of the first two transmembrane domain regions (Fig. 2). Interestingly, a myosin motor domain is found in Chs5, which is present in chitin synthases of other fungal species, and mediates interactions with the actin cytoskeleton (51).
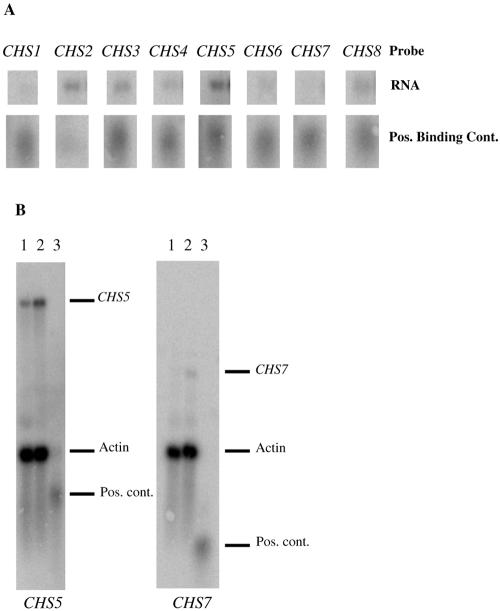
Relative expression levels of CHS genes under vegetative growth conditions.
In order to gain some insight into the expression levels of each of the chitin synthase genes in H99 during vegetative growth, the mRNA levels for each chitin synthase gene in H99 (Fig. 3A and data not shown) were compared by Northern blot analysis. These data indicated that CHS2, CHS3, CHS5, and CHS8 are transcribed at the highest levels. CHS1 and CHS4 had lower levels of expression. The CHS6 transcript was barely detectable, and CHS7 was undetectable. BLAST analysis of the expressed sequence tag (EST) library from The Institute for Genomic Research generated from RNAs isolated from JEC21 cells (var. neoformans) grown under a variety of conditions (28) revealed that although ESTs could be found that corresponded to seven of eight of the CHS genes, there was no EST for CHS7, suggesting that CHS7 is unexpressed in another serotype as well.
FIG. 3.
Chitin synthase expression analysis. A. Relative chitin synthase mRNA expression levels in vegetatively growing H99 (wild type) cells. The top row of bands is the same quantity of H99 RNA probed with each of the chitin synthase genes (labeled at the top). The second row of bands consists of equal amounts of positive binding controls for each chitin synthase (see Materials and Methods). These results are representative of two independent experiments. B. Northern blots showing the increase in transcription of two of the chitin synthase genes in a chs3Δ mutant. Lane 1 is H99 RNA, lane 2 is chs3Δ RNA, and lane 3 is the positive binding control ladder. The blot on the left was probed with CHS5 and actin, and the blot on the right was probed with CHS7 and actin. The dashes on the left indicate where the different transcripts and controls migrate. Pos., positive; cont., control.
Three putative homologs of the S. cerevisiae SKT5 chitin synthase regulator genes were identified in C. neoformans.
Analysis of JEC21 revealed three putative chitin synthase regulator (CSR) genes (Table 1), and each of these genes was identified in the H99 genome sequence. Based on proposed amino acid content, these genes code for proteins similar to the S. cerevisiae Skt5p protein (also known as Chs4p), a direct regulator of Chs3p in S. cerevisiae (39). BLAST analysis of the putative Csr1, Csr2, and Csr3 proteins revealed that the three homologues are 20%, 27%, and 21% identical to the Skt5p found in S. cerevisiae, respectively. Sequence analysis reveals that each of these potential synthase regulators contains one to three SEL1 domains (SMART accession number SM00671; http://smart.embl.de/), a stretch thought to be important for protein-protein interaction, as well as the C-A-A-X box, a possible site for prenylation of each Csr protein (60) (Fig. 2).
None of the chitin synthase and chitin synthase regulator genes are essential for viability.
In order to determine the importance of the chitin synthase genes as well as the putative chitin synthase regulator genes, deletion strains of each of the CHS and CSR coding regions were made using PCR generated gene-specific deletion constructs (see Materials and Methods as well as supplemental material). The regions deleted are shown in Fig. 2. Deletion mutations were made in each chitin synthase and chitin synthase regulator gene. Each of the resultant strains was viable, demonstrating that no single gene was essential for viability.
Slight compensatory RNA expression was detected in the chs3Δ strain.
Because C. neoformans has eight chitin synthases, we explored the possibility that deletion of one gene would cause others to be up-regulated. Each of the deletion strains, as well as H99, was grown under vegetative conditions, and the transcripts for each of the chitin synthases was measured by Northern analysis. There was no up-regulation in expression of any of the chitin synthases in any of the deletion strains (data not shown) except for chs3Δ (Fig. 3B). A slight increase in CHS5 transcript was detected, and transcripts for CHS7, which had previously been undetectable, were now observed. These data suggests that transcriptional regulation of chitin synthases may be regulated through a feedback mechanism.
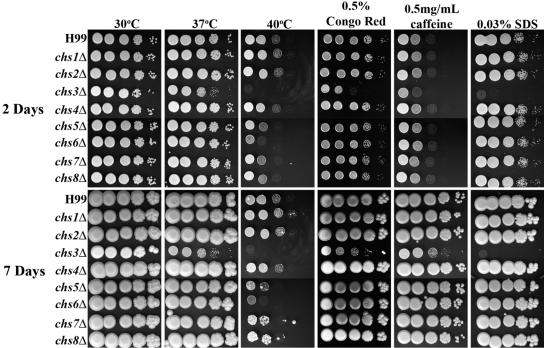
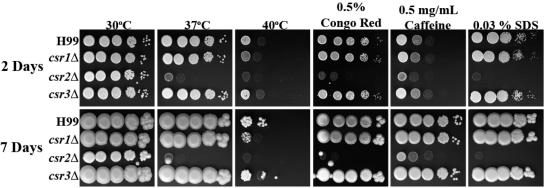
The chs3Δ and csr2Δ strains are sensitive to cell wall inhibitors and temperature.
Strains were assessed for in vitro phenotypes on YPD medium supplemented with cell wall stressors as well as for any temperature sensitivity (Fig. 4 and 5). The chs6Δ strain grew slower than H99 at 40°C, even though it was similar in growth to wild-type at 37°C. The chs5Δ, chs7Δ, chs8Δ, and csr1Δ strains, which grew similarly to wild-type at 37°C, showed only a slight growth defect at 40°C. The chs1Δ, chs2Δ, chs4Δ, and csr3Δ strains showed no growth defects when subjected to any of the cell wall stressors or high temperature. Interestingly, growth defects were not detected for any of the chs or csr strains when plated on YPD supplemented with calcofluor white (data not shown), which binds to chitin and causes cell wall stress.
FIG. 4.
Analysis of H99 and chitin synthase deletion strains for in vitro sensitivity to cell wall stressors. Strains were grown in YPD medium to mid-log phase, serially diluted 10-fold (starting with 1 × 105 cells in the left column of each panel), and plated on YPD solid medium and YPD solid medium supplemented with various cell wall stressors. Strains plated on YPD medium were also tested for high-temperature sensitivity. Time of incubation is indicated at the far left. Strains are indicated at the left of the panels. Incubation conditions are indicated at the top of the panels. All plates containing cell wall stressors were incubated at 30°C.
FIG. 5.
Analysis of H99 and chitin synthase regulator deletion strains for in vitro sensitivity to cell wall stressors. The experiment was done as described for Fig. 4.
Dramatic phenotypes were shared between the chs3Δ and csr2Δ strains (Fig. 4 and 5). For instance, both chs3Δ and csr2Δ were sensitive to YPD supplemented with 0.03% SDS or with 0.5 mg/ml caffeine or with 0.5% Congo red. SDS has been used extensively to test cell wall integrity, caffeine has been used to test signal transduction and cell integrity phenotypes in S. cerevisiae, and Congo red is a dye that inhibits microfibril assembly of the cell wall (55). Some differences were most notable after 7 days of growth, including a slightly slower growth at 30°C and the caffeine sensitivity. Furthermore, chs3Δ and csr2Δ were temperature sensitive at 40°C (Fig. 4 and 5). Interestingly, although the mutants initially grew at 37°C, their growth slowed with longer incubation. After 2 days at 37°C, only minimal growth for the chs3Δ strain and no growth for the csr2Δ strain was seen when the cells were shifted to 30°C and incubated for an additional 7 days (data not shown), indicating that the cells were inviable following prolonged incubation at 37°C. The chs3Δ and csr2Δ strains were unable to survive growth in liquid YPD at 37°C with shaking (data not shown).
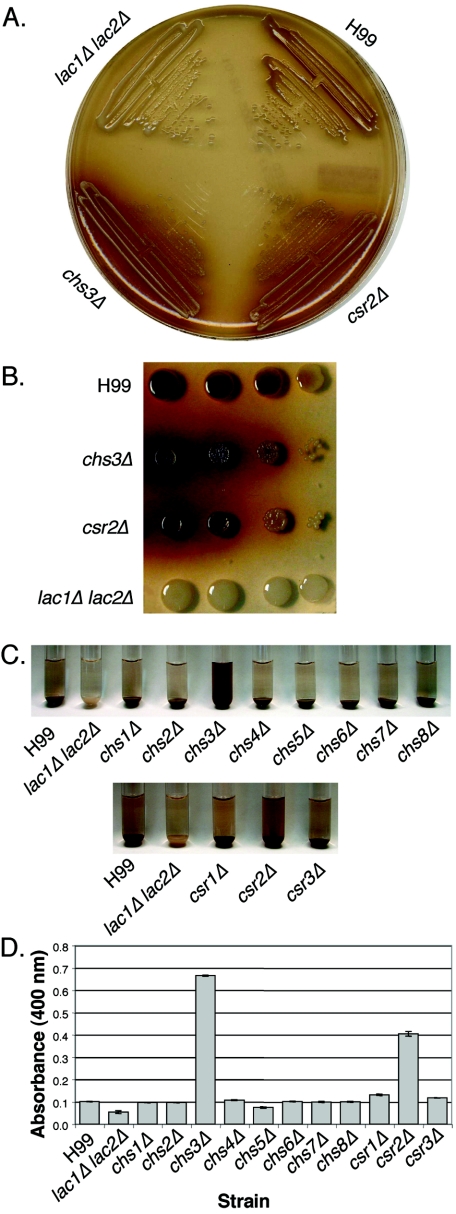
Melanin production is altered in chs3Δ and csr2Δ strains.
The mutant strains were streaked onto bird seed agar plates, which contain phenolic compounds that are oxidized by the C. neoformans laccase enzymes to produce melanin. Melanin is localized to the cell wall (58) and gives the colonies a brown appearance. However, both chs3Δ and csr2Δ cells appear to produce melanin but seem to have lost the ability to retain the substance in the cell wall, as is apparent by the halo of pigment surrounding the colonies (Fig. 6A). This phenotype is similar to a Cryptococcus gattii mutant with an insertion in the CHS3 gene (56). All other deletion strains appeared to have melanin production and retention properties similar to the wild type.
FIG. 6.
Melanin production of H99, chitin synthase deletion strains, and chitin synthase regulator deletion strains. A lac1Δ lac2Δ strain was used as a negative control. A. Cells from chs3Δ and csr2Δ strains were streaked out onto bird seed agar medium and allowed to grow at 30°C for 3 days. B. Serial dilutions of cells spotted onto L-DOPA medium. The spots on the left represent 104 cells, and 10-fold serial dilutions were plated. The strain is indicated to the left of the plate. C. Cells were resuspended to identical optical density in glucose-free asparagine medium containing L-DOPA and allowed to shake at 30°C. Cells were centrifuged to visualize the secreted pigment and the color of the pellets. Pictures were taken at 24 h. The strain name is indicated at the bottom of the panel. D. Quantitation of the OD400 of the supernatants from the cultures as shown in panel C.
The “leaky” phenotype for chs3Δ and csr2Δ was also apparent when cells were resuspended in glucose-free asparagine medium containing another laccase substrate, L-DOPA (Fig. 6C). The supernatant was darker for these mutant strains than for the wild type (Fig. 6D).
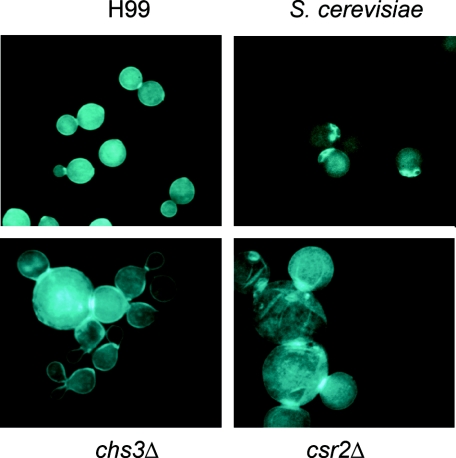
The morphology of chs3Δ and csr2Δ strains is altered.
Strains were grown in YPD medium at 30°C and stained with calcofluor white, a fluorescent brightener that binds to chitin, and examined using UV microscopy (Fig. 7). Unlike S. cerevisiae, the wild-type H99 appears to stain brightly over the entire surface of the cells, with some increased staining at the bud neck, but bud scars cannot be distinguished. This may reflect the higher chitin content and/or different distribution of chitin in C. neoformans. Once again, chs3Δ and csr2Δ strains both showed dramatic differences from the wild type. First of all, cells of these strains were much greater in size; many were two to three times larger than the wild type, and a greater proportion of the csr2Δ cells were large compared to chs3Δ cells. The cells of these strains were also not as uniform in shape as the wild type and seemed to have large scar-like regions across the cell. The budding cells also appeared to have difficulty in completion of cleavage, as displayed by the large groups of cells with greatly enhanced staining of the septae between budding cells. In general, cell staining was not as uniform for the chs3Δ and csr2Δ strains as in the wild type. Staining patterns of all other deletion strains were similar to the wild type.
FIG. 7.
Calcofluor white staining of S. cerevisiae, C. neoformans H99, and deletion strains. Cells were grown in YPD at 30°C, formaldehyde fixed, and stained with calcofluor white, a dye that binds to chitin. The strain is indicated at the left of each photo. All photos are at the same magnification (1,000×).
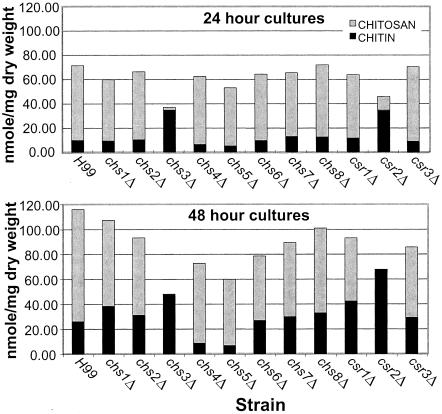
The chs3Δ and csr2Δ deletion strains have reduced chitosan and increased chitin content.
In order to assess the role that each chitin synthase and putative chitin synthase regulator has on the levels of cellular chitin and chitosan, wild-type H99 as well as the chs and csr deletion strains were assayed for chitin and chitosan content at 24 and 48 h of growth in liquid YPD (Fig. 8). chs3Δ and csr2Δ once again had similar phenotypes, with both strains containing greatly reduced amounts of chitosan at 24 h and no detectable chitosan at 48 h. However, chitin levels were approximately two- to threefold higher than those of H99. Further, chs4Δ and chs5Δ strains had 50% less chitin than the wild type. All other chsΔ strains and csrΔ strains had chitin and chitosan levels similar to H99.
FIG. 8.
Chitin and chitosan content of Cryptococcus neoformans H99 and mutant strains. Measurements were taken at 24-h (top panel) and 48-h (bottom panel) time points. The strains are indicated at the bottom of each graph, and the time of growth is indicated at the top of each graph. Chitin and chitosan content are indicated at the left of each panel.
DISCUSSION
We have found that vegetatively growing C. neoformans has significant levels of chitin in the cell wall, and the majority of the chitin is deacetylated to chitosan. This result differs from what has been observed for most other fungi, although it is unclear whether the detection methods used in previous studies would have specifically distinguished chitin from chitosan or if chitosan is actually absent in these fungi. However, it is clear that C. albicans and S. cerevisiae contain no chitosan during vegetative growth. In S. cerevisiae, chitosan has only been detected in the ascospore (11).
Results indicate that the composition of the cell wall of C. neoformans is highly dependent on phase of growth. When cells are actively dividing, there is little change in the amount of chitin and chitosan in the cell wall, but both increase as cells progress from late-log into stationary phase. This validates the idea that the cell wall is a dynamic structure that is changing over time. It is possible that this phenomenon could impact results from related studies and should be considered during experimental design.
C. neoformans has eight chitin synthases, which is similar to other fungi that elaborate complex structures such as Aspergillus spp., Ustilago maydis (35), or the zygomycete Phycomyces blakesleeanus (34). This could reflect a requirement for different synthases at different developmental stages. C. neoformans not only grows as a budding yeast but also forms conjugation tubes, hyphae with clamp connections, basidia, and basidiospores (1). This is readily testable using the molecular reagents that have been generated as part of this study. By measuring mRNA levels in mid-log-phase yeast, it is clear that different chitin synthases are likely to be important. We observed relatively abundant transcripts for CHS2, CHS3, CHS5, and CHS8 and did not detect transcript for CHS7. It is quite possible that at alternate developmental stages, at different growth phases, or growth in various culture conditions or host environments, each chitin synthase could play a different role.
Class V chitin synthases with myosin motor-like domains have been found in filamentous fungi, such as Exophiala (Wangiella) and Aspergillus. It has been shown that the myosin domain is important for polarized filamentous growth via interaction with the actin cytoskeleton (51). Further studies of this class V chitin synthase of C. neoformans might shed light on its function, perhaps for hyphal growth during sexual reproduction. However, it is clear that CHS5 also plays a role in growth of budding yeast since it is transcribed, and deletion of this gene causes slight temperature sensitivity.
One very interesting phenotype that was shared by chs3Δ and csr2Δ strains was “leaky” melanin. Both laccase, the enzyme that is required for melanin production, and melanin are found in the cell wall (15, 33, 58). Although the chs3Δ and csr2Δ cells clearly retained some melanin, it was also evident that pigment was secreted into the surrounding medium. There are three obvious possibilities for this phenomenon. First, the cell wall composition, perhaps the lack of chitosan, reduces the wall's ability to bind melanin, so that much of it leaks into the medium. Another possibility is that laccase is mislocalized due to wall defects and no longer deposits the melanin precursors in the same location. Finally, the black pigment secreted into the medium may not be melanin, although it is produced from two distinct substrates (bird seed agar and L-DOPA); therefore, it will be important to characterize the secreted substance.
Chs3 is clearly the most important chitin synthase for vegetative growth. It is the most abundantly transcribed, and the phenotypes associated with deletion of CHS3 were the most severe. The phenotypes of the csr2Δ mutant were strikingly similar to chs3Δ phenotypes. These parallel phenotypes suggest that Chs3 and Csr2 are complexed in a similar manner as Chs3p and Skt5p in S. cerevisiae (14, 39, 52). It has been shown that Chs3p requires association with Skt5p in order to synthesize chitin. Because the magnitude of the parallel phenotypes were so similar between csr2Δ and chs3Δ, we hypothesize that Csr2 is the specific regulator of Chs3. C. neoformans contains two additional Skt5p homologs, Csr1 and Csr3, but the specific partnerships of these two putative chitin synthase regulators remain to be discovered.
The most unexpected phenotype that we observed for both the chs3Δ and csr2Δ mutants was the dramatic reduction (Fig. 8) of chitosan. This observation leads to the model that the chitin deacetylases are not randomly distributed throughout the cell wall and available to deacetylate chitin made by other chitin synthases. The restriction of chitosan being predominantly the product of deacetylating chitin synthesized by the Chs3-Csr2 complex suggests direct interaction with chitin deacetylase(s). This would also be an efficient mechanism for making chitosan, deacetylating chitin as it is extruded through the plasma membrane before individual chitin chains bond together to form insoluble chitin fibers. Elevated chitin levels were also found for the chs3Δ and csr2Δ strains, further indicating, along with the increase of transcription of CHS5 and CHS7, that there is compensational synthesis by other chitin synthases. That there is additional chitin synthesis without apparent deacetylation also supports our hypothesis that the Chs3-Csr2 complex specifically recruits chitin deacetylases.
We have presented evidence for eight chitin synthases being encoded by C. neoformans genes. There is, however, an “Achilles' heel” to the system: Chs3 and Csr2. Deletion of either gene leads to cell death at 37°C, which strongly suggests that targeting either protein or the complex to disrupt function will also be lethal at mammalian host body temperature. However, it is still unclear if the cell wall defect is caused by the lack of conversion of chitin to chitosan or is by the lack of the specific chitin synthesized by the Chs3-Csr2 complex. We are pursuing the deletion of the four chitin deacetylase genes of C. neoformans to provide an answer to whether targeting chitin deacetylase function could be an equivalent point of weakness in its cell wall biosynthesis.
Supplementary Material
Acknowledgments
We thank Tricia A. Missall, Carlos E. Soto, and Samantha N. Piper for technical support and Lorina Baker for critical reading of the manuscript. We also thank the C. neoformans H99 sequencing project, Duke Center for Genome Technology (http://cgt.genetics.duke.edu), the Broad Institute (www.broad.mit.edu/annotation/fungi/cryptococcus_neoformans), and the Genome Sequence Centre, BC Cancer Research Centre (http://www.bcgsc.bc.ca/), as well as the C. neoformans serotype D Genome Project, Stanford Genome Technology Center, funded by the NIAID/NIH under cooperative agreement U01 AI47087, and The Institute for Genomic Research, funded by the NIAID/NIH under cooperative agreement U01 AI48594. We thank the C. neoformans cDNA sequencing project at the University of Oklahoma (http://www.genome.ou.edu/cneo.html), funded by NIH/NIAID AI147079.
This work was supported by NIH/NIAID grant RO1-AI50184 to J.K.L. and R01AI025780 to S.M.L.
REFERENCES
- 1.Alspaugh, J. A., R. C. Davidson, and J. Heitman. 2000. Morphogenesis of Cryptococcus neoformans. Contrib. Microbiol. 5:217-238. [DOI] [PubMed] [Google Scholar]
- 2.Araki, Y., and E. Ito. 1975. A pathway of chitosan formation in Mucor rouxii. Enzymatic deacetylation of chitin. Eur. J. Biochem. 55:71-78. [DOI] [PubMed] [Google Scholar]
- 3.Bartnicki-Garcia, S., and E. Lippman.1969. Fungal morphogenesis: cell wall construction in Mucor rouxii. Science 165:302-304. [DOI] [PubMed] [Google Scholar]
- 4.Bartnicki-Garcia, S., and W. J. Nickerson. 1962. Isolation, composition, and structure of cell walls of filamentous and yeast-like forms of Mucor rouxii. Biochim. Biophys. Acta 58:102-119. [DOI] [PubMed] [Google Scholar]
- 5.Bowen, A. R., J. L. Chen-Wu, M. Momany, R. Young, P. J. Szaniszlo, and P. W. Robbins. 1992. Classification of fungal chitin synthases. Proc. Natl. Acad. Sci. USA 89:519-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bulawa, C. E. 1992. CSD2, CSD3, and CSD4, genes required for chitin synthesis in Saccharomyces cerevisiae: the CSD2 gene product is related to chitin synthases and to developmentally regulated proteins in Rhizobium species and Xenopus laevis. Mol. Cell. Biol. 12:1764-1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6a.Bulik, D. A., M. Olczak, H. A. Lucero, B. C. Osmond, P. W. Robbins, and C. A. Specht. 2003. Chitin synthesis in Saccharomyces cerevisiae in response to supplementation of growth medium with glucosamine and cell wall stress. Eukaryot. Cell 2:886-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cabib, E., S. J. Silverman, and J. A. Shaw. 1992. Chitinase and chitin synthase 1: counterbalancing activities in cell separation of Saccharomyces cerevisiae. J. Gen. Microbiol. 138:97-102. [DOI] [PubMed] [Google Scholar]
- 8.Cabib, E., A. Sburlati, B. Bowers, and S. J. Silverman. 1989. Chitin synthase 1, an auxiliary enzyme for chitin synthesis in Saccharomyces cerevisiae. J. Cell Biol. 108:1665-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Casadevall, A., and J. R. Perfect. 1998. Cryptococcus neoformans. ASM Press, Washington, D.C.
- 10.Choi, W. J., B. Santos, A. Duran, and E. Cabib. 1994. Are yeast chitin synthases regulated at the transcriptional or the posttranslational level? Mol. Cell. Biol. 14:7685-7694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christodoulidou, A., V. Bouriotis, and G. Thireos. 1996. Two sporulation-specific chitin deacetylase-encoding genes are required for the ascospore wall rigidity of Saccharomyces cerevisiae. J. Biol. Chem. 271:31420-31425. [DOI] [PubMed] [Google Scholar]
- 12.Cohen, E. 1987. Chitin biochemistry: synthesis and inhibition. Annu. Rev. Entomol. 32:71-93. [Google Scholar]
- 13.Davidson, R. C., J. R. Blankenship, P. R. Kraus, M. de Jesus Berrios, C. M. Hull, C. D'Souza, P. Wang, and J. Heitman. 2002. A PCR-based strategy to generate integrative targeting alleles with large regions of homology. Microbiology 148:2607-2615. [DOI] [PubMed] [Google Scholar]
- 14.DeMarini, D. J., A. E. Adams, H. Fares, C. De Virgilio, G. Valle, J. S. Chuang, and J. R. Pringle. 1997. A septin-based hierarchy of proteins required for localized deposition of chitin in the Saccharomyces cerevisiae cell wall. J. Cell Biol. 139:75-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eisenman H. C., J. D. Nosanchuk, J. B. Webber, R. J. Emerson, T. A. Camesano, and A. Casadevall. 2005. Microstructure of cell wall-associated melanin in the human pathogenic fungus Cryptococcus neoformans. Biochemistry 44:3683-3693. [DOI] [PubMed] [Google Scholar]
- 16.Fromtling, R. A., H. J. Shadomy, and E. S. Jacobson. 1982. Decreased virulence in stable, acapsular mutants of Cryptococcus neoformans. Mycopathologia 79:23-29. [DOI] [PubMed] [Google Scholar]
- 17.Fujimura, H., and Y. Sakuma. 1993. Simplified isolation of chromosomal and plasmid DNA from yeasts. BioTechniques 14:538-540. [PubMed] [Google Scholar]
- 18.Garcera-Teruel, A., B. Xoconostle-Cazares, R. Rosas-Quijano, L. Ortiz, C. Leon-Ramirez, C. A. Specht, R. Sentandreu, and J. Ruiz-Herrera. 2004. Loss of virulence in Ustilago maydis by Umchs6 gene disruption. Res. Microbiol. 155:87-97. [DOI] [PubMed] [Google Scholar]
- 19.Gerik, K. J., M. J. Donlin, C. E. Soto, A. M. Banks, I. R. Banks, M. A. Maligi, C. P. Selitrennikoff, and J. K. Lodge. 2005. Cell wall integrity is dependent on the PKC1 signal transduction pathway in Cryptococcus neoformans. Mol. Microbiol. 58:393-408. [DOI] [PubMed] [Google Scholar]
- 20.Hua, J., J. D. Meyer, and J. K. Lodge. 2000. Development of positive selectable markers for the fungal pathogen Cryptococcus neoformans. Clin. Diagn. Lab. Immunol. 7:125-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Igual, J. C., A. L. Johnson, and L. H. Johnston. 1996. Coordinated regulation of gene expression by the cell cycle transcription factor Swi4 and the protein kinase C MAP kinase pathway for yeast cell integrity. EMBO J. 15:5001-5013. [PMC free article] [PubMed] [Google Scholar]
- 22.Kafetzopoulos, D., A. Martinou, and V. Bouriotis. 1993. Bioconversion of chitin to chitosan: purification and characterization of chitin deacetylase from Mucor rouxii. Proc. Natl. Acad. Sci. USA 90:2564-2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klis, F. M. 1994. Review: cell wall assembly in yeast. Yeast 10:851-869. [DOI] [PubMed] [Google Scholar]
- 24.Kollar R., E. Petrakova, G. Ashwell, P. W. Robbins, and E. Cabib. 1995. Architecture of the yeast cell wall. The linkage between chitin and beta(1->3)-glucan. J. Biol. Chem. 270:1170-1178. [DOI] [PubMed] [Google Scholar]
- 25.Kollar R., B. B. Reinhold, E. Petrakova, et al. 1997. Architecture of the yeast cell wall. Beta(1->6)-glucan interconnects mannoprotein, beta(1->)3-glucan, and chitin. J. Biol. Chem. 272:17762-17775. [DOI] [PubMed] [Google Scholar]
- 26.Krishnarao, T. V., and J. N. Galgiani. 1997. Comparison of the in vitro activities of the echinocandin LY303366, the pneumocandin MK-0991, and fluconazole against Candida species and Cryptococcus neoformans. Antimicrob. Agents Chemother. 41:1957-1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu, H., S. Kauffman, J. M. Becker, and P. J. Szaniszlo. 2004. Wangiella (Exophiala) dermatitidis WdChs5p, a class V chitin synthase, is essential for sustained cell growth at temperature of infection. Eukaryot. Cell 3:40-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loftus, B. J., E. Fung, P. Roncaglia, et al. 2005. The genome of the basidiomycetous yeast and human pathogen Cryptococcus neoformans. Science 307:1321-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Madrid, M. P., A. Di Pietro, and M. I. Roncero. 2003. Class V chitin synthase determines pathogenesis in the vascular wilt fungus Fusarium oxysporum and mediates resistance to plant defence compounds. Mol. Microbiol. 47:257-266. [DOI] [PubMed] [Google Scholar]
- 30.Maligie, M. A., and C. P. Selitrennikoff. 2005. Cryptococcus neoformans resistance to echinocandins: (1,3)beta-glucan synthase activity is sensitive to echinocandins. Antimicrob. Agents Chemother. 49:2851-2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McDade, H. C., and G. M. Cox. 2001. A new dominant selectable marker for use in Cryptococcus neoformans. Med. Mycol. 39:151-154. [DOI] [PubMed] [Google Scholar]
- 32.Minke R., and J. Blackwell. 1978. The structure of alpha-chitin. J. Mol. Biol. 120:167-181. [DOI] [PubMed] [Google Scholar]
- 33.Missall, T. A., J. M. Moran, J. A. Corbett, and J. K. Lodge. 2005. Distinct stress responses of two functional laccases in Cryptococcus neoformans are revealed in the absence of the thiol-specific antioxidant Tsa1. Eukaryot. Cell 4:202-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miyazaki, A., and T. Ootaki. 1997. Multiple genes for chitin synthase in the zygomycete fungus Phycomyces blakesleeanus. J. Gen. Appl. Microbiol. 43:333-340. [DOI] [PubMed] [Google Scholar]
- 35.Munro, C. A., and N. A. R. Gow. 2001. Chitin synthesis in human pathogenic fungi. Med. Mycol. 39:41-53. [PubMed] [Google Scholar]
- 36.Munro, C. A., K. Winter, A. Buchan, K. Henry, J. M. Becker, A. J. Brown, C. E. Bulawa, and N. A. Gow. 2001. Chs1 of Candida albicans is an essential chitin synthase required for synthesis of the septum and for cell integrity. Mol. Microbiol. 39:1414-1426. [DOI] [PubMed] [Google Scholar]
- 37.Nagahashi, S., M. Sudoh, N. Ono, R. Sawada, E. Yamaguchi, Y. Uchida, T. Mio, M. Takagi, M. Arisawa, and H. Yamada-Okabe. 1995. Characterization of chitin synthase 2 of Saccharomyces cerevisiae. Implication of two highly conserved domains as possible catalytic sites. J. Biol. Chem. 270:13961-13967. [DOI] [PubMed] [Google Scholar]
- 38.Nelson, R. T., B. A. Pryor, and J. K. Lodge. 2003. Sequence length required for homologous recombination in Cryptococcus neoformans. Fungal Genet. Biol. 38:1-9. [DOI] [PubMed] [Google Scholar]
- 39.Ono, N., T. Yabe, M. Sudoh, T. Nakajima, T. Yamada-Okabe, M. Arisawa, and H. Yamada-Okabe. 2000. The yeast Chs4 protein stimulates the trypsin-sensitive activity of chitin synthase 3 through an apparent protein-protein interaction. Microbiology 146:385-391. [DOI] [PubMed] [Google Scholar]
- 40.Orlowski, M. 1991. Mucor dimorphism. Microbiol. Rev. 55:234-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pringle, J. R., R. A. Preston, A. E. Adams, T. Stearns, D. G. Drubin, B. K. Haarer, and E. W. Jones. 1989. Fluorescence microscopy methods for yeast. Methods Cell Biol. 31:357-435. [DOI] [PubMed] [Google Scholar]
- 42.Pukkila-Worley, R., Q. D. Gerrald, P. R. Kraus, M. J. Boily, M. J. Davis, S. S. Giles, G. M. Cox, J. Heitman, and J. A. Alspaugh. 2005. Transcriptional network of multiple capsule and melanin genes governed by the Cryptococcus neoformans cyclic AMP cascade. Eukaryot. Cell 4:190-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reese, A. J., and T. L. Doering. 2003. Cell wall alpha-1,3-glucan is required to anchor the Cryptococcus neoformans capsule. Mol. Microbiol. 50:1401-1409. [DOI] [PubMed] [Google Scholar]
- 44.Richmond, T. 2000. Higher plant cellulose synthases. Genome Biol. 1:3001-3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ruiz-Herrera, J., J. M. Gonzalez-Prieto, and R. Ruiz-Medrano. 2002. Evolution and phylogenetic relationships of chitin synthases from yeasts and fungi. FEMS Yeast Res. 1:247-256. [DOI] [PubMed] [Google Scholar]
- 46.Sambrook, J., and D. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 47.Sanz, M., L. Carrano, C. Jimenez, G. Candiani, J. A. Trilla, A. Duran, and C. Roncero. 2005. Candida albicans strains deficient in CHS7, a key regulator of chitin synthase III, exhibit morphogenetic alterations and attenuated virulence. Microbiology 151:2623-2636. [DOI] [PubMed] [Google Scholar]
- 48.Shaw, J. A., P. C. Mol, B. Bowers, S. J. Silverman, M. H. Valdivieso, A. Duran, and E. Cabib. 1991. The function of chitin synthases 2 and 3 in the Saccharomyces cerevisiae cell cycle. J. Cell Biol. 114:111-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Soulie, M. C., A. Piffeteau, M. Choquer, M. Boccara, and A. Vidal-Cros. 2003. Disruption of Botrytis cinerea class I chitin synthase gene Bcchs1 results in cell wall weakening and reduced virulence. Fungal Genet. Biol. 40:38-46. [DOI] [PubMed] [Google Scholar]
- 50.Specht, C. A., Y. Liu, P. W. Robbins, C. E. Bulawa, N. Iartchouk, K. R. Winter, P. J. Riggle, J. C. Rhodes, C. L. Dodge, D. W. Culp, and P. T. Borgia. 1996. The chsD and chsE genes of Aspergillus nidulans and their roles in chitin synthesis. Fungal Genet. Biol. 20:153-167. [DOI] [PubMed] [Google Scholar]
- 51.Takeshita, N., A. Ohta, and H. Horiuchi. 2005. CsmA, a class V chitin synthase with a myosin motor-like domain, is localized through direct interaction with the actin cytoskeleton in Aspergillus nidulans. Mol. Biol. Cell 16:1961-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Trilla, J. A., T. Cos, A. Duran, and C. Roncero. 1997. Characterization of CHS4 (CAL2), a gene of Saccharomyces cerevisiae involved in chitin biosynthesis and allelic to SKT5 and CSD4. Yeast 13:795-807. [DOI] [PubMed] [Google Scholar]
- 53.Toffaletti, D. L., T. H. Rude, S. A. Johnston, D. T. Durack, and J. R. Perfect. 1993. Gene transfer in Cryptococcus neoformans by use of biolistic delivery of DNA. J. Bacteriol. 175:1405-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Valdivieso, M. H., P. C. Mol, J. A. Shaw, E. Cabib, and A. Duran. 1991. CAL1, a gene required for activity of chitin synthase 3 in Saccharomyces cerevisiae. J. Cell Biol. 114:101-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vannini, G. L., F. Poli, A. Donini, and S. Pancaldi. 1983. Effects of Congo red on wall synthesis and morphogenesis in Saccharomyces cerevisiae. Plant Sci. Lett. 31:9-17. [Google Scholar]
- 56.Walton, F. J., A. Idnurm, and J. Heitman. 2005. Novel gene functions required for melanization of the human pathogen Cryptococcus neoformans. Mol. Microbiol. 57:1381-1396. [DOI] [PubMed] [Google Scholar]
- 57.Wang, Y., P. Aisen, and A. Casadevall. 1995. Cryptococcus neoformans melanin and virulence: mechanism of action. Infect. Immun. 63:3131-3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Williamson, P. R. 1994. Biochemical and molecular characterization of the diphenol oxidase of Cryptococcus neoformans: identification as a laccase. J. Bacteriol. 176:656-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yabe, T., T. Yamada-Okabe, T. Nakajima, M. Sudoh, M. Arisawa, and H. Yamada-Okabe. 1998. Mutational analysis of chitin synthase 2 of Saccharomyces cerevisiae. Identification of additional amino acid residues involved in its catalytic activity. Eur. J. Biochem. 258:941-947. [DOI] [PubMed] [Google Scholar]
- 60.Zhang, F. L., and P. J. Casey. 1996. Protein prenylation: molecular mechanisms and functional consequences. Annu. Rev. Biochem. 65:241-269. [DOI] [PubMed] [Google Scholar]
- 61.Ziman, M., J. S. Chuang, and R. W. Schekman. 1996. Chs1p and Chs3p, two proteins involved in chitin synthesis, populate a compartment of the Saccharomyces cerevisiae endocytic pathway. Mol. Biol. Cell 7:1909-1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.