Abstract
The yeast Mcm1 protein is a member of the MADS box family of transcription factors that interacts with several cofactors to differentially regulate genes involved in cell-type determination, mating, cell cycle control and arginine metabolism. Residues 18 to 96 of the protein, which form the core DNA-binding domain of Mcm1, are sufficient to carry out many Mcm1-dependent functions. However, deletion of residues 2 to 17, which form the nonessential N-terminal (NT) arm, confers a salt-sensitive phenotype, suggesting that the NT arm is required for the activation of salt response genes. We used a strategy that combined information from the mutational analysis of the Mcm1-binding site with microarray expression data under salt stress conditions to identify a new subset of Mcm1-regulated genes. Northern blot analysis showed that the transcript levels of several genes encoding associated with the cell wall, especially YGP1, decrease significantly upon deletion of the Mcm1 NT arm. Deletion of the Mcm1 NT arm results in a calcofluor white-sensitive phenotype, which is often associated with defects in transcription of cell wall genes. In addition, the deletion makes cells sensitive to CaCl2 and alkaline pH. We found that the defect caused by removal of the NT arm is not due to changes in Mcm1 protein level, stability, DNA-binding affinity, or DNA bending. This suggests that residues 2 to 17 of Mcm1 may be involved in recruiting a cofactor to the promoters of these genes to activate transcription.
The MADS box family of proteins contains a highly conserved DNA-binding and dimerization domain, named after the founding members, Mcm1, Agamous, Deficiens, and serum response factor (SRF) (34, 36, 46, 51). MADS box proteins have been identified in most eukaryotic organisms, and these proteins are involved in the transcriptional regulation of a wide array of cellular and developmental processes (3, 26, 34, 46, 49). Many MADS box proteins are able to bind DNA as dimers with high affinity on their own in vitro. However, these proteins often interact with other DNA-binding cofactors to regulate transcription in vivo. Their interaction with different cofactors helps specify the DNA target sites bound by these proteins and thus the genes that they regulate. The conserved 80-amino-acid MADS box, DNA-binding, and dimerization domain is often sufficient for the interaction with these cofactors (6, 10, 37, 45). However, many MADS box proteins also appear to use regions outside of the conserved domain to interact with cofactors. For example, the 28-amino-acid-long myocyte enhancer factor (MEF) domain of MEF2 proteins in animals is found immediately C terminal to the MADS box domain and enables these proteins to heterodimerize with other members of the MEF2 family (32). In plants, the I region and K box outside the MADS box are involved in homo- and heterodimerization (11, 16, 39).
Mcm1 is a MADS box transcriptional regulatory protein in the yeast Saccharomyces cerevisiae. The 286-amino-acid-long Mcm1 protein binds to a 10-bp palindromic sequence, frequently called the CArG box [CC(A/T)6GG] (31). In combination with different cofactors, Mcm1 bound to CArG sites activates or represses the transcription of a variety of genes, including those involved in cell type determination, arginine metabolism, pheromone response, mating, and cell cycle progression (20, 27, 29, 36). It is also involved in Ty-mediated transcription and minichromosome maintenance (36, 52). Mcm1 was recently found to bind to yeast autonomous replicating sequences (designated ARS) and initiate replication (8, 9). Mcm1 is an essential protein in yeast, and the conserved MADS box domain (residues 18 to 96; designated Mcm1-18-96) is sufficient to maintain viability, DNA binding, dimerization, interaction with several cofactors, and transcriptional regulation of many of its target genes (6, 10, 28). However, deletion of the nonessential N-terminal extension (residues 2 to 17) of Mcm1 causes cells to be sensitive to high-salt levels, indicating that regions outside the conserved MADS box are important for this function (24).
In this paper, we investigate the role of the Mcm1 N-terminal (NT) arm in regulating cellular osmotolerance. We have found that deletion of the NT arm confers strong sensitivity to CaCl2. Deletion of the arm also confers sensitivity to cell wall-disrupting agents like calcofluor white (CFW) and alkaline pH. Importantly, we show here that the NT arm of Mcm1 is required for the proper regulation of a subset of genes that encode components of the cell wall, such as YGP1 (for yeast glycoprotein 1). Our results suggest that the NT arm of Mcm1 may be needed for interaction with another cofactor that is required for the proper regulation of these cell wall genes.
MATERIALS AND METHODS
Plasmids and strains.
Yeast strain JM01 (MATα leu2-3,112 trp1-1 ura3 his3-11,15 ade2-1 can1-100 mcm1::LEU2/pSL1574-CEN/ARS MCM1 URA3) was used for all the in vivo assays (28). Plasmids with different mcm1 mutations were transformed into strain JM01 and selected on synthetic medium lacking histidine. Colonies that had lost the wild-type MCM1 on plasmid pSL1574 were selected by growth on 5-fluoroorotic acid plates. Mutations of the putative Mcm1 and Rlm1 sites in the YGP1-lacZ reporters were constructed by site-directed mutagenesis using the Stratagene QuikChange kit. Plasmids used in this study are listed in Table 1. Sequences of the oligonucleotides used for creating probes, construction of plasmids, and site-directed mutagenesis are available on request.
TABLE 1.
Plasmids used in this study
| Plasmid | Description | Reference or source |
|---|---|---|
| pJM231 | MCM1 CEN HIS3 | 1 |
| pDA126 | mcm1-Δ2-17 CEN HIS3 | This study |
| pDA135 | C-terminally-V5 tagged MCM1 CEN HIS3 | This study |
| pDA136 | C-terminally-V5 tagged mcm1-Δ2-17 CEN HIS3 | This study |
| pDA135-T35A | C-terminally-V5 tagged mcm1-T35A CEN HIS3 | This study |
| pDA181 | 700 bp of YGP1 promoter lacZ reporter | This study |
| pDA181-r1 | pDA181 with mutation in site R1 | This study |
| pDA181-m2 | pDA181 with mutation in site M2 | This study |
| pDA181-m3 | pDA181 with mutation in site M3 | This study |
| pDA181-m4 | pDA181 with mutation in site M4 | This study |
| pDA181-r1m2 | pDA181 with mutation in sites R1, M2 | This study |
| pDA181-m2m3 | pDA181 with mutation in sites M2, M3 | This study |
| pDA181-m1m2m3 | pDA181 with mutation in sites M1, M2, M3 | This study |
| pDA181-r1m2m3 | pDA181 with mutation in sites R1, M2, M3 | This study |
| pDA181-m1m2m3m4 | pDA181 with mutation in sites M1, M2, M3, M4 | This study |
| pDA181-r1m1m2m3 | pDA181 with mutation in sites R1, M1, M2, M3 | This study |
| pDA192 | R1M2M3 site cloned into pGD579 | This study |
| pTBA22 | Mcm1-18-96 fused to maltose-binding protein | 2 |
| pTBA25 | Mcm1-1-96 fused to maltose-binding protein | 2 |
Biological assays.
JM01 strains transformed with wild-type or mutant mcm1 on HIS3 plasmids were grown overnight in liquid SD-His-Leu medium (28). The concentration of the cells in each culture was normalized after their optical density at 600 nm was measured. Threefold serial dilutions (each, 3 μl) of the normalized strains were spotted onto yeast extract-peptone-dextrose (YEPD) medium supplemented with the indicated concentrations of calcofluor white, KCl, sorbitol, dithiothreitol (DTT), and CaCl2. The pH of alkaline YEPD medium was adjusted to 8.5 with NaOH before being autoclaved. The β-galactosidase activities of YGP1 promoter-lacZ fusion reporters were determined in strain JM01 cotransformed with wild-type or mutant derivatives of pJM231 (MCM1 HIS3 CEN) and the different lacZ reporter plasmids (2μ URA3). Cells were grown to log phase in minimal medium lacking leucine, histidine, and uracil; liquid β-galactosidase assays were performed as described previously (20).
Protein purification.
The Mcm1 proteins used in the in vitro DNA-binding and DNA-bending studies were purified from Escherichia coli BL21 cells transformed with pTBA25 or pTBA22 or maltose-binding protein fusion expression vectors with sequences encoding Mcm1 residues 1 to 96 and 18 to 96, respectively. The proteins were purified from 50-ml cultures as described previously (2). Protein concentrations were determined by Bradford assays, normalized, and verified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
Northern blot analysis.
Yeast strain JM01 was transformed with wild-type and mutant derivatives of pJM231 (MCM1 HIS3 CEN) or pAB76 (MCM1 HIS3 2μ), and cultures were grown at 30°C in SD-His-Leu medium to mid-log phase (optical density at 600 nm, ∼0.6). The cells were split into two aliquots. One half was resuspended in dropout medium supplemented with 1.4 M NaCl and grown at 30°C for 1.5 h, while the other half was harvested after growth in medium without a high salt level. Total RNA was extracted from both samples by hot acid-phenol extraction (5). RNA was separated by molecular weight on a 1.5% agarose gel containing formaldehyde and transferred to a Nytran membrane (Schleicher and Schuell) by capillary transfer. The membrane was prehybridized in a 10-ml hybridization solution (50% formamide, 5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 50 mM NaPO4 [pH 7.0], 1× Denhardt's reagent, 0.1% SDS, 100-μg/ml sheared single-stranded salmon sperm DNA). DNA probes to open reading frames (ORFs) were generated by PCR and labeled with [α-32P]dCTP by random priming (MegaPrime DNA Labeling Kit, Amersham). After overnight hybridization to labeled DNA probes and washing with 2× SSC and 0.1% SDS at 42°C, specific mRNAs were detected by exposure to a phosphorscreen and imaging on a Molecular Dynamics phosphorimager.
Western blot analysis.
Cultures of yeast strain JM01 transformed with C-terminal V5 epitope tagged full-length MCM1 and mcm1-Δ2-17 were grown to mid-log phase in SD-His-Leu medium. Proteins were extracted by glass bead lysis, and the protein concentration was determined with the Bio-Rad Protein Assay kit. Proteins were separated on 12% SDS-polyacrylamide gel, transferred to a nitrocellulose membrane, probed with mouse antibody directed against the V5 epitope (Invitrogen), and detected with sheep anti-mouse immunoglobulin G horseradish peroxidase (Amersham Pharmacia). The blot was stripped and probed with rabbit anti-Rpa1 antibodies (a gift from Steve Brill) and goat anti-rabbit immunoglobulin G horseradish peroxidase (Bio-Rad). Proteins were detected with the ECL Western blot detection kit (Amersham Pharmacia Biotech). Protein levels were quantitated with FluorChem software. To determine the rate of degradation of Mcm1, cells were grown to mid-log phase, and protein synthesis was arrested by the addition of cycloheximide to a final concentration of 20 μg/ml (14). Cells were harvested every 30 min for 4 h after the addition of cycloheximide. Protein extraction and detection were performed as described above.
Electrophoretic mobility shift assay (EMSA).
Oligonucleotides with P(PAL) or YGP1 M1M2 sites were end labeled with [γ-32P]ATP using polynucleotide kinase and purified using QIAGEN nucleotide removal columns. The labeled oligonucleotides were incubated at 90°C for 5 min with a threefold excess of the matching strand and allowed to anneal by being cooled slowly to 25°C. The purified Mcm1-1-96 and Mcm1-18-96 proteins were incubated for 1 h at room temperature with the labeled DNA probes in 20 mM Tris (pH 7.6), 5 mM MgCl2, 0.1 mM EDTA, 0.1% NP-40, 5% glycerol, 10-mg/ml bovine serum albumin, 10-mg/ml single-stranded DNA, and labeled oligonucleotide pairs (300 cpm/μl) in a total volume of 24 μl at room temperature for 1 h. All protein dilutions were made in 50 mM Tris (pH 7.6), 500 mM NaCl, 1 mM EDTA, 10 mM β-mercaptoethanol, and 1-mg/ml bovine serum albumin. Samples were loaded on a 6% polyacrylamide gel (run in 0.5× Tris-borate-EDTA buffer for 1 h at 200 V). Gels were dried after electrophoresis, exposed to a phosphor screen, and scanned with a Molecular Dynamics Phosphorimager.
Circular permutation assay.
A circularly permuted plasmid, pDA192, with a 80-bp fragment containing the R1, M2, and M3 sites from the YGP1 promoter cloned into the SacI-XbaI polylinker site of pGD579 (2) was digested with BamHI, NheI, HindIII, or EcoRI to yield probes with the R1-M2-M3 region located at different positions along the length of each DNA fragment. EMSAs were performed using position-permuted fragments and purified Mcm1-1-96 and Mcm1-18-96. The bend angle for each protein-DNA complex was calculated using the Thompson and Landy relationship (48).
ChIP assay.
Transformants of strain JM01 with wild-type and mutant derivatives of MCM1 with or without the V5 epitope tag were used for chromatin immunoprecipitation (ChIP) assays. Proteins bound to DNA were cross-linked by the addition of 1.35 ml of 37% formaldehyde to 50 ml of log-phase cultures and shaking at room temperature for 15 min. The cells were then harvested and lysed with glass beads. The crude lysate was sonicated to shear the protein-bound DNA to approximately 500-bp-long fragments. After cell debris was removed by centrifugation, the total chromatin was precleared by nutation with protein G-agarose for 1 h and centrifugation. The supernatant was then incubated with the anti-V5 antibody overnight at 4°C. Protein G-agarose was added to immunoprecipitate the sample. Reversal of formaldehyde cross-links was carried out as described previously (22). The DNA fragments precipitated with the V5-tagged Mcm1 proteins were detected by PCR using primers that annealed to the target promoter regions. Each PCR used three sets of primers: one set to amplify the positive control for Mcm1 binding, another for the test promoter, and a third for the negative control.
RESULTS
Identification of genes requiring the NT arm of Mcm1 for activation.
Deletion of the NT arm of Mcm1 (residues 2 to 17) makes cells osmosensitive (24). We therefore reasoned that the NT arm may be required for the transcriptional activation of genes required for tolerance to high salt levels. To identify potential Mcm1-activated salt response genes, we scanned the promoter regions of genes that showed increased expression under high-salt conditions for sites that are similar to the consensus sequence ccywwwxxrg (where y = T/C, w = A/T, x = any base, and r = A/G) that was derived from mutational analysis of an Mcm1-binding site (2, 50). Among the 570 salt-induced genes that were induced twofold or more in the microarray analysis, 163 genes had one or more putative Mcm1-binding sites in the 500-bp region upstream of their start codon (50). Among this group, we focused on genes that contained multiple Mcm1-binding sites or that were known to be involved in the osmotic stress response. To test if the NT arm of Mcm1 was required for the regulation of the selected salt-induced genes, we compared their expression by Northern blot analysis in strains containing wild-type Mcm1 or Mcm1-Δ2-17 grown under high- and low-salt conditions. Deletion of Mcm1 residues 2 to 17 resulted in a decrease in the mRNA levels of YGP1, PNC1, DAK1, and YHR087W (Fig. 1). The most dramatic effect was observed for YGP1, in which both the noninduced and salt-induced transcription decreased to <10% of wild-type levels. In addition to causing a decrease in expression for some genes, the deletion of the NT arm caused an increase in the expression of URA1 and SIT1. This suggests that the NT arm might be required for Mcm1 to function as a repressor at these loci. In contrast to YGP1, the mRNA level of CAR2, a salt-induced arginine catabolic gene that is activated by the Mcm1-Arg80 complex, was unaffected by deletion of the NT arm (Fig. 1) (4). Deletion of the NT arm also did not affect expression of CLB2, a target of the Mcm1-Fkh1 complex, or STE3, an α-specific gene activated by the Mcm1-α1 complex (Fig. 1 and data not shown) (21, 43). Taken together, these results indicate that Mcm1 residues 2 to 17 are required for the activation of a specific subset of genes that are induced by salt.
FIG. 1.
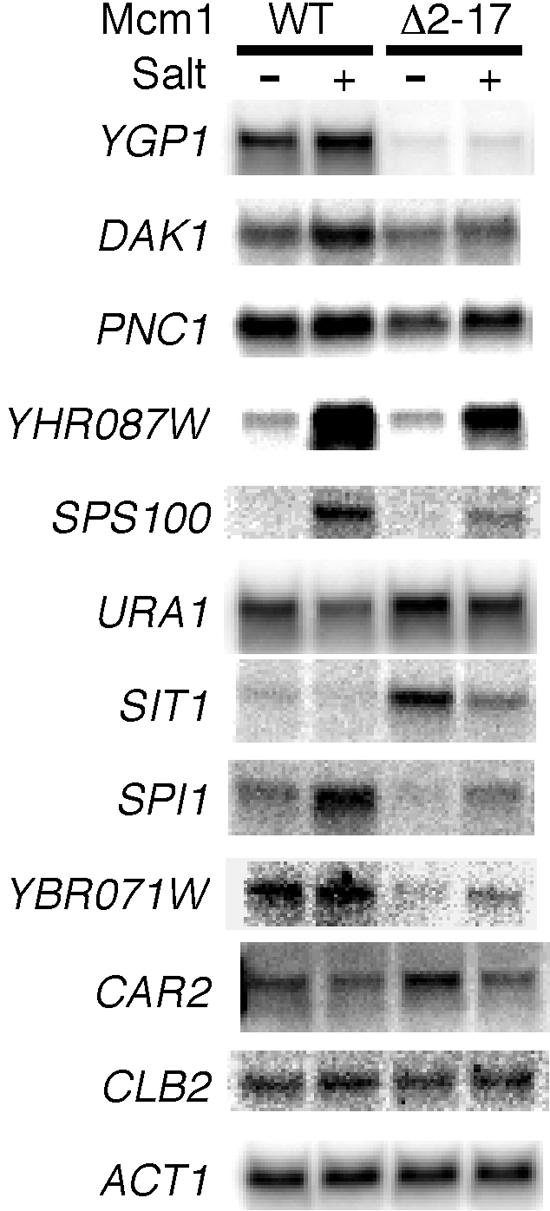
The N-terminal arm of Mcm1 is required for the regulation of a specific subset of salt-induced and cell wall genes. The mRNA levels of genes with putative Mcm1-binding sites were measured under low- and high-salt conditions by Northern blot analysis of strain JM01 transformed with either wild-type MCM1 (pDA135) or mcm1-Δ2-17 (pDA136). The CAR2 and CLB2 genes are regulated by Mcm1 but do not require the NT arm for activation. ACT1 is shown as a loading control.
Among the genes that were assayed, deletion of the NT arm had the greatest effect on YGP1, which encodes a protein that is secreted from protoplasts during cell wall regeneration. Ygp1 is highly glycosylated, and its synthesis is induced in response to glucose, nitrogen, phosphate starvation, and cell wall disruptions (12, 35). We therefore wanted to determine if other stress-induced genes that were coinduced with YGP1 under these conditions also required the Mcm1 NT arm for activation. YGP1 is one of 25 genes that are regulated by Rlm1, another MADS box transcription factor (18). Twenty of these genes had putative Mcm1 sites in the 1-kb region upstream of their start sites. However, Northern blot analysis showed that only the salt-induced expression of SPS100 was dependent on the presence of the Mcm1 NT arm (Fig. 1 and data not shown). Since YGP1 is induced in response to disruption of the cell wall, we also examined the promoters of genes that are induced in response to various cell wall stresses (25). Among the 80 genes categorized as the cell wall compensatory cluster, 38 had putative Mcm1-binding sites in their promoters. The expression of 15 of these genes was assayed under high-salt conditions in Mcm1 and Mcm1-Δ2-17 strains by Northern blotting. Among them, only SPI1 and YBR071W expression was dependent on the presence of the NT arm by Northern analysis (Fig. 1 and data not shown).
Deletion of the NT arm causes cell wall defects.
Deletion of the Mcm1 NT arm causes sensitivity to increased NaCl levels (24). Since the Mcm1 NT arm is required for the proper regulation of a subset of cell wall biosynthesis genes, we were interested in determining if this mutation affected sensitivity to other cell wall related stresses. While the mcm1-Δ2-17 strain was very sensitive to YEPD medium with 0.35 M CaCl2 (Fig. 2B), it was only mildly sensitive to 1.3 M KCl (Fig. 2C) and hardly sensitive to 0.4 M LiCl (data not shown).
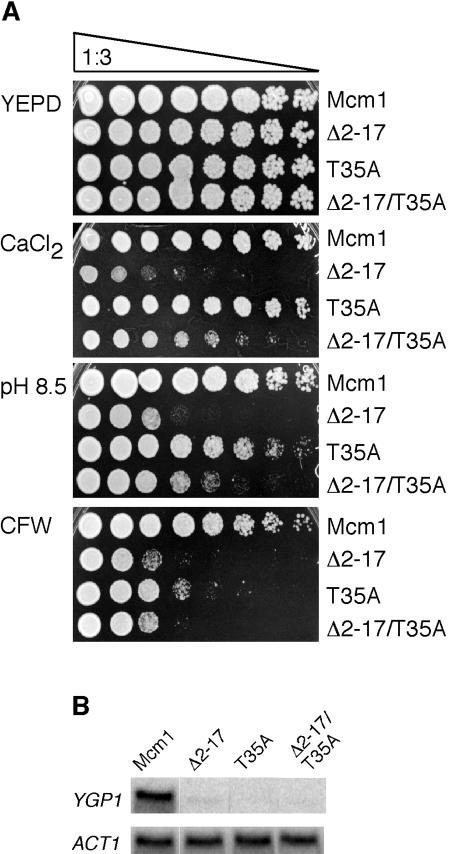
FIG. 2.
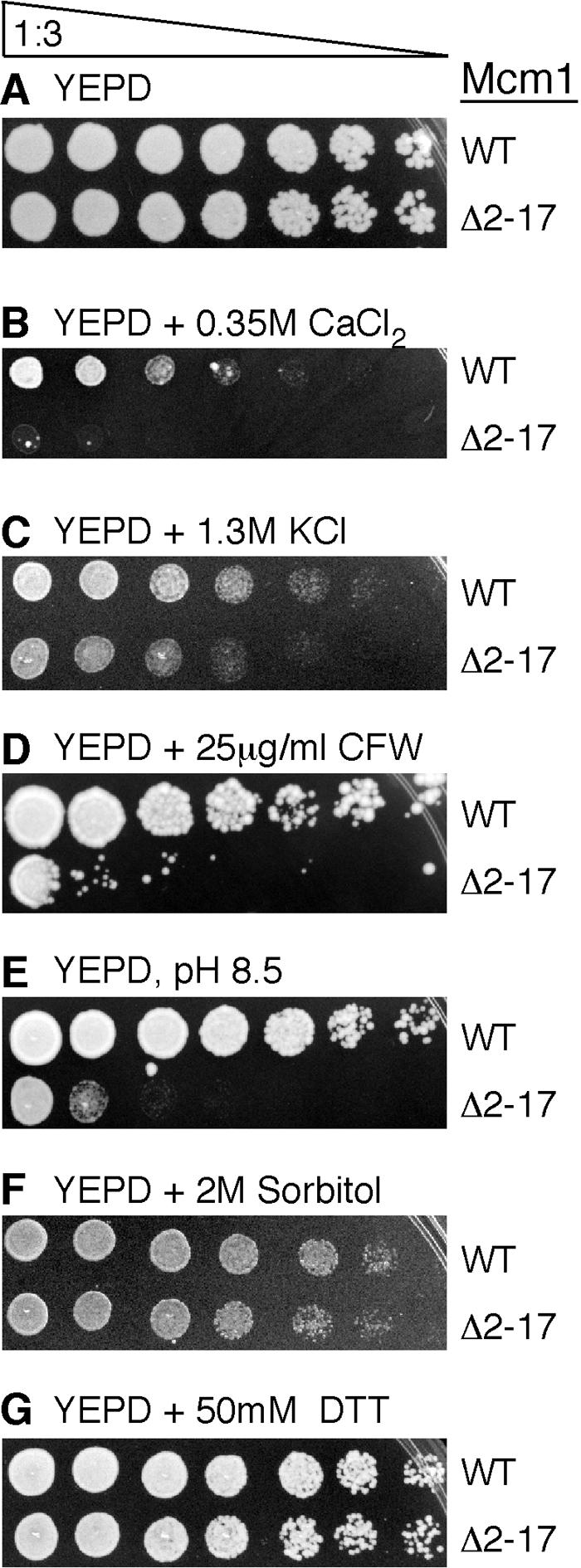
Strains containing mcm1-Δ2-17 are sensitive to CaCl2, KCl, CFW, and pH 8.5, but not sorbitol or DTT. Serial dilutions (threefold) of yeast strain JM01 transformed with plasmids containing MCM1 or mcm1-Δ2-17 were spotted onto the indicated media and grown at 30°C for 2 to 5 days.
Since the NT arm of Mcm1 was required for the proper expression of genes that encode components of the cell wall, we tested cells with mcm1-Δ2-17 for sensitivity to several cell wall-disrupting agents. The drug CFW is a fluorescent dye that binds to chitin in the cell wall and is thought to weaken the cell wall by preventing microfibril formation (15, 33, 38). CFW sensitivity is commonly used as an indicator of defects in cell wall biogenesis. Deletion of the Mcm1 NT arm conferred a strong sensitivity to CFW (Fig. 2D). Deletion of the Mcm1 NT arm also caused the cells to be sensitive to alkaline pH (Fig. 2E). However, the mutant was not sensitive to other agents that are commonly used to monitor defects in the cell wall, such as 2 M sorbitol, 50 mM DTT, and acidic pH (Fig. 2F and G and data not shown). These data suggest that Mcm1 is required for the transcriptional regulation of a subset of cell wall genes that are required for CFW and high pH resistance but not DTT or sorbitol resistance.
CFW interacts with chitin in the yeast cell wall, and exposure to CFW induces chitin synthesis (40-42). CHS1, CHS2, and CHS3 encode the catalytic subunits of chitin synthetases. There are three putative Mcm1-binding sites in the 1-kb region upstream of the CHS2 start codon. However, Northern analysis showed that CHS1, CHS2, and CHS3 mRNA levels were not altered by the Mcm1 NT arm deletion, suggesting that the CFW-sensitive phenotype is not caused by improper expression of these genes (data not shown).
Effect of Mcm1 point mutations on YGP1 transcription and CFW and alkaline pH tolerance.
Although there are multiple phosphorylated isoforms of Mcm1, residues S2 and T8 in the Mcm1 NT arm are the two major phosphorylation sites in the protein (24). Under normal conditions, residue S2 in the NT arm is phosphorylated with nearly 100% efficiency and a serine-to-alanine mutation at position S2, which mimics the constitutively dephosphorylated form of S2, has a salt-sensitive phenotype. This result suggested that phosphorylation of S2 was required for the induction of Mcm1-dependent salt response genes. Therefore, it is possible that phosphorylation of residues in the NT arm also has a role in YGP1 regulation. We tested this hypothesis by making alanine (A) and aspartate (D) substitutions of S2, T8, and T10 to mimic constitutive dephosphorylation and phosphorylation, respectively, of these residues. If differential phosphorylation states of the NT arm were important for the activation of YGP1, either A or D substitutions would be expected to have antagonistic effects on its transcription. However, Northern analysis showed that both the A and D substitutions of residues in the NT arm only resulted in slightly lower levels of the YGP1 transcript (Fig. 3A). Different combinations of these mutations also did not significantly affect YGP1 expression. In contrast, point mutations (V34A, T35A, S37A, K40A, and T66A) in the MADS box domain of Mcm1 that were previously shown to decrease DNA-binding affinity or bending and thus affect transcriptional activation were defective in YGP1 transcription (see Fig. 5B and data not shown).
FIG. 3.
The presence of the NT arm rather than its phosphorylation state regulates YGP1 transcription. Northern blots are shown for the expression of YGP1 and ACT1 (loading control) for strains containing wild-type MCM1 and the indicated mcm1 mutants.
FIG. 5.
(A) The T35A mutation in Mcm1 suppresses the growth defect of Mcm1-Δ2-17 on media containing CFW, pH 8.5, and CaCl2. Serial dilutions (threefold) of yeast strain JM01 transformed with plasmids containing MCM1, mcm1-Δ2-17, mcm1-T35A, or mcm1-Δ2-17,T35A were spotted onto the indicated media and grown at 30°C for 2 to 5 days. (B) The T35A mutation does not suppress the defective YGP1 transcription of Mcm1-Δ2-17. Northern blots are shown for the expression of YGP1 and ACT1 (loading control) for strains containing wild-type MCM1 and the indicated mcm1 mutants.
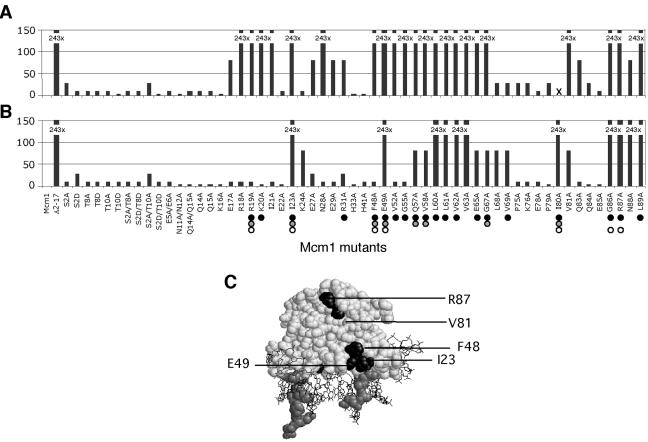
We next tested the effects of the amino acid substitutions in the NT arm and MADS box domain on resistance to cell wall-disrupting agents. Although deletion of the Mcm1 NT arm conferred a pronounced growth defect on rich medium supplemented with CaCl2 (Fig. 2B), single amino acid substitutions in the NT arm or MADS box domain were not sensitive to CaCl2 (data not shown). However, although significantly less than the 243-fold decrease in growth caused by deletion of the NT arm, A and D substitutions of residues S2, T8, and T10 caused 3- to 27-fold decreases in growth on medium with CFW or alkaline pH (Fig. 4A and B). Alanine replacements of other residues in the arm that are not phosphorylated also made the cells sensitive to CFW. Since deletion of the NT arm showed a much stronger phenotype than individual point mutations in the arm, it appears that the entire arm is important for its function.
FIG. 4.
Effects of amino acid substitutions in Mcm1 on CFW and alkaline pH tolerance. Each of the indicated mutants was assayed for growth on YEPD medium with 25-mg/ml CFW (A) and alkaline pH 8.5 (B). Bars represent the fold decrease in growth of the Mcm1 mutations indicated at the bottom, relative to wild-type MCM1. X, a mutant that was not tested. Mutants that affect activation in complex with α1 (dark circles), Ste12 (gray circles), or Mcm1 on its own (open circles) are shown for comparison (28). (C) Model of the Mcm1 dimer bound to DNA and positions of mutations (highlighted in black) that result in significant growth defects on CFW and alkaline pH. Residues 15 to 22 that form part of the NT arm are shaded gray. The protein is shown in spacefill and the DNA is shown as a wireframe.
We also examined the relative contribution of different residues in the MADS box domain toward CFW and alkaline pH tolerance. A comparison of the mutant strains showed that the same regions in Mcm1 are important for tolerance to both CFW (Fig. 4A) and alkaline pH (Fig. 4B). In general, the mutations had a more severe growth defect on CFW than high pH, underscoring the sensitivity of the CFW phenotype over the pH phenotype. Interestingly, the mutations that were the most defective in these assays mapped to three locations on the crystal structure of the Mcm1 dimer bound to DNA. One class of mutants, exemplified by L60, L61, V62, and V63, consists of residues along the dimer interface. It is not surprising that mutations of these residues affect Mcm1-mediated functions, most likely by adversely affecting Mcm1 homodimerization. The second class of mutants, represented by V81A, G86A, and R87A, form a hydrophobic groove on the surface of Mcm1 (Fig. 4C). In the crystal structure of the Mcm1-α2-DNA complex, this region of Mcm1 interacts with residues in the α2 protein and it is possible that Mcm1 uses these same residues to interact with another cofactor required for induction of the cell wall-associated genes identified (47). The third class of mutants, I23A, F48A, and E49A, are positioned near the NT arm in the crystal structure (Fig. 4D). These mutants have previously been shown to affect the expression of promoters driven by the Mcm1-α1 and Mcm1-Ste12 complexes, but not by Mcm1 alone or repression by the Mcm1-α2 complex (28). The sensitivity of the mutants belonging to the second and third categories suggests that Mcm1 may be interacting with a cofactor to activate the transcription of genes required for resistance to CFW and alkaline pH.
Mutations that suppress the CFW, alkaline pH, and CaCl2 sensitivity to deletion of the Mcm1 NT arm do not suppress the transcriptional defect of YGP1.
The T35A mutation in the MADS box domain of Mcm1 caused cells to grow better than the wild-type strain in the presence of high salt levels (23). This mutation also suppressed the effect of the NT arm deletion. We therefore wanted to determine if the T35A mutation was able to suppress the effects of the Mcm1 NT arm deletion on sensitivity to cell wall-disrupting agents. On its own, the T35A Mcm1 mutant grew as well as the wild-type MCM1 strain on medium with CaCl2 (Fig. 5). In contrast, this mutant was defective in growth on plates with CFW and alkaline pH, although not to the same extent as mcm1-Δ2-17. As was observed for the NaCl-sensitive phenotype, the T35A mutant was able to partially suppress the CaCl2-sensitive phenotype of the Mcm1 NT arm deletion. Similarly, the T35A mutation partially suppressed the growth defect of mcm1-Δ2-17 on alkaline pH and very weakly on CFW.
Since YGP1 transcription was induced with high salt levels and was adversely affected by the deletion of the Mcm1 NT arm, we wondered if the T35A mutation suppressed the effect of Mcm1-Δ2-17 on expression of the gene. However, Northern blot analysis showed that both Mcm1-T35A and Mcm1-Δ2-17,T35A caused decreases in YGP1 expression (Fig. 5B). Taken together, these data suggest that the mechanism of Mcm1-dependent regulation of genes required for growth on CFW and with alkaline pH and CaCl2 is different than Mcm1 regulation of YGP1.
The YGP1 promoter is regulated by two MADS box proteins, Mcm1 and Rlm1.
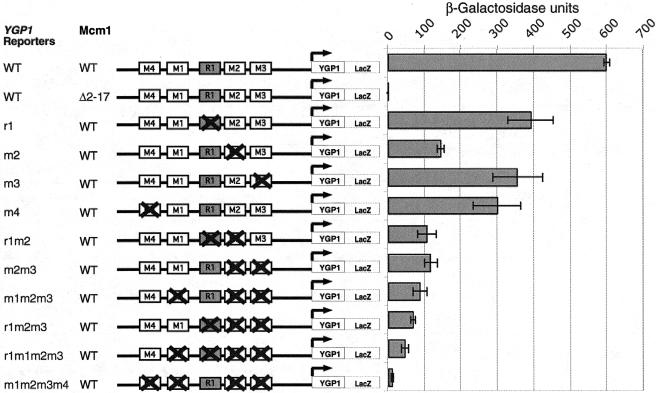
Since deletion of the Mcm1 NT arm showed the largest effect on the transcription of YGP1, we decided to analyze the YGP1 promoter to understand the mode of regulation of this gene by Mcm1. To monitor expression of the gene, we fused the 700-bp promoter region 5′ to the YGP1 ORF to the lacZ gene. The reporter was strongly expressed in a wild-type strain, but its expression was decreased almost 300 fold in an mcm1-Δ2-17 strain (Fig. 6). This result showed that the regulatory elements that responded to the deletion of the Mcm1 NT arm were located in this region of the YGP1 promoter. Sequence inspection of the YGP1 reporter predicted four potential Mcm1-binding sites, which we called M1, M2, M3, and M4. To determine if the putative Mcm1-binding sites were functional, these sites were mutated individually and in combination in the context of the YGP1-lacZ reporter, and their effect on YGP1 expression was measured by assaying β-galactosidase activity in cells with wild-type Mcm1. Individual disruption of each of these sites caused a partial decrease in expression of the reporter, indicating that these sites are required for regulation of YGP1 and are likely to be functional Mcm1 targets. The relative effect of each of the mutants varied, suggesting that these sites may be bound by Mcm1 with differing affinities. Mutation of all four Mcm1-binding sites (M1, M2, M3, and M4) resulted in a significant decrease in activation, similar to β-galactosidase levels obtained by deletion of the NT arm.
FIG. 6.
The putative Mcm1-binding sites (M1, M2, M3, and M4) and the Rlm1-binding site (R1) in the YGP1 promoter are activating sequences. A schematic representation of the known and putative upstream regulatory motifs in the 700-bp region upstream of the YGP1 ORF is shown. The crosses indicate mutations at those sites in the promoter. The reporters were assayed for lacZ expression in a wild-type MCM1 or mcm1-Δ2-17 background. The bars indicate the average values of expression for three independent transformants and the standard deviations for the samples are shown.
In addition to the Mcm1-binding sites, there is a putative binding site for the Rlm1 MADS box protein in the YGP1 promoter. Rlm1 is activated through phosphorylation by the Mpk1 MAP kinase in response to cell wall integrity signaling (13). Northern blot analysis of a strain with constitutively activated Rlm1 showed that YGP1 expression was decreased, suggesting that Rlm1 functions as a repressor of this gene (18). However, mutation of the Rlm1-binding site in the YGP1-lacZ fusion reporter caused a slight decrease in expression both on its own and in combination with mutations in the Mcm1 sites (Fig. 6). This result suggests that Rlm1 may have a direct role in activating YGP1 expression under the noninducing growth condition used.
Deletion of the Mcm1 NT arm does not affect the level or stability of Mcm1 protein.
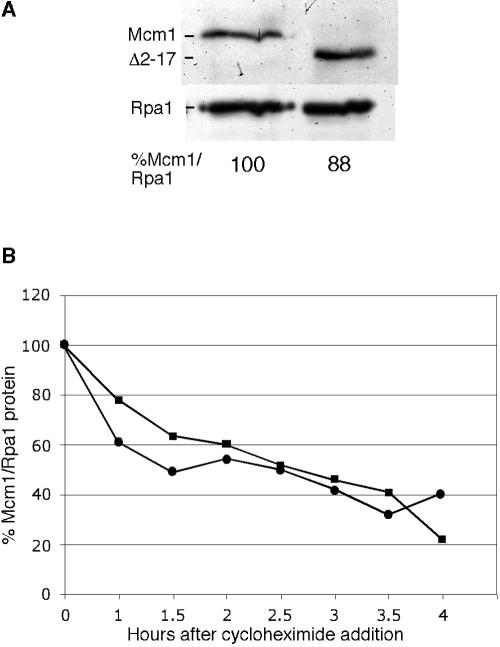
It is possible that the deletion of the NT arm alters the expression or stability of the Mcm1 protein such that it affects the expression of cell wall genes but not other Mcm1 targets. However, quantitative Western blot analysis showed that the level of N-terminally V5 epitope-tagged Mcm1-Δ2-17 was not significantly different from that of wild-type Mcm1-V5 when normalized to the levels of Rpa1 (Fig. 7A). To compare the relative degradation rates of Mcm1-Δ2-17-V5 to that of Mcm1-V5, protein translation was arrested by the addition of cycloheximide to yeast strains containing the two forms of Mcm1. The rate of degradation of the V5-tagged wild-type Mcm1 and Mcm1-Δ2-17 proteins was followed by quantitative Western blots of samples at different time points after cycloheximide treatment. Both Mcm1-Δ2-17-V5 and Mcm1-V5 were degraded at approximately the same rate, with a half-life of approximately 2 h (Fig. 7B). This shows that defects in YGP1 transcription and the biological phenotypes associated with the deletion of Mcm1 residues 2 to 17 are not a result of changes in the amount of Mcm1 protein or its stability.
FIG. 7.
Deletion of the Mcm1 NT arm does not change the level or stability of the protein. (A) A quantitative Western blot of V5-tagged Mcm1 and Mcm1-Δ2-17 using anti-V5 antibodies is shown. Anti-Rpa1 was used to detect Rpa1 protein, a loading control. (B) Plot of quantitative Western blot of V5-tagged Mcm1 proteins. Protein synthesis was arrested by the addition of cycloheximide to log phase cultures, and aliquots of cells from the indicated time points were assayed for Mcm1 and Rpa1 levels by Western blotting. The ratios of Mcm1:Rpa1 (circles) and Mcm1Δ2-17:Rpa1 (squares) proteins were plotted at the different time points after the addition of cycloheximide.
Deletion of the Mcm1 NT arm does not affect the in vitro and in vivo DNA-binding affinity of Mcm1.
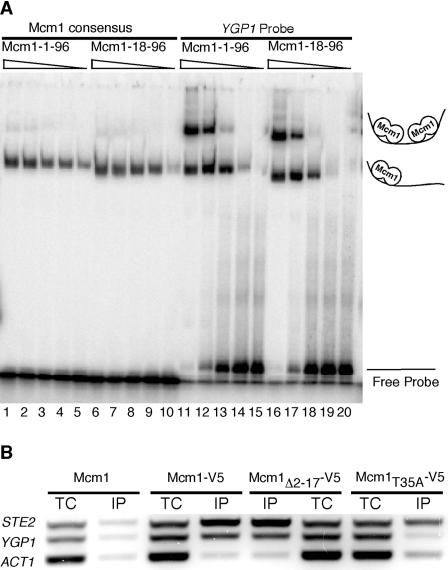
Mutations in Mcm1 that decrease its DNA-binding affinity for its binding site are known to decrease transcriptional activation of Mcm1-regulated genes (1). We therefore examined the DNA-binding affinity of bacterially expressed Mcm1-1-96 and Mcm1-18-96 for a probe with a consensus Mcm1-binding site and a second probe that includes the M2 and M3 sites from the YGP1 promoter. The protein with NT arm deletion bound to the consensus P(PAL) and YGP1 probes with the same affinity as the wild-type protein (Fig. 8A). This indicates that the arm is not required for binding to these sites in vitro.
FIG. 8.
Deletion of the Mcm1 NT arm does not change its in vitro or in vivo DNA-binding affinity for the YGP1 promoter. (A) EMSA analysis of the DNA-binding affinity of Mcm1-1-96 and Mcm1-18-96 to a 26 bp P(PAL) Mcm1 consensus probe and a YGP1 probe containing the M2 and M3 Mcm1-binding sites. The concentrations of the purified Mcm1-1-96 and Mcm1-18-96 proteins were normalized and binding by the protein (fivefold serial dilutions) is shown. (B) ChIP assays were performed on strain JM01 transformed with untagged Mcm1, Mcm1-V5, Mcm1-Δ2-17-V5, and Mcm1-T35A-V5 using anti-V5 antibody. Both total chromatin (TC) and immunoprecipitated (IP) DNA were assayed by PCR for the presence of STE2 (positive control), YGP1 (test), and ACT1 (negative control) promoters.
Although deletion of the NT arm did not affect Mcm1 binding in vitro, it was possible that the mutations affect the ability of the protein to bind its target sites in vivo. We therefore tested the in vivo DNA-binding affinity of Mcm1-Δ2-17-V5 and Mcm1-V5 to the YGP1 promoter by ChIP assays in which Mcm1-bound DNA was immunoprecipitated with the α-V5 antibody. We probed for the presence of Mcm1-regulated STE2 and YGP1 promoters and, as a control, the Mcm1-independent ACT1 promoter using primers sets that were specific to each promoter (Fig. 8B). The deletion of Mcm1 residues 2 to 17 did not affect DNA binding to Mcm1-regulated genes that do or do not require the NT arm for transcriptional regulation. In contrast, the T35A mutation of Mcm1 caused a dramatic decrease in binding of Mcm1 to the YGP1 promoter (Fig. 8B). This accounts for the poor transcription of this gene in this mutant background (Fig. 5B). The ChIP data also showed that Mcm1 is bound to the YGP1 promoter in the absence of salt stress. This indicates that the Mcm1 NT arm is not required for binding to the YGP1 promoter in vivo.
Deletion of the Mcm1 NT arm does not affect the in vitro DNA-bending angle of YGP1 DNA.
Mcm1 binding to DNA sites in the promoters of some Mcm1-regulated genes is not sufficient to activate their transcription (2). When the Mcm1 dimer binds to DNA, it also induces a bend in the DNA that is important for activation (2, 7, 44). Mutations that change the bending angle result in a decrease in transcriptional activation. It is therefore possible that deletion of the NT arm affects DNA bending by the protein and thereby decreases the expression of the NT arm-dependent genes. However, using EMSAs with circularly permuted fragments, we determined that the apparent bending angles induced by Mcm1-1-96 and Mcm1-18-96 binding to the R1M2M3 YGP1 probe were almost identical, suggesting that the decrease in transcriptional activation of YGP1 was not due to changes in the DNA-bending angle (Fig. 9).
FIG. 9.
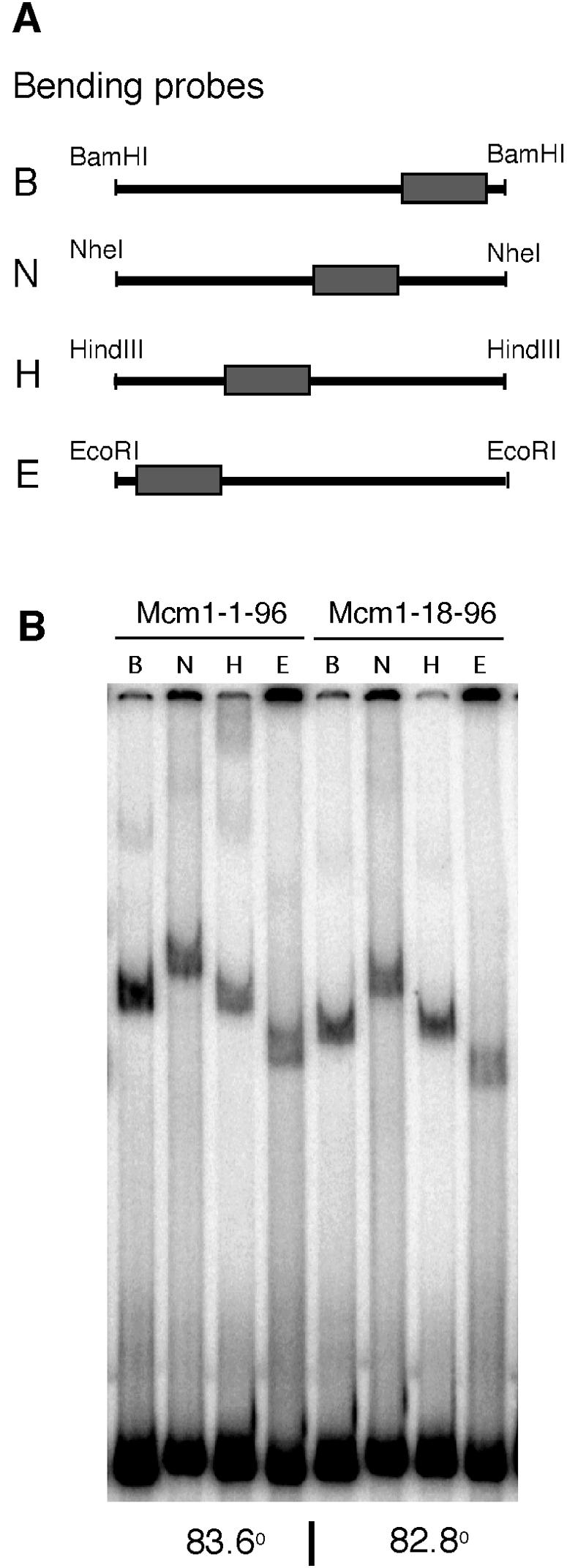
Deletion of Mcm1 residues 1 to 17 does not change the apparent bending angle of YGP1 DNA. (A) A circularly permuted probe with the R1-M2-M3 fragment, shown by a rectangle, from the YGP1 promoter was digested with BamHI, NheI, HindIII, or EcoRI to yield probes designated B, N, H, and E, respectively, so that the R1-M2-M3 fragment is located at different positions along the length of each probe. (B) EMSAs were performed using position-permuted fragments and purified Mcm1-1-96 and Mcm1-18-96. The bend angle for each protein-DNA complex was calculated using the Thompson and Landy relationship (48).
DISCUSSION
The N-terminal arm of Mcm1 has a role in regulating genes that are required for tolerance to NaCl (24). In addition to the NaCl-sensitive phenotype previously associated with deletion of the arm, we have observed other biological phenotypes, including sensitivity to CaCl2, high pH, and CFW. We also determined that the NT arm of Mcm1 is required for transcriptional activation of a set of genes, including cell wall biogenesis and transport genes that were not earlier known to be regulated by Mcm1. In addition to the requirement for the NT arm, mutations in the MADS box domain that are defective in the transcriptional activation of other subsets of Mcm1-regulated genes also affect the transcription of many of these newly identified Mcm1 targets. Among the genes that we identified, only the promoter region of YGP1 appeared to be bound by Mcm1 by genome-wide location analysis (17). Four of these genes, YBR071W, SPI1, PNC1, and YGP1, are induced under various cell wall-damaging conditions (25). YBR071W is an uncharacterized ORF that appears to be part of the cell wall integrity pathway (25). YGP1 and SPI1 are components of the cell wall (12, 19). SPS100, which has 50% overall identity to YGP1, is a spore wall component but is also induced under stress. The increased sensitivity of the mcm1-Δ2-17 strain to NaCl, CaCl2, high pH, and CFW is likely caused by decreased transcription of these and other cell wall genes that are regulated by Mcm1.
The Mcm1 protein has strong in vitro DNA-binding affinity for the promoters of a subset of cell wall- and membrane-associated genes, such as GFA1, HSP150, PMA1, and PIS1 (23). Given that we identified other cell wall-associated genes that were sensitive to the Mcm1 NT arm deletion, it was possible that these genes were regulated by a similar mechanism. However, Northern blot analysis indicated that expression of these genes was not affected by deletion of the Mcm1 NT arm (data not shown). Thus, the Mcm1 arm-dependent genes we identified are distinct from other cell wall- and membrane-associated genes that may be regulated by Mcm1. It is likely that transcriptional regulation of the two groups of genes, although both mediated by Mcm1, occur through distinct mechanisms.
Most of the genes that we have identified showed a decrease in expression in strains containing the Mcm1 NT arm deletion. This result suggests that Mcm1 is functioning as an activator of these genes. However, two of the NT arm-dependent genes we identified, SIT1 and URA1, showed an increase in expression in the deletion mutant, suggesting that Mcm1 is functioning as a repressor of these genes. It is possible that depending on the promoter, Mcm1 can function either as an activator or a repressor of the salt induced genes. This would be similar to regulation by the Mcm1-ArgR complex, which functions to both activate arginine catabolic genes and repress anabolic genes in the presence of arginine in the medium (30). However, we were unable to detect the direct binding of either Mcm1 or Mcm1-Δ2-17 to these promoters by ChIP analysis, suggesting that Mcm1 may indirectly regulate the expression of these genes through the Mcm1 NT arm-dependent expression of a repressor protein that targets these genes (data not shown).
The decreased transcription of YGP1 in the mcm1-Δ2-17 mutant was not due to a decrease in Mcm1 protein level or stability. The deletion of the Mcm1 arm also did not decrease DNA-binding affinity or change the apparent DNA-bending angle, both of which contribute to transcriptional activation by Mcm1 (1). Based on our data, we hypothesize that the Mcm1 NT arm may be required to recruit or stabilize binding of another transcription cofactor that binds to the YGP1 promoter to activate its transcription.
Transcription of YGP1 was earlier shown to be regulated by Rlm1, a type II MADS box protein (18). In a strain with constitutively activated Rlm1 there was a decrease in YGP1 expression, leading to the conclusion that Rlm1 was a repressor of this gene (18). However, mutational analysis of the regulatory sites in the YGP1 promoter suggests that Rlm1 may also have a minor role in the activation of YGP1. This model is supported by Northern analysis, which showed that deletion of RLM1 resulted in a decrease in YGP1 transcript compared to the wild-type strain (18). We have shown here that Mcm1, a type I MADS box protein, has a direct role in regulating the expression of this gene. Interestingly, this is the first report of a gene that is jointly regulated by both type I and type II MADS box proteins in yeast. Since Rlm1 is known to regulate YGP1 transcription, there is a possibility that Rlm1 may interact with the NT arm of Mcm1. Among the 25 genes that are known to be regulated by Rlm1, 20 also have putative Mcm1-binding sites (18; our data). While the expression of some of these transcripts appear to be affected by mutations in the Mcm1 MADS box domain that decrease Mcm1 DNA-binding affinity, we found that only YGP1 and SPS100 transcription are dependent on the NT arm (Fig. 1 and data not shown). If the Mcm1 NT arm is important for interaction with its cofactor, this result suggests that Rlm1 is unlikely to be the cofactor that Mcm1 is interacting with at the YGP1 promoter. Also, since mutagenesis of the Rlm1-binding site, R1, in the YGP1-lacZ reporter showed only a minor decrease in activation (Fig. 6), there is little evidence for the role of Rlm1 as the cofactor recruited by the Mcm1 NT arm.
Residues S2 and T8 in the Mcm1 NT arm are heavily phosphorylated and a mutation of S2 to alanine, which partially mimics the unphosphorylated form of these residues, had a salt-sensitive phenotype that was similar to deletion of the NT arm (24). Since the deletion of the NT arm also causes cells to be sensitive to CFW, high pH, and CaCl2, we expected that amino acid replacements of residues S2, T8, and T10 may have similar phenotypes on these media. However, none of the point mutations affected growth on CaCl2 and had relatively mild effects on growth on CFW and high pH in comparison to the NT arm deletion. Interestingly, mutations of these residues to either alanine or aspartate, which partially mimic constitutive dephosphorylation or phosphorylation, respectively, showed similar sensitivity to CFW and alkaline pH (Fig. 4A and B). This suggests that the phosphorylation state of these residues may have little to do with resistance to CFW and alkaline pH. Alternatively, it is also possible that the aspartate mutation does not effectively mimic the phosphorylated form of these residues.
The T35A mutation in the MADS box domain of Mcm1 suppresses the growth defect of the NT arm deletion under high-salt conditions (23). We found that this mutation had a similar effect on suppressing the NT arm-sensitive phenotypes on media with CFW, high pH, and CaCl2 but did not restore growth to the level of wild-type Mcm1. Residue T35 of Mcm1 contacts the phosphate backbone of the DNA and a T35A mutation decreases Mcm1 in vitro DNA-binding affinity (1, 45). As expected from the decreased DNA-binding affinity, the T35A mutation caused a significant decrease in YGP1 transcript level. Although the T35A mutation partially suppressed the growth defects caused by the deletion of the Mcm1 NT arm, it did not cause an increase in YGP1 expression (Fig. 5). Therefore, the ability to grow in medium with CFW, high pH, and CaCl2 is not simply due to restoring expression of YGP1. This result implies that the Mcm1 NT arm may have different mechanisms of regulating YGP1 and other genes required for resistance to various cell wall stresses. This suggests that there are other Mcm1 NT arm-dependent genes that are required for resistance to these growth conditions. It is unclear how the T35A mutation suppresses the sensitivity to cell wall stresses caused by deletion of the NT arm. It is possible the T35A mutation prevents interaction with other Mcm1 cofactors or decreases Mcm1 binding to weak sites in the genome, causing an increase in the level of Mcm1 that is available to interact with cofactors at the cell wall gene promoters.
The Mcm1 point mutations in the NT arm and the MADS box domain appear to be more sensitive to CFW than growth under high-pH conditions, while hardly affecting sensitivity to CaCl2. One model to account for this differential sensitivity would be that genes conferring resistance to high pH and CaCl2 have other activators in addition to Mcm1. For example, Mcm1 may be the primary activator for NT arm-dependent genes that confer CFW resistance, causing cells with the NT arm deletion to be very sensitive to the presence of this compound. On the other hand, genes required for growth under high-pH conditions may be regulated by other activators in addition to Mcm1 and are therefore less sensitive to particular Mcm1 mutations.
While deletion of the NT arm made cells sensitive to CaCl2, in contrast to the CFW and high pH phenotypes, growth on CaCl2 was not sensitive to any of the point mutations in the MADS box domain, suggesting that a very different mechanism may be at play in regulating genes that confer resistance to this growth condition. It is possible that another cofactor that binds strongly to these promoters and may be tethering Mcm1 to the DNA mainly via interactions with its NT arm. Thus, DNA-binding defects in Mcm1 may not have significant effects on activation of these genes as long as Mcm1 is tethered to the promoter through interactions with this cofactor.
Acknowledgments
We thank Steve Brill for the anti-Rpa1 antibodies and members of the Vershon laboratory for plasmids and helpful discussions.
This work was supported by the Charles and Johanna Busch Fellowship (Rutgers University) to D.S.A. and by a grant from the National Institutes of Health (GM49265) to A.K.V.
REFERENCES
- 1.Acton, T. B., J. Mead, A. M. Steiner, and A. K. Vershon. 2000. Scanning mutagenesis of Mcm1: residues required for DNA binding, DNA bending, and transcriptional activation by a MADS-box protein. Mol. Cell. Biol. 20:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Acton, T. B., H. Zhong, and A. K. Vershon. 1997. DNA-binding specificity of Mcm1: operator mutations that alter DNA-bending and transcriptional activities by a MADS box protein. Mol. Cell. Biol. 17:1881-1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Affolter, M., J. Montagne, U. Walldorf, J. Groppe, U. Kloter, M. LaRosa, and W. J. Gehring. 1994. The Drosophila SRF homolog is expressed in a subset of tracheal cells and maps within a genomic region required for tracheal development. Development 120:743-753. [DOI] [PubMed] [Google Scholar]
- 4.Amar, N., F. Messenguy, M. El Bakkoury, and E. Dubois. 2000. ArgRII, a component of the ArgR-Mcm1 complex involved in the control of arginine metabolism in Saccharomyces cerevisiae, is the sensor of arginine. Mol. Cell. Biol. 20:2087-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ausubel, F. M. 1987. Current protocols in molecular biology, vol. 1. Greene Publishing Associates and Wiley-Interscience, New York, N.Y.
- 6.Bruhn, L., J.-J. Hwang-Shum, and G. F. Sprague, Jr. 1992. The N-terminal 96 residues of MCM1, a regulator of cell type-specific genes in Saccharomyces cerevisiae, are sufficient for DNA binding, transcription activation, and interaction with α1. Mol. Cell. Biol. 12:3563-3572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carr, E. A., J. Mead, and A. K. Vershon. 2004. Alpha1-induced DNA bending is required for transcriptional activation by the Mcm1-alpha1 complex. Nucleic Acids Res. 32:2298-2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang, V. K., J. J. Donato, C. S. Chan, and B. K. Tye. 2004. Mcm1 promotes replication initiation by binding specific elements at replication origins. Mol. Cell. Biol. 24:6514-6524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang, V. K., M. J. Fitch, J. J. Donato, T. W. Christensen, A. M. Merchant, and B. K. Tye. 2003. Mcm1 binds replication origins. J. Biol. Chem. 278:6093-6100. [DOI] [PubMed] [Google Scholar]
- 10.Christ, C., and B. K. Tye. 1991. Functional domains of the yeast transcription/replication factor MCM1. Genes Dev. 5:751-763. [DOI] [PubMed] [Google Scholar]
- 11.Davies, B., M. Egea-Cortines, E. de Andrade Silva, H. Saedler, and H. Sommer. 1996. Multiple interactions amongst floral homeotic MADS box proteins. EMBO J. 15:4330-4343. [PMC free article] [PubMed] [Google Scholar]
- 12.Destruelle, M., H. Holzer, and D. J. Klionsky. 1994. Identification and characterization of a novel yeast gene: the YGP1 gene product is a highly glycosylated secreted protein that is synthesized in response to nutrient limitation. Mol. Cell. Biol. 14:2740-2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dodou, E., and R. Treisman. 1997. The Saccharomyces cerevisiae MADS-box transcription factor Rlm1 is a target for the Mpk1 mitogen-activated protein kinase pathway. Mol. Cell. Biol. 17:1848-1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.El Alami, M., F. Messenguy, B. Scherens, and E. Dubois. 2003. Arg82p is a bifunctional protein whose inositol polyphosphate kinase activity is essential for nitrogen and PHO gene expression but not for Mcm1p chaperoning in yeast. Mol. Microbiol. 49:457-468. [DOI] [PubMed] [Google Scholar]
- 15.Elorza, M. V., H. Rico, and R. Sentandreu. 1983. Calcofluor white alters the assembly of chitin fibrils in Saccharomyces cerevisiae and Candida albicans cells. J. Gen. Microbiol. 129:1577-1582. [DOI] [PubMed] [Google Scholar]
- 16.Fan, H. Y., Y. Hu, M. Tudor, and H. Ma. 1997. Specific interactions between the K domains of AG and AGLs, members of the MADS domain family of DNA binding proteins. Plant J. 12:999-1010. [DOI] [PubMed] [Google Scholar]
- 17.Harbison, C. T., D. B. Gordon, T. I. Lee, N. J. Rinaldi, K. D. Macisaac, T. W. Danford, N. M. Hannett, J. B. Tagne, D. B. Reynolds, J. Yoo, E. G. Jennings, J. Zeitlinger, D. K. Pokholok, M. Kellis, P. A. Rolfe, K. T. Takusagawa, E. S. Lander, D. K. Gifford, E. Fraenkel, and R. A. Young. 2004. Transcriptional regulatory code of a eukaryotic genome. Nature 431:99-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jung, U. S., and D. E. Levin. 1999. Genome-wide analysis of gene expression regulated by the yeast cell wall integrity signalling pathway. Mol. Microbiol. 34:1049-1057. [DOI] [PubMed] [Google Scholar]
- 19.Kapteyn, J. C., B. ter Riet, E. Vink, S. Blad, H. De Nobel, H. Van Den Ende, and F. M. Klis. 2001. Low external pH induces HOG1-dependent changes in the organization of the Saccharomyces cerevisiae cell wall. Mol. Microbiol. 39:469-479. [DOI] [PubMed] [Google Scholar]
- 20.Keleher, C. A., C. Goutte, and A. D. Johnson. 1988. The yeast cell-type-specific repressor alpha 2 acts cooperatively with a non-cell-type-specific protein. Cell 53:927-936. [DOI] [PubMed] [Google Scholar]
- 21.Kumar, R., D. M. Reynolds, A. Shevchenko, S. D. Goldstone, and S. Dalton. 2000. Forkhead transcription factors, Fkh1p and Fkh2p, collaborate with Mcm1p to control transcription required for M-phase. Curr. Biol. 10:896-906. [DOI] [PubMed] [Google Scholar]
- 22.Kuo, M. H., and C. D. Allis. 1999. In vivo cross-linking and immunoprecipitation for studying dynamic protein:DNA associations in a chromatin environment. Methods 19:425-433. [DOI] [PubMed] [Google Scholar]
- 23.Kuo, M. H., and E. Grayhack. 1994. A library of yeast genomic MCM1 binding sites contains genes involved in cell cycle control, cell wall and membrane structure, and metabolism. Mol. Cell. Biol. 14:348-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuo, M. H., E. T. Nadeau, and E. J. Grayhack. 1997. Multiple phosphorylated forms of the Saccharomyces cerevisiae Mcm1 protein include an isoform induced in response to high salt concentrations. Mol. Cell. Biol. 17:819-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lagorce, A., N. C. Hauser, D. Labourdette, C. Rodriguez, H. Martin-Yken, J. Arroyo, J. D. Hoheisel, and J. Francois. 2003. Genome-wide analysis of the response to cell wall mutations in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 278:20345-20357. [DOI] [PubMed] [Google Scholar]
- 26.Lilly, B., S. Galewsky, A. B. Firulli, R. A. Schulz, and E. N. Olson. 1994. D-MEF2: a MADS box transcription factor expressed in differentiating mesoderm and muscle cell lineages during Drosophila embryogenesis. Proc. Natl. Acad. Sci. USA 91:5662-5666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lydall, D., G. Ammerer, and K. Nasmyth. 1991. A new role for MCM1 in yeast: cell cycle regulation of SW15 transcription. Genes Dev. 5:2405-2419. [DOI] [PubMed] [Google Scholar]
- 28.Mead, J., A. R. Bruning, M. K. Gill, A. M. Steiner, T. B. Acton, and A. K. Vershon. 2002. Interactions of the Mcm1 MADS box protein with cofactors that regulate mating in yeast. Mol. Cell. Biol. 22:4607-4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Messenguy, F., and E. Dubois. 1993. Genetic evidence for a role for MCM1 in the regulation of arginine metabolism in Saccharomyces cerevisiae. Mol. Cell. Biol. 13:2586-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Messenguy, F., and E. Dubois. 2003. Role of MADS box proteins and their cofactors in combinatorial control of gene expression and cell development. Gene 316:1-21. [DOI] [PubMed] [Google Scholar]
- 31.Minty, A., and L. Kedes. 1986. Upstream regions of the human cardiac actin gene that modulate its transcription in muscle cells: presence of an evolutionarily conserved repeated motif. Mol. Cell. Biol. 6:2125-2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Molkentin, J. D., B. L. Black, J. F. Martin, and E. N. Olson. 1996. Mutational analysis of the DNA binding, dimerization, and transcriptional activation domains of MEF2C. Mol. Cell. Biol. 16:2627-2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murgui, A., M. V. Elorza, and R. Sentandreu. 1985. Effect of papulacandin B and calcofluor white on the incorporation of mannoproteins in the wall of Candida albicans blastospores. Biochim. Biophys. Acta 841:215-222. [DOI] [PubMed] [Google Scholar]
- 34.Norman, C., M. Runswick, R. Pollock, and R. Treisman. 1988. Isolation and properties of cDNA clones encoding SRF, a transcription factor that binds to the c-fos serum response element. Cell 55:989-1003. [DOI] [PubMed] [Google Scholar]
- 35.Pardo, M., L. Monteoliva, J. Pla, M. Sanchez, C. Gil, and C. Nombela. 1999. Two-dimensional analysis of proteins secreted by Saccharomyces cerevisiae regenerating protoplasts: a novel approach to study the cell wall. Yeast 15:459-472. [DOI] [PubMed] [Google Scholar]
- 36.Passmore, S., G. T. Maine, R. Elble, C. Christ, and B. K. Tye. 1988. Saccharomyces cerevisiae protein involved in plasmid maintenance is necessary for mating of MAT alpha cells. J. Mol. Biol. 204:593-606. [DOI] [PubMed] [Google Scholar]
- 37.Primig, M., H. Winkler, and G. Ammerer. 1991. The DNA binding and oligomerization domain of MCM1 is sufficient for its interaction with other regulatory proteins. EMBO J. 10:4209-4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ram, A. F., A. Wolters, R. Ten Hoopen, and F. M. Klis. 1994. A new approach for isolating cell wall mutants in Saccharomyces cerevisiae by screening for hypersensitivity to calcofluor white. Yeast 10:1019-1030. [DOI] [PubMed] [Google Scholar]
- 39.Riechmann, J. L., B. A. Krizek, and E. M. Meyerowitz. 1996. Dimerization specificity of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA, and AGAMOUS. Proc. Natl. Acad. Sci. USA 93:4793-4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roncero, C., and A. Duran. 1985. Effect of Calcofluor white and Congo red on fungal cell wall morphogenesis: in vivo activation of chitin polymerization. J. Bacteriol. 163:1180-1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roncero, C., M. H. Valdivieso, J. C. Ribas, and A. Duran. 1988. Effect of Calcofluor white on chitin synthases from Saccharomyces cerevisiae. J. Bacteriol. 170:1945-1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roncero, C., M. H. Valdivieso, J. C. Ribas, and A. Duran. 1988. Isolation and characterization of Saccharomyces cerevisiae mutants resistant to Calcofluor white. J. Bacteriol. 170:1950-1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sengupta, P., and B. H. Cochran. 1990. The PRE and PQ box are functionally distinct yeast pheromone response elements. Mol. Cell. Biol. 10:6809-6812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sharrocks, A. D., and P. Shore. 1995. DNA bending in the ternary nucleoprotein complex at the c-fos promoter. Nucleic Acids Res. 23:2442-2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sharrocks, A. D., F. von Hesler, and P. E. Shaw. 1993. The identification of elements determining the different DNA binding specificities of the MADS box proteins p67SRF and RSRFC4. Nucleic Acids Res. 21:215-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sommer, H., J. P. Beltran, P. Huijser, H. Pape, W. E. Lonnig, H. Saedler, and Z. Schwarz-Sommer. 1990. Deficiens, a homeotic gene involved in the control of flower morphogenesis in Antirrhinum majus: the protein shows homology to transcription factors. EMBO J. 9:605-613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tan, S., and T. J. Richmond. 1998. Crystal structure of the yeast MATalpha2/MCM1/DNA ternary complex. Nature 391:660-666. [DOI] [PubMed] [Google Scholar]
- 48.Thompson, J. F., and A. Landy. 1988. Empirical estimation of protein-induced DNA bending angles: applications to lambda site-specific recombination complexes. Nucleic Acids Res. 16:9687-9705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trobner, W., L. Ramirez, P. Motte, I. Hue, P. Huijser, W. E. Lonnig, H. Saedler, H. Sommer, and Z. Schwarz-Sommer. 1992. GLOBOSA: a homeotic gene which interacts with DEFICIENS in the control of Antirrhinum floral organogenesis. EMBO J. 11:4693-4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yale, J., and H. J. Bohnert. 2001. Transcript expression in Saccharomyces cerevisiae at high salinity. J. Biol. Chem. 276:15996-16007. [DOI] [PubMed] [Google Scholar]
- 51.Yanofsky, M. F., H. Ma, J. L. Bowman, G. N. Drews, K. A. Feldmann, and E. M. Meyerowitz. 1990. The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature 346:35-39. [DOI] [PubMed] [Google Scholar]
- 52.Yu, G., and J. S. Fassler. 1993. SPT13 (GAL11) of Saccharomyces cerevisiae negatively regulates activity of the MCM1 transcription factor in Ty1 elements. Mol. Cell. Biol. 13:63-71. [DOI] [PMC free article] [PubMed] [Google Scholar]