Abstract
Extracellular nucleotides play many biological roles, including intercellular communication and modulation of nucleotide receptor signaling, and are dependent on the phosphorylation state of the nucleotide. Regulation of nucleotide phosphorylation is necessary, and a specialized class of enzymes, nucleotide pyrophosphatases/phosphodiesterases (E-NPPs), has been identified in mammals to perform this function. Although the E-NPP class is conserved among complex eukaryotes, this system has not yet been identified in Saccharomyces cerevisiae. Using genetic and biochemical experiments, we show that two orthologs of the E-NPP family, referred to as Npp1p and Npp2p, exist in budding yeast and can perform nucleotide phosphate hydrolysis. This activity is enhanced during phosphate starvation, where hydrolyzed phosphates can be imported from extracellular sources and utilized to overcome phosphate starvation through the activity of the Pho5p acid phosphatase. The added compensatory effect by Pho5p is also a newly established role for Pho5p. This study demonstrates that extracellular nucleotide phosphate metabolism appears to be controlled by at least two independent regulatory mechanisms, uniting phosphate starvation with extracellular nucleotide regulation.
The phosphorylation state of extracellular nucleotides is critical for maintaining intercellular communication. Nucleotide phosphates are also involved in activation of many cell surface receptors for initiation of various intracellular processes. It is critical to regulate the phosphorylation state of extracellular nucleotides, as well as to control extracellular pyrophosphate concentrations, and it must be tightly synchronized with dynamic environmental conditions. A regulatory mechanism to control extracellular nucleotide hydrolysis consists of a multigene nucleotide pyrophosphatase/phosphodiesterase (E-NPP) family (14). Five members of this family have been identified in humans (NPP1 to -5), each with distinct functional roles (10, 13, 19, 22). NPP1 to -3 are found in nearly all human tissue types, and these enzymes are categorized to contain an alkaline ecto-nucleotide pyrophosphatase/phosphodiesterase type 1 domain (12). These enzymes catalyze hydrolysis of pyrophosphate and phosphodiester bonds from nucleotide sources. All three are involved in regulating nucleotide recycling. In addition, NPP1 and NPP2 may regulate pathological mineralization, and NPP2 and NPP3 are involved in regulating cell motility (33, 43). NPP4 and NPP5 are additional members of this family; however, little is known about their activity. Several disorders have been found to correlate with misregulation or altered activity of E-NPP family members, including diabetes (5), chrondocalcinosis deposition disease (21), and tumor migration (48). Thus, although the NPP family performs considerable physiological roles in humans, the discovery of cellular roles of E-NPPs has not been previously established for Saccharomyces cerevisiae.
Another regulatory mechanism in yeast involves phosphate sensing and acquisition from the extracellular medium. Inorganic phosphate (Pi) is an essential nutrient for the biosynthesis of many cellular components. The availability of nutrients in the environment is critical in determining the cellular decision to undergo processes such as growth and proliferation. When phosphate supplies are limited, cells induce a response to acquire inorganic phosphate from multiple sources for cellular uptake (PHO response) (24). This includes expression of several genes, such as those encoding nonspecific scavenger phosphatases, to generate inorganic phosphates from various phosphate-containing molecules (6, 32). Free inorganic phosphate is subsequently transported and localized into the cytoplasm as a source for metabolic processes (47).
A hallmark of high-affinity phosphate transport pathway activation is the transcriptional upregulation of two classes of genes: acid phosphatases and phosphate symporters (2). The PHO response upregulates three genes (PHO5, PHO11, and PHO12) encoding repressible acid phosphatases (APases) when phosphate is scarce. These phosphatases are localized to either the periplasmic space or the cell wall and are responsible for phosphate scavenging by working in conjunction with high-affinity transporters to acquire phosphate when Pi concentrations in the environment are low (31, 41). Of the three phosphatases, Pho5p is responsible for >90% of APase activity (44). Whereas the APases are believed to utilize many different substrates for phosphate scavenging, it remains unclear whether they play a role in extracellular nucleotide recycling through phosphate hydrolysis.
The other key components of the PHO transport pathway are phosphate symporters. Two classes of symporters have been identified for phosphate uptake: a low-affinity and a high-affinity transport system (35). The low-affinity system is believed to be constitutively expressed, regardless of extracellular phosphate concentrations. It includes Pho87p, Pho90p, and Pho91p (2) and can import extracellular phosphate at a Km of 770 μM (45). The high-affinity phosphate regulatory system mediates specific uptake of inorganic phosphate from extracellular sources under phosphate starvation conditions when external phosphate concentrations fall below 100 μM (28). Under low-phosphate conditions, expression of the high-affinity transporters, Pho84p and Pho89p, is rapidly increased. This correlates with the decrease in expression of low-affinity phosphate transporters. These transporters have an approximately 10- to 100-fold-lower Km value for extracellular phosphates than their low-affinity counterparts (27, 34, 38).
In this report, we show that genes encoding putative E-NPPs (NPP1 and NPP2) can catalyze NPPase reactions and contribute to extracellular nucleotide-derived phosphate hydrolysis, demonstrating the existence and function of E-NPPs in budding yeast. Their levels of protein expression and enzymatic activity are enhanced during phosphate starvation. We demonstrate that nucleotide phosphate hydrolysis can also be performed by repressible Pho5p activity. Together, Npp1p, Npp2p, and Pho5p are the major contributors to NPPase activity. All three components are likely to work in conjunction with the high-affinity phosphate transporters to import scavenged phosphates once hydrolyzed from extracellular nucleotides. Regulation of extracellular nucleotide phosphatase activity in S. cerevisiae is distinct from that of multicellular eukaryotes, where E-NPPs and the PHO response share overlapping roles in yeast for phosphate acquisition and may implicate shared functions in extracellular nucleotide regulation.
MATERIALS AND METHODS
Reagents.
Adenosine [α-32P]triphosphate, adenosine [γ-32P]triphosphate, adenosine [32P]monophosphate, and [2-3H]adenosine were obtained from Amersham Biosciences. Lactate dehydrogenase, phosphoenol pyruvate, NADH, morpholinepropanesulfonic acid (MOPS), MgCl2, Tris, and MgSO4 were obtained from Sigma Aldrich. Concentrated aqueous ammonia was purchased from EMD Chemicals. Pyruvate kinase was obtained from U.S. Biologicals. 4-Nitrophenyl phosphate was obtained from Fluka. Bacto-yeast extract and Bacto-peptone were purchased from Becton Dickinson.
Strains and media.
The S. cerevisiae strains used in this study were derived from the Saccharomyces Genome Deletion Project (46) and are described in Table 1. All strains used in this study had comparable growth rates over a 24-h growth period. The strains were grown at 25°C unless otherwise specified, and standard techniques for yeast manipulation were performed (1). Yeast extract-peptone-dextrose (YPD) medium was prepared as previously described (40) and is referred to as high-phosphate medium with an inorganic phosphate concentration of 1 to 10 mM. Low-phosphate medium was prepared by combining reactants of 20 ml of 1 M MgSO4 and 20 ml of 30% aqueous ammonia in a molar excess of phosphate from YP medium (1% Bacto-yeast extract and 2% Bacto-peptone) per liter. The majority of inorganic phosphate from YP medium was precipitated as MgNH4PO4 for 30 min at room temperature, and the precipitate was removed using a Whatman no. 1 filter. The pH of the medium was then adjusted with HCl to 5.8, and 2% glucose was added. The solution was then sterilized by autoclaving.
TABLE 1.
Saccharomyces cerevisiae strains used in this study
| Strain | Relevant genotype |
|---|---|
| LPY06494 | MATα his3-1 leu2-0 lys2-0 met15-0 ura3-0 |
| LPY10198 | MATα his3-1 leu2-0 lys2-0 met15-0 ura3-0 npp2Δ::KANMX |
| LPY10199 | MATahis3-1 leu2-0 lys2-0 met15-0 ura3-0 npp1Δ::KANMX |
| LPY10200 | MATahis3-1 leu2-0 lys2-0 met15-0 ura3-0 npp1Δ::KANMX npp2Δ::KANMX |
| LPY10203 | MATα his3-1 leu2-0 lys2-0 met15-0 ura3-0 pho5Δ::KANMX |
| LPY10565 | MATahis3-1 leu2-0 lys2-0 met15-0 ura3-0 PHO5::GFP |
| LPY10807 | MATahis3-1 leu2-0 lys2-0 met15-0 ura3-0 NPP1::TAP |
| LPY10808 | MATahis3-1 leu2-0 lys2-0 met15-0 ura3-0 NPP2::TAP |
| LPY10883 | MATα his3-1 leu2-0 lys2-0 met15-0 ura3-0 npp1Δ::KANMX npp2Δ::KANMX pho5Δ::KANMX |
Extracellular radioprobe uptake experiments.
Yeast strains were grown to stationary phase in YPD medium and used to inoculate YPD or modified medium enriched with 1 mM ATP to an initial optical density at 600 nm (OD600) of 0.05. Radioprobes were added to each culture at 1 μCi (6,000 Ci/mmol) for 32P-labeled probes and 3H-labeled probes. After either a series of time points or a 24-h growth period, cultures were pelleted, washed three times with YPD, and resuspended in YPD. Each time point correlates with distinct stages of log-phase growth, where 8 h corresponds to early log phase (OD600 = 0.2 to 0.4), 16 h corresponds to mid-log phase (OD600 = 0.6 to 0.8), and 24 h corresponds to late log phase (OD600 = 1.0 to 1.3). The cells were broken with glass beads, and the supernatant was collected. The radioactivity of each of the fractions was determined by liquid scintillation counting (Beckman Coulter).
Sequence homology for potential E-NPPs.
BLAST, PSI-BLAST, and RPS-BLAST searches were performed where each member of the human E-NPP family was independently queried against the S. cerevisiae genome. From these searches, two yeast genes, NPP1 (YCR026c) and NPP2 (YEL016c), were identified. Additionally, the active site amino acid sequences (see Fig. 2, where 17 residues are highlighted) from known human E-NPPs were queried by BLAST searches against the S. cerevisiae genome, and the same two genes were identified. Both genes are uncharacterized, hypothetical open reading frames with predicted type I phosphodiesterase/nucleotide pyrophosphatase motifs as determined by the Conserved Domains Database (26). These genes were also used to perform BLAST searches against the fission yeast Schizosaccharomyces pombe. Two putative, uncharacterized open reading frames were identified in the search that were also predicted to have type I phosphodiesterase/nucleotide pyrophosphatase motifs. Protein sequences for the following genes were retrieved from GenBank (see Fig. 2): S. cerevisiae NPP1, AAT92706; S. cerevisiae NPP2, P25353; Homo sapiens NPP1, AAF36094; H. sapiens NPP2, BAA08260; H. sapiens NPP3, AAC51813; H. sapiens NPP4, BAA74902; H. sapiens NPP5, CAB56566, Rattus norvegicus NPP1, AF320054; R. norvegicus NPP3, BAA06333; and fowlpox virus NPP, AAF44374.
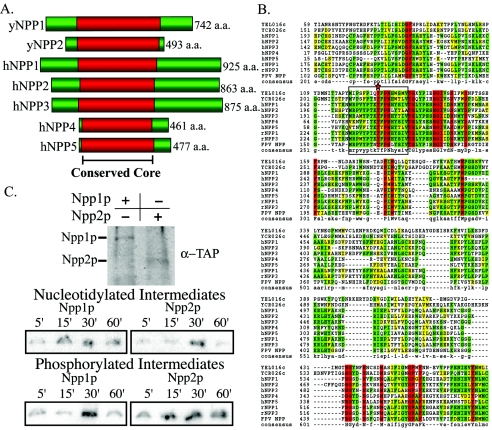
FIG. 2.
E-NPPs are conserved across species, including S. cerevisiae. (A) Alignment of two yeast genes with predicted NPPase activity. NPP1 and NPP2 were compared to members of the human E-NPP gene family. Alignments are centered on the conserved core residues (red) demonstrated to participate in catalytic activity in vertebrates. (B) Sequences of the two predicted E-NPPs in yeast are aligned with the five E-NPPs in H. sapiens (h, human), along with R. norvegicus (r, rat) and fowlpox virus (FPV, fowlpox virus) E-NPP genes. Red residues represent those conserved among all genes, green represents identical residues, and yellow represents similar residues between compared sequences. Boxed residues highlight the highly conserved active site of the E-NPP family, with the catalytic threonine residue is marked by a star. Protein sequences were retrieved from GenBank and were aligned with CLUSTALW. (C) Western analysis demonstrates isolation and purification of immunoprecipitated Npp1p and Npp2p TAP fusions utilized for the trapped intermediates and autophosphorylation experiments. Covalently trapped nucleotidylated intermediates were monitored over 60 min. Npp1p and Npp2p could perform the classic NPPase reaction where AMP was covalently attached to the catalytic threonine during catalysis. Autophosphorylation states were also monitored over 60 min as an indicator of autoinhibition. Npp2p was autophosphorylated at the earliest time point and persisted throughout the time course.
Assay of NPP activity.
Strains expressing tandem affinity purification (TAP)-tagged fusion proteins of Npp1p and Npp2p were grown overnight in YPD. Cells were lysed in 20 mM Tris-HCl (pH 7.5), 0.5 mM phenylmethylsulfonyl fluoride, 0.5 mM benzamidine, 0.3 M NaCl, and 0.2% Triton X-100. After centrifugation, the TAP tag fusion proteins were immunoprecipitated from the supernatant with TAP tag monoclonal antibodies (Open Biosystems) and protein A-agarose (Upstate). Immunoprecipitates were washed five times, resuspended in 50 mM HEPES (pH 7.5), and assayed for enzymatic activity. Activity was measured at 30°C in 50 mM Tris HCl (pH 9.0) with 0.9 mM p-nitrophenyl dTMP as a substrate. The reaction was stopped by 10-fold dilution in 3% trichloroacetic acid (Sigma Aldrich). The amount of p-nitrophenyl formed was quantified at λ = 410 nm after the addition of a 0.1 volume equivalent of 5 M NaOH. One unit of NPPase activity was defined as the amount of enzyme required to hydrolyze 1 μmol of substrate/min under the assay conditions used.
Isolation of the NPP-nucleotidylated intermediate.
The trapped intermediate was performed according to Blytt et al. (4) with minor changes (42). Purified Npp1p or Npp2p (1 U/ml) was incubated in 0.57 M imidazole-formate, pH 4.0, and 260 μM [α-32P]ATP. At specific time points, 0.3-mg/ml bovine serum albumin and 10% trichloroacetic acid were added. After 20 min on ice, the precipitated proteins were pelleted by centrifugation and washed three times in 50 mM Tris HCl, pH 6.8. The pellet was then boiled in 25 mM Tris HCl (pH 6.8), 2% sodium dodecyl sulfate (SDS), 10% glycerol, and 250 mM 2-mercaptoethanol (electrophoresis buffer). The trapped intermediates were visualized by autoradiography following SDS-polyacrylamide gel electrophoresis (PAGE) (7.5%). As a negative control, identical 60-min reactions were set up using [γ-32P]ATP as a substrate to verify that no radiolabeled trapped intermediates were generated under these conditions.
Isolation of the NPP phosphorylated intermediate.
Phosphorylation was performed by incubating of 200-mU/ml of Npp1p or Npp2p in 50 mM HEPES, pH 7.5, with 50 μM [γ-32P]ATP. At specific time points, the reaction was stopped by the addition of electrophoresis buffer. The phosphorylated intermediates were visualized by autoradiography following SDS-PAGE (7.5%). As a negative control, an identical 60-min reaction containing [α-32P]ATP was run to verify that phosphorylated intermediates were not generated under these conditions.
TAP-tagged protein expression and Western blot immunodetection.
Strains with chromosomally expressed genes of interest with either TAP fusions for Npp1p and Npp2p or green fluorescent protein (GFP) fusions for Pho5p were obtained (Open Biosystems). Cells from stationary-growth cultures were used to inoculate cultures in either YPD or low-phosphate medium with an initial OD600 of 0.05. Cultures were grown for various time points and normalized for 5 OD600 equivalents to be prepared per lysate. Cell lysates were prepared by bead beating in 1% SDS, 8 M urea, 10 mM MOPS, pH 6.8, and 10 mM EDTA. Normalized total cell lysates were loaded per lane on an 8% PAGE gel, electrophoresed, and transferred to a nitrocellulose membrane. Anti-TAP antibody or anti-GFP antibody was used as a primary antibody (Open Biosystems and Sigma, respectively). Horseradish peroxidase-conjugated antibodies against rabbit immunoglobulin G (Santa Cruz Biotechnology) were used as secondary antibodies. For protein loading controls, mouse anti-414 antibody (obtained from D. Forbes) was used to detect a constitutively expressed nuclear protein, Nap62p. Western blotting was performed using the ECL detection system protocol (Perkin-Elmer).
NPPase activity assays with ATP substrates.
Cells grown to late log phase were used to inoculate either YPD or modified medium to an initial OD600 of 0.05. Cultures were grown for the times indicated, pelleted, washed three times with Tris-EDTA, and resuspended in 0.5 ml 50 mM Tris-HCl, pH 8.0. A kinetic activity assay was adapted to spectrophotometrically measure activity with ATP as a substrate (9). Cells were then combined with a reaction mixture containing 100 mM MOPS, pH 8.0, 10 mM MgCl2, 1 mM phosphoenol pyruvate, 12 U lactate dehydrogenase, 15 U pyruvate kinase, and 140 μg NADH. The absorbance was zeroed at λ = 340 nm, and then 10 μl of 100 mM ATP was added. ATP hydrolysis to ADP was monitored as a function of NADH oxidation over 1-min elapsed reaction time by measuring the decease in absorbance at λ = 340 nm (Beckman-Coulter DU 640). NADH consumption (in micromoles per minute) was determined by the equation [(A340(t = 0) − A340(t = 1 min))/(ɛNADH)]/1 min.
RESULTS
Cellular uptake of extracellular nucleotide-derived phosphates is dependent on extracellular phosphate concentration.
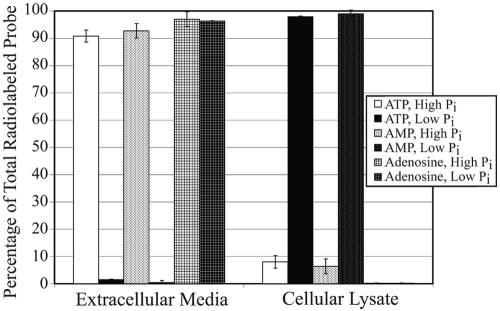
Ecto-nucleotide phosphatase activity had yet not been established to exist in yeast. To ascertain whether this activity was present in budding yeast, extracellular ATP hydrolysis was measured. If E-NPP activity occurs, hydrolyzed phosphates will be imported during cell growth to maintain proper intracellular phosphate concentrations. Cultures were grown in the presence of radiolabeled nucleotide substrates, and the level of imported radiolabel was monitored over time. This was measured in both low- and high-phosphate medium from early- to late-log-phase growth (Fig. 1). Nutrient-enriched YPD medium was utilized for high-phosphate conditions, whereas YPD medium with depleted inorganic phosphate provided low-phosphate conditions. Cells were grown in both conditions in the presence of 1 μCi [γ-32P]ATP. At each time point, cells were lysed and the amount of imported γ-phosphate was determined by comparison to that remaining in the medium. When grown in high-phosphate medium, <10% of the radiolabel could be detected within the cellular lysate. However, when phosphate was depleted from the growth medium, >95% of the radiolabeled phosphate was detected in the cellular lysate. To demonstrate that the observed hydrolysis is a regulated cellular event, NPPase activity was monitored over time under normal and low-phosphate conditions, illustrating induced NPPase activity during phosphate starvation over 24 h (see Fig. 4). To establish whether phosphate hydrolysis is limited to the gamma phosphate, the same experiment was performed with radiolabeled AMP. Cellular import of radiolabeled phosphate could not be detected in high-phosphate medium, yet nearly all of it was imported when extracellular phosphate was depleted. Thus, phosphate hydrolysis activity could clearly be detected for both ATP and AMP substrates. To establish that this hydrolysis event was occurring extracellularly, it was necessary to eliminate the possibility that nucleotides could be passively or actively imported to an appreciable level. The importation of extracellular adenosine was measured under the same conditions using [2-3H] adenosine. Regardless of the extracellular phosphate concentration, no adenosine could be detected within the cellular lysate. Together, these observations clearly demonstrate that inorganic phosphate is hydrolyzed from extracellular nucleotides under low-phosphate conditions and that all three phosphates can be utilized as substrates for hydrolysis.
FIG. 1.
Nucleotide-derived phosphate hydrolysis and uptake from extracellular ATP and AMP sources are dependent on extracellular phosphate concentrations. Radiolabeled phosphate from ATP and AMP (1 μCi) was tested for uptake in both low- and high-phosphate media to determine if all three phosphates could be hydrolyzed from adenosine during phosphate starvation. Cells were grown for 24 h in the presence of radiolabeled compounds prior to uptake analysis. Tritiated adenosine uptake was evaluated as a control to establish that phosphates were hydrolyzed from the nucleotide base prior to cellular import.
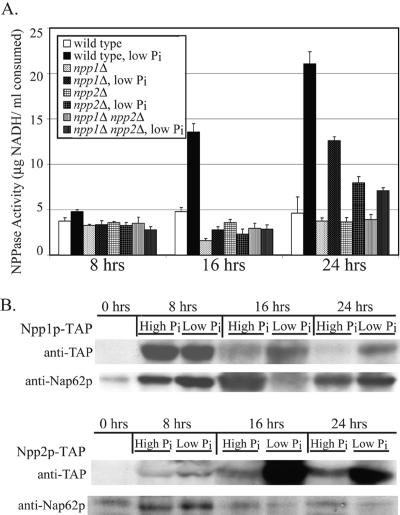
FIG. 4.
E-NPP activity depends on NPP1 and NPP2. (A) E-NPP activity was measured in strains lacking NPP1 and/or NPP2. Nucleotide phosphate hydrolysis was measured by a coupled enzyme assay where ATP hydrolysis was monitored as a function of oxidation of NADH. Cultures were grown over a 24-h time course in regular or phosphate-depleted medium. Lack of both genes caused a significant deficiency in E-NPP activity under low-phosphate conditions. (B) Protein expression levels were monitored over a 24-h time course. Cellular lysates were prepared at each time point, and TAP fusion proteins were detected from whole-cell lysates through immunoblotting. Protein expression of Npp1p and Npp2p increased during log-phase growth, and this expression was enhanced for cells grown under low-phosphate conditions during the same phase of growth. Detection of the constitutively expressed Nap62p was used a protein-loading control.
Identification of two putative E-NPP genes, NPP1 and NPP2.
In several eukaryotes, the E-NPP multigene family controls a complex mechanism of extracellular nucleotide regulation and metabolism. Despite their diversity, the family members share many physiological roles involving nucleotide phosphate hydrolysis. As demonstrated above, extracellular nucleotide-derived phosphate hydrolysis occurs in S. cerevisiae, yet the enzyme(s) involved in this activity has not been previously identified. Based upon sequence comparisons to this family of enzymes, two candidates for genes encoding E-NPPs were identified in S. cerevisiae, neither of which is individually required for viability. These genes share approximately 30% similarity and 15 to 20% identity to the E-NPP family within the type I phosphodiesterase-nucleotide pyrophosphatase motif and have BLAST scores with E-values as significant as 10−79 compared to human homologs with demonstrated E-NPP activity (Fig. 2). Notably, in regions containing catalytically significant residues including the active site, nearly all residues were identical. Based on their sequence similarities and functional characterization (described below), we have named these genes NPP1 (YCR026c) and NPP2 (YEL016c), in accordance with the Saccharomyces Genome Database naming guidelines (7).
To ascertain whether these genes encode functional NPPs in yeast, Npp1p and Npp2p were purified and assayed with the same tests used for characterization of human NPPs. The primary trademark of the NPP family is to catalyze phosphate hydrolysis from nucleotide triphosphates, producing an intermediate where the nucleotide is covalently attached to the catalytic threonine (12). In the second step of this reaction, the nucleotide monophosphate is released. Previous characterization of other members of the NPP family illustrated that the nucleotidylated catalytic intermediate could be trapped under acidic conditions in the presence of imidazole using the substrate [α-32P]ATP. Imidazole inhibits substrate release after nucleotide phosphate hydrolysis, leading to stabilization of the intermediate state. Npp1p and Npp2p were isolated by immunoprecipitation and verified by Western blotting analysis (Fig. 2C). The semipurified proteins were tested to determine whether they could perform the NPP reaction mechanism. Under these reaction conditions, the catalytic activity of Npp1p and Npp2p were measured over a 60-min time course (Fig. 2C). For both enzymes, the nucleotidylated intermediates were observed. As a negative control, the same experimental setup using [γ-32P]ATP was performed to verify that nonspecific labeling did not occur under these conditions (data not shown). The difference in the kinetics of each intermediate trapping profile demonstrates that each enzyme has unique activity, and may be differentially regulated. A possible explanation for this observation is through autoinhibition of catalytic activity.
Some members of the NPP family can undergo autophosphorylation and autodephosphorylation of the catalytic threonine as a mechanism of catalytic regulation (42). To establish whether these two enzymes could undergo autophosphorylation, the phosphorylation state of each enzyme was measured over time (Fig. 2C). Npp1p and Npp2p were able to undergo autophosphorylation. Distinct profiles for autophosphorylation were observed, suggesting that their catalytic activities are differentially autoregulated. Both enzymes demonstrated the characteristic qualities of NPPases, signifying that NPPs are conserved in yeast.
NPP1 and NPP2 are important for cellular uptake of extracellular nucleotide-derived phosphates.
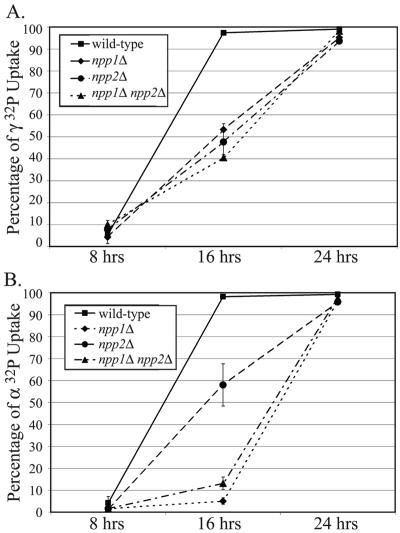
To determine if these genes were responsible for the observed extracellular nucleotide phosphate hydrolysis, null mutants were tested for their ability to perform hydrolysis under low-phosphate conditions. Experiments were conducted parallel to those in Fig. 1 over a 24-h time course monitoring hydrolysis at early-, mid-, and late-log-phase growth. Using both [γ-32P]ATP and [α-32P] ATP, the mutant strains were assayed for levels of imported radiolabeled phosphate (Fig. 3A). When γ-32P uptake was monitored, strains lacking either NPP1 or NPP2 could import only 50% of the radiolabeled phosphate by 16 h. In the double mutant, the defect in phosphate hydrolysis was slightly more pronounced. These defects are in stark comparison to the wild type, which could import nearly the entire radiolabeled probe by this time point. Defects in these mutant strains were also seen when α-32P uptake was measured, which correlates with complete phosphate hydrolysis from the nucleotide moiety (Fig. 3B). While nearly 60% of the radiolabel was imported in the npp2Δ mutant, only 5% of the radiolabel was imported in the npp1Δ mutant at 16 h. The decreased rate of α-phosphate hydrolysis may simply be due to catalytic rates of hydrolysis, where more Npp2p was found to be in an autoinhibited, phosphorylated state than Npp1p. Yet this notable result also demonstrates that NPP1 may preferentially utilize substrates at the α-phosphate position to perform phosphodiester hydrolysis. The hydrolysis defect was comparable in a double null mutant where about 10% of the radiolabel was detected in the cellular lysate. However, by 24 h, nearly all of the α-32P and γ-32P could be imported in all three mutant strains, indicating that the absence of activity from these genes may be compensated from another source. Accordingly, Npp1p and Npp2p demonstrate conserved E-NPP activity in yeast and partially contribute to nucleotide-derived phosphate hydrolysis during phosphate starvation.
FIG. 3.
Effects of yeast E-NPPs on phosphate uptake and extracellular nucleotide-derived phosphate hydrolysis. (A) Strains lacking NPP1 and/or NPP2 were evaluated for their contributions to nucleotide phosphate hydrolysis as a function of detected phosphate uptake. Extracellular [γ-32P]ATP was utilized as a substrate, and cultures were grown over a 24-h time course in phosphate-depleted medium. A loss of either or both genes caused a defect in phosphate uptake. (B) Extracellular [α-32P]ATP was utilized to measure phosphate hydrolysis at the proximal position. Loss of either or both genes enhanced the defect of phosphate hydrolysis; the effect was most severe in strains lacking both genes.
E-NPPs can hydrolyze nucleotide-derived phosphates, particularly during phosphate starvation.
The absence of either or both genes encoding E-NPPs caused a substantial defect in measured phosphate uptake, indicating that both gene products can perform extracellular nucleotide hydrolysis. To establish that these gene products are directly utilizing nucleotide substrates for phosphate hydrolysis, mutant strains lacking NPP1 and/or NPP2 were tested for their ability to hydrolyze nucleotide phosphates in a direct NPPase assay using ATP as a substrate in a coupled enzymatic assay where ATP hydrolysis was detected as a function of NADH oxidation (Fig. 4A). Under normal phosphate conditions, a low level of NPPase activity was present in all strains at levels comparable to that of a wild-type strain. When grown under low-phosphate conditions, NPPase activity in the wild-type strain was greatly enhanced by 16 and 24 h. Strains lacking either NPP1 or NPP2 had no increased NPPase activity by 16 h but had a minor increase in activity by 24 h, although activity was <50% of that in the wild type. The double mutant was slightly worse, where NPPase activity levels were approximately 30% of the wild type. Thus, Npp1p and Npp2p can hydrolyze extracellular phosphate from ATP, and this activity is greatly enhanced when extracellular phosphate concentrations are low.
Since activity depends on phosphate starvation conditions, protein expression levels of Npp1p and Npp2p were monitored over a 24-h time course from early to late log phase under both normal and low-phosphate growth conditions (Fig. 4B). At each time point, cell lysates were prepared, and expression levels of TAP-tagged Npp1p and Npp2p were detected by immunoblotting. Protein expression levels were nearly undetectable at 0 h (stationary-growth phase), but levels were increased during active log-phase growth. In correlation with enhanced enzymatic activity, protein expression levels for both proteins were significantly augmented under low-phosphate conditions, particularly for Npp2p at later time points. These protein expression profiles may implicate Npp1p as an early responder for NPPase activity, and Npp2p may subsequently assist in sustaining the response over time by delayed expression. This may also demonstrate that under low-phosphate conditions, Npp1p and Npp2p play complementary roles for regulation of extracellular nucleotide phosphate hydrolysis during phosphate starvation.
Pho5p, a secreted acid phosphatase, partially mediates nucleotide-derived phosphate hydrolysis.
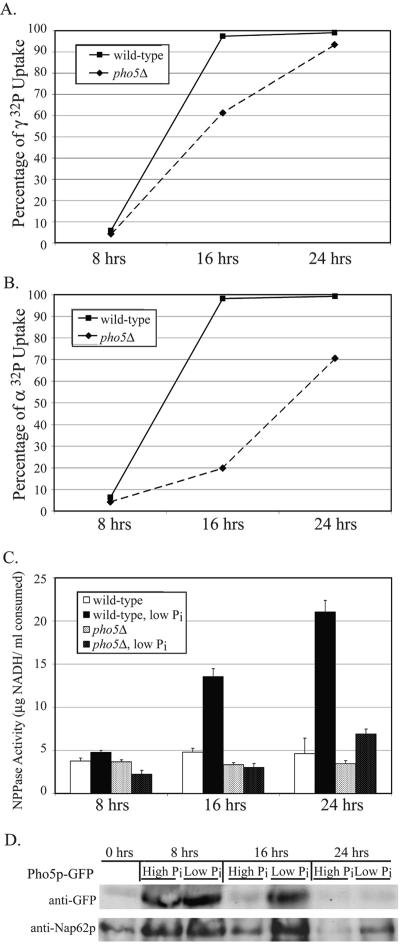
While Npp1p and Npp2p appear to be highly activated during phosphate starvation, the possibility that an element of the PHO response may be involved in extracellular nucleotide phosphate hydrolysis was also investigated. The prospect seemed likely, since this activity is strongly phosphate dependent and only a 50% reduction in NPPase activity was seen in the absence of NPP1 and NPP2. When the PHO response system is activated, the repressible acid phosphatase encoded by PHO5 is upregulated and plays a major role in extracellular phosphate scavenging (47). Although two other acid phosphatases are also upregulated during this response (Pho11p and Pho12p), >90% of the detectable phosphatase activity was due to Pho5p. To determine if PHO5 contributes to phosphate hydrolysis of extracellular nucleotides, a pho5Δ mutant was assayed for its ability to internalize phosphate. Both [α-32P]- and [γ-32P]ATP were tested as potential substrates (Fig. 5A and B). After 16 h of growth, the wild-type cells imported nearly all of the radiolabeled phosphate. However, the pho5Δ strain imported 60% of the γ-32P and only 20% of the α-32P at the same time point. By 24 h, the pho5Δ mutant approached the wild-type level of imported γ-32P but still lagged in importing α-32P. Although nucleotide phosphate hydrolysis still occurred in the pho5Δ mutant, the absence of Pho5p caused a significant decrease in the rate of hydrolysis. The defect was most pronounced in the instance with AMP as a substrate. In addition to Npp1p and Npp2p, Pho5p appears to play a significant role in phosphate-mediated uptake from extracellular nucleotide-derived phosphoryl substrates, and this function is distinct from its previously characterized activities.
FIG. 5.
Nucleotide phosphate hydrolysis and uptake is also affected by Pho5p, a component of the PHO response. (A) The ability to import extracellular phosphate from nucleotide sources was determined in pho5-null mutants. Cells were grown in low-phosphate medium containing [γ-32P]ATP, and the rate of imported phosphate was compared to a wild-type strain over 24 h. The mutant strain was unable to hydrolyze and subsequently import phosphate as effectively as the wild type. (B) Hydrolysis of [α-32P] from ATP was measured over a 24-h time course. The pho5Δ mutant had a pronounced shortcoming in phosphate hydrolysis, as demonstrated by the lack of imported phosphate at 16 and 24 h, but still less severe than the npp1Δ npp2Δ double mutant. (C) Utilization of ATP as an extracellular phosphate substrate was tested by comparing a pho5Δ mutant to a wild-type strain grown in low-phosphate medium containing ATP. E-NPP activity was measured over a 24-h time course, where utilization of ATP as a substrate for APase activity was measured as a function of oxidized NADH. The lack of activity measured in the pho5Δ mutant demonstrated that ATP is a substrate of Pho5p. (D) Protein expression levels of Pho5p were measured over 24 h from cultures grown in YPD or phosphate-depleted medium. Protein levels were detected by immunoblotting against Pho5p-GFP fusions. Pho5p expression is enhanced during phosphate starvation.
Pho5p can utilize nucleotide-derived phosphates as substrates.
Since the absence of PHO5 caused a notable reduction in nucleotide-derived phosphate cleavage, this raised the possibility that Pho5p may also directly utilize extracellular ATP as a substrate. This model was tested using a coupled enzyme assay to monitor ATP hydrolysis (Fig. 5C). Both wild-type and pho5Δ strains were grown in low-phosphate medium, and APase activity was determined as a function of time. As cultures reached mid- and late-log-phase growth, the rate of ATP hydrolysis was significantly upregulated in the wild type. The pho5Δ mutant demonstrated no upregulation by early- or mid-log-phase growth. Even by late-log-phase growth, the level of ATP hydrolysis by the pho5Δ mutant only slightly increased, and this level of defective hydrolysis was comparable to the rate seen with the npp1Δ npp2Δ double mutant. The defect in the pho5Δ mutant strongly indicates that Pho5p plays a significant role alongside Npp1p and Npp2p in performing phosphate hydrolysis with extracellular ATP.
The observed loss of nucleotide phosphate hydrolysis in the absence of PHO5 should correlate with enhanced enzymatic activity. To establish that Pho5p expression is increased under these growth conditions, Pho5p expression levels were monitored over a 24-h time course under high- and low-phosphate conditions (Fig. 5D). Protein levels increased during log-phase growth and were enhanced under phosphate starvation conditions. Pho5p demonstrated the greatest increase of expression by 8 h, indicating that Pho5p is rapidly expressed alongside Npp1p. Thus, Pho5p and Npp1p may perform some redundancy in performing early NPPase activity, and this precedes the time-delayed response of Npp2p. This evidence links the increased nucleotide phosphate hydrolysis activity demonstrated in a wild-type strain through primary enhanced expression levels of Npp1p and Pho5p, followed by the secondary enhancement Npp2p, to collectively perform NPPase activity during phosphate starvation.
NPP1, NPP2, and PHO5 are principal components of NPPase activity in S. cerevisiae.
Three separate genes from two distinct gene classes were identified to perform NPP activity in yeast. All three genes appear to play a large role in overall activity, although their function appears to be redundant. It remains unknown whether there are any other genes that can additionally contribute to NPP activity and whether NPP activity is even a necessary cellular event. To address these questions, a strain lacking NPP1, NPP2, and PHO5 was generated. The strain was used to determine the extent of defectiveness in extracellular nucleotide phosphate hydrolysis. The rate of imported γ-32P from extracellular ATP was measured over 24 h (Fig. 6A). By the end of the time course, the triple-null strain could intracellularly import only about half of the total γ-32P, compared to nearly 100% in either the npp1Δ npp2Δ or pho5Δ strains. To determine whether the triple-null strain could fully dephosphorylate all three phosphate groups from ATP, α-32P ATP was provided extracellularly. Phosphate hydrolysis and import were subsequently measured under low-phosphate conditions (Fig. 6B). In this case, only about 20% of the total α-32P was hydrolyzed by 24 h. This is in stark contrast to the npp1Δ npp2Δ or pho5Δ strains, where hydrolysis reached nearly 100% or 70%, respectively, throughout the same growth period. This clearly demonstrates that all three genes are major components of the NPPase activity.
FIG. 6.
NPP1, NPP2, and PHO5 are the major contributors for NPPase activity. (A) Extracellular phosphate hydrolysis by the triple-null strain was monitored through measuring phosphate import over 24 h. Extracellular [γ-32P]ATP was utilized as a substrate. At the end of the time course, a 50% defect in phosphate uptake was measured in the triple-null strain. (B) Extracellular [α-32P]ATP was utilized to test phosphate hydrolysis at the proximal position. The absence of all three genes caused a nearly complete loss of phosphate hydrolysis at the α-position of ATP. (C) NPPase activity was measured in the triple-null strain using the coupled enzyme assay with ATP as a substrate. The triple-null strain demonstrated nominal NPPase activity throughout the entire time course.
The NPPase activity of the triple-null strain was further tested by directly monitoring ATP hydrolysis over time (Fig. 6C). Using the coupled enzyme assay, ATP hydrolysis was measured at various time points after cells were grown in either YPD or phosphate-depleted medium. At each time point, the triple-null strain demonstrated diminished NPPase activity and was slightly more defective than the npp1Δ npp2Δ or pho5Δ strains alone. The level of NPPase activity measured was comparable to basal activity demonstrated under normal YPD growth conditions. This demonstrates that NPP1, NPP2, and PHO5 are collectively the major contributors to NPPase activity in yeast and interconnects the possibility that NPPase activity and the PHO response may share overlapping regulatory roles in S. cerevisiae.
DISCUSSION
Yeast homologs of the E-NPP family can hydrolyze nucleotide phosphates.
In many eukaryotes, E-NPPs are directly responsible for phosphate hydrolysis from extracellular nucleotides. Based on sequence criteria, we identified two candidate E-NPPs in the S. cerevisiae genome. They were both evaluated for their roles during phosphate starvation, and both demonstrated the capability of performing extracellular phosphate hydrolysis from nucleotide substrates. We have demonstrated that both Npp1p and Npp2p are functional nucleotide pyrophosphatase/phosphodiesterases. Both enzymes catalyzed nucleotide phosphate hydrolysis to produce the trapped nucleotide monophosphate intermediate. These enzymes can subsequently catalyze hydrolysis of the monophosphate to produce the nucleotide base. Additionally, this is the first demonstration that NPP1 and NPP2 are upregulated via phosphate-regulated transcription. Distinct profiles of catalytic regulation for each enzyme parallels the functional diversity seen among NPPs specialized for particular functions in humans. This is the first documentation of E-NPP activity in yeast and highlights the notion that functional diversity is conserved among the NPP gene family.
E-NPPs may mediate intercellular signaling.
In humans, the fundamental role of the E-NPP family is to regulate extracellular nucleotide metabolism and nucleotide-derived intercellular signaling. While budding yeast is a unicellular organism, intercellular signaling has been documented to coordinate events such as aggregation, differentiation, sexual reproduction, and sporulation (18). During sporulation, differentiating yeast cells cooperate to maintain high concentrations of extracellular purines and produce adenosine 5′-tetraphosphate and adenosine 5′-pentaphosphate to mark ascospore formation (17). Intriguingly, NPP2 expression is strongly induced during sporulation (8). Taken together, this may implicate a novel intercellular signaling role for E-NPPs by regulating extracellular nucleotide metabolism to mediate sporulation and possibly other intercellular signaling events.
Yeast can derive essential phosphate from extracellular nucleotides.
Importing phosphate from extracellular sources during periods of phosphate starvation can be critical for cellular viability. If extracellular phosphate is scarce, sequestering phosphate from diverse chemical substrates would be advantageous for survival. Enzymes responsible for obtaining phosphates from polyinorganic phosphate chains and/or pyrophosphates have been identified in yeast (15, 23, 25, 29, 30, 39). In principle, hydrolysis of phosphates from nucleotide sources should provide an additional nutrient source. The import of phosphate from extracellular nucleotide substrates demonstrates the diversity of substrates utilized to overcome phosphate starvation. This finding is also compelling in that it unites E-NPP regulation with the phosphate starvation response. The PHO response is a powerful mechanism in many unicellular organisms that allows them to adapt to an ever-changing environment. It is intriguing that the repressible expression of several genes that specialize in acquisition and import of available phosphates can bridge its activity with extracellular nucleotide regulation.
A new function for Pho5p as a nucleotide phosphatase.
Since extracellular nucleotide-derived phosphate hydrolysis was significantly upregulated during periods of phosphate starvation, several components of the PHO response were investigated for their contributions. When the PHO response is activated, the secreted acid phosphatase Pho5p is the most active acid phosphatase for scavenging and hydrolyzing phosphates. Our results demonstrate that nucleotide phosphate hydrolysis performed by Pho5p is intertwined with the PHO response during phosphate. It is noteworthy that another component of the PHO response is high-affinity phosphate uptake. In conjunction with Pho5p, phosphate transporters are critical components for generating and importing extracellular free inorganic phosphate to maintain viability during periods of starvation. Additionally, nucleotide dephosphorylation has profound effects in cellular regulation of many mammalian systems. Furthermore, the fact that this activity coincides with the PHO response system may implicate an integrated scheme of regulation during phosphate starvation through multiple modes of cellular regulation.
A model of nucleotide-derived phosphate acquisition.
Under high-phosphate conditions, low levels of nucleotide phosphates are hydrolyzed by constitutive E-NPP activity composed primarily of NPP1, NPP2, and PHO5. Such basal levels of inorganic phosphate stimuli can be detected by phosphate transporters to define an adequate level of available inorganic phosphate for consumption by various biological processes (3). If extracellular phosphate becomes depleted, then phosphate starvation is sensed. This leads to the upregulation of a robust, multicomponent system of proteins known as the PHO response. This response system involves dual upregulation of phosphatases to generate free inorganic phosphate and phosphate symporters to import the generated inorganic phosphate. Our results provide the first evidence that nucleotide-derived phosphates can be hydrolyzed extracellularly in budding yeast by Npp1p, Npp2p, and Pho5p during phosphate starvation. Once free inorganic phosphate has been generated extracellularly, the repressible phosphate symporters Pho84p and Pho89p are presumably the major contributors to phosphate import to maintain cellular viability (Fig. 7).
FIG. 7.
Regulation of nucleotide-derived phosphate hydrolysis. Under low-phosphate conditions, the PHO response upregulates expression of a collection of repressible, phosphate-responsive genes. This includes the high-affinity transporters Pho84p and Pho89p, as well as the major repressible acid phosphatase Pho5p. The phosphate transporters and acid phosphatases are either subsequently transported to the plasma membrane or secreted to scavenge and import extracellular phosphate. The E-NPP proteins are constitutively expressed at low levels but have increased expression during active cellular growth. Npp1p and Npp2p have a predicted extracellular domain and can carry out nucleotide-derived phosphate hydrolysis. In conjunction with Pho5p, Npp1p and Npp2p can hydrolyze ATP to produce ADP, AMP, adenosine, and free inorganic phosphate. Under low-phosphate conditions, the high-affinity phosphate transporters can subsequently import the generated pools of inorganic phosphate. This provides a means of overcoming phosphate starvation and may point to regulation of extracellular nucleotides, uniting the PHO response system with E-NPP regulation. Circles bearing the letter A represent the adenosine nucleotide.
In summary, our results establish that the E-NPP family is conserved in yeast and can perform extracellular nucleotide phosphate hydrolysis. Extracellular nucleotides, particularly adenosine, have been found to play many related regulatory roles in cell signaling in multicellular organisms, including platelet aggregation (36), differentiation (20), cell proliferation (11), and apoptosis (16, 37). NPP1 and NPP2 remain conserved in S. cerevisiae and possibly in several other diverse fungal species, raising the possibility that these genes may also perform critical biological roles in yeast. We also establish a new role for Phop5p in sharing overlapping activity with the E-NPP family. These studies establish a mechanism for extracellular nucleotide regulation that is distinct from multicellular organisms by uniting its function with the phosphate starvation response. It will be important to investigate whether the regulation and biological roles of extracellular nucleotides by the E-NPP family are conserved among unicellular and multicellular organisms or identify what divergent roles have been adapted over evolution. It will also be exciting to establish whether the PHO response plays a larger regulatory role in extracellular nucleotide regulation and further to ascertain how these distinct regulatory mechanisms complement the function of the other.
Acknowledgments
We thank Olga Savinova for her contributions to other investigative aspects of this project not presented in this work. We also thank Douglass Forbes for generously providing the anti-Nap62p antibody for Western blotting controls. We appreciate critical comments from Johan Thevelein, Paul Insel, Susan Taylor, Tom Huxford, Renee Garza, Randy Hampton, Mike Burkart, and members of the Pillus laboratory.
This work has been supported by the Minority Access to Science, Engineering and Mathematics Predoctoral Fellowship (NSF), a Cellular and Molecular Genetics Training Grant (NIH), and an American Heart Association Predoctoral Fellowship for E.J.K.; funding from NIH for the laboratories of L.P. and G.G.; and funding from the Human Frontier Science Program for the laboratory of G.G.
REFERENCES
- 1.Amberg, D. C., D. J. Burke, and J. N. Strathern. 2005. Methods in yeast genetics: a Cold Spring Harbor Laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 2.Auesukaree, C., T. Homma, Y. Kaneko, and S. Harashima. 2003. Transcriptional regulation of phosphate-responsive genes in low-affinity phosphate-transporter-defective mutants in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 306:843-850. [DOI] [PubMed] [Google Scholar]
- 3.Auesukaree, C., T. Homma, H. Tochio, M. Shirakawa, Y. Kaneko, and S. Harashima. 2004. Intracellular phosphate serves as a signal for the regulation of the PHO pathway in Saccharomyces cerevisiae. J. Biol. Chem. 279:17289-17294. [DOI] [PubMed] [Google Scholar]
- 4.Blytt, H. J., J. E. Brotherton, and L. Butler. 1985. Assay of covalent intermediate of 5′-nucleotide phosphodiesterase. Anal. Biochem. 147:517-520. [DOI] [PubMed] [Google Scholar]
- 5.Canani, L. H., D. P. Ng, A. Smiles, J. J. Rogus, J. H. Warram, and A. S. Krolewski. 2002. Polymorphism in ecto-nucleotide pyrophosphatase/phosphodiesterase 1 gene (ENPP1/PC-1) and early development of advanced diabetic nephropathy in type 1 diabetes. Diabetes 51:1188-1193. [DOI] [PubMed] [Google Scholar]
- 6.Carroll, A. S., and E. K. O'Shea. 2002. Pho85 and signaling environmental conditions. Trends Biochem. Sci. 27:87-93. [DOI] [PubMed] [Google Scholar]
- 7.Cherry, J. M. 1995. Genetic nomenclature guide. Saccharomyces cerevisiae. Trends Genet. Mar.:11-12. [PubMed] [Google Scholar]
- 8.Chu, S., J. DeRisi, M. Eisen, J. Mulholland, D. Botstein, P. O. Brown, and I. Herskowitz. 1998. The transcriptional program of sporulation in budding yeast. Science 282:699-705. [DOI] [PubMed] [Google Scholar]
- 9.Cook, P. F., M. E. Neville, Jr., K. E. Vrana, F. T. Hartl, and R. Roskoski, Jr. 1982. Adenosine cyclic 3′,5′-monophosphate dependent protein kinase: kinetic mechanism for the bovine skeletal muscle catalytic subunit. Biochemistry 21:5794-5799. [DOI] [PubMed] [Google Scholar]
- 10.Funakoshi, I., H. Kato, K. Horie, T. Yano, Y. Hori, H. Kobayashi, T. Inoue, H. Suzuki, S. Fukui, M. Tsukahara, et al. 1992. Molecular cloning of cDNAs for human fibroblast nucleotide pyrophosphatase. Arch. Biochem. Biophys. 295:180-187. [DOI] [PubMed] [Google Scholar]
- 11.Gendaszewska-Darmach, E., M. Maszewska, M. Zaklos, and M. Koziolkiewicz. 2003. Degradation of extracellular nucleotides and their analogs in HeLa and HUVEC cell cultures. Acta Biochim. Pol. 50:973-984. [PubMed] [Google Scholar]
- 12.Gijsbers, R., H. Ceulemans, and M. Bollen. 2003. Functional characterization of the non-catalytic ectodomains of the nucleotide pyrophosphatase/phosphodiesterase NPP1. Biochem. J. 371:321-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gijsbers, R., H. Ceulemans, W. Stalmans, and M. Bollen. 2001. Structural and catalytic similarities between nucleotide pyrophosphatases/phosphodiesterases and alkaline phosphatases. J. Biol. Chem. 276:1361-1368. [DOI] [PubMed] [Google Scholar]
- 14.Goding, J. W., B. Grobben, and H. Slegers. 2003. Physiological and pathophysiological functions of the ecto-nucleotide pyrophosphatase/phosphodiesterase family. Biochim. Biophys. Acta 1638:1-19. [DOI] [PubMed] [Google Scholar]
- 15.Gonzalez, M. A., M. R. Webb, K. M. Welsh, and B. S. Cooperman. 1984. Evidence that catalysis by yeast inorganic pyrophosphatase proceeds by direct phosphoryl transfer to water and not via a phosphoryl enzyme intermediate. Biochemistry 23:797-801. [DOI] [PubMed] [Google Scholar]
- 16.Hashikawa, T., S. W. Hooker, J. G. Maj, C. J. Knott-Craig, M. Takedachi, S. Murakami, and L. F. Thompson. 2004. Regulation of adenosine receptor engagement by ecto-adenosine deaminase. FASEB J. 18:131-133. [DOI] [PubMed] [Google Scholar]
- 17.Jakubowski, H. 1986. Sporulation of the yeast Saccharomyces cerevisiae is accompanied by synthesis of adenosine 5′-tetraphosphate and adenosine 5′-pentaphosphate. Proc. Natl. Acad. Sci. USA 83:2378-2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jakubowski, H., and E. Goldman. 1988. Evidence for cooperation between cells during sporulation of the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 8:5166-5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin-Hua, P., J. W. Goding, H. Nakamura, and K. Sano. 1997. Molecular cloning and chromosomal localization of PD-Iβ (PDNP3), a new member of the human phosphodiesterase I genes. Genomics 45:412-415. [DOI] [PubMed] [Google Scholar]
- 20.Johnson, K., J. Goding, D. Van Etten, A. Sali, S. I. Hu, D. Farley, H. Krug, L. Hessle, J. L. Millan, and R. Terkeltaub. 2003. Linked deficiencies in extracellular PP(i) and osteopontin mediate pathologic calcification associated with defective PC-1 and ANK expression. J. Bone Miner. Res. 18:994-1004. [DOI] [PubMed] [Google Scholar]
- 21.Johnson, K., and R. Terkeltaub. 2005. Inorganic pyrophosphate (PPI) in pathologic calcification of articular cartilage. Front. Biosci. 10:988-997. [DOI] [PubMed] [Google Scholar]
- 22.Kawagoe, H., O. Soma, J. Goji, N. Nishimura, M. Narita, J. Inazawa, H. Nakamura, and K. Sano. 1995. Molecular cloning and chromosomal assignment of the human brain-type phosphodiesterase I/nucleotide pyrophosphatase gene (PDNP2). Genomics 30:380-384. [DOI] [PubMed] [Google Scholar]
- 23.Kumble, K. D., and A. Kornberg. 1996. Endopolyphosphatases for long chain inorganic polyphosphate in yeast and mammals. J. Biol. Chem. 271:27146-27151. [DOI] [PubMed] [Google Scholar]
- 24.Lenburg, M. E., and E. K. O'Shea. 1996. Signaling phosphate starvation. Trends Biochem. Sci. 21:383-387. [PubMed] [Google Scholar]
- 25.Lichko, L. P., N. A. Andreeva, T. V. Kulakovskaya, and I. S. Kulaev. 2003. Exopolyphosphatases of the yeast Saccharomyces cerevisiae. FEMS Yeast Res. 3:233-238. [DOI] [PubMed] [Google Scholar]
- 26.Marchler-Bauer, A., J. B. Anderson, C. DeWeese-Scott, N. D. Fedorova, L. Y. Geer, S. He, D. I. Hurwitz, J. D. Jackson, A. R. Jacobs, C. J. Lanczycki, C. A. Liebert, C. Liu, T. Madej, G. H. Marchler, R. Mazumder, A. N. Nikolskaya, A. R. Panchenko, B. S. Rao, B. A. Shoemaker, V. Simonyan, J. S. Song, P. A. Thiessen, S. Vasudevan, Y. Wang, R. A. Yamashita, J. J. Yin, and S. H. Bryant. 2003. CDD: a curated Entrez database of conserved domain alignments. Nucleic Acids Res. 31:383-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martinez, P., and B. L. Persson. 1998. Identification, cloning and characterization of a derepressible Na+-coupled phosphate transporter in Saccharomyces cerevisiae. Mol. Gen. Genet. 258:628-638. [DOI] [PubMed] [Google Scholar]
- 28.Martinez, P., R. Zvyagilskaya, P. Allard, and B. L. Persson. 1998. Physiological regulation of the derepressible phosphate transporter in Saccharomyces cerevisiae. J. Bacteriol. 180:2253-2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neef, D. W., and M. P. Kladde. 2003. Polyphosphate loss promotes SNF/SWI- and Gcn5-dependent mitotic induction of PHO5. Mol. Cell. Biol. 23:3788-3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ogawa, N., J. DeRisi, and P. O. Brown. 2000. New components of a system for phosphate accumulation and polyphosphate metabolism in Saccharomyces cerevisiae revealed by genomic expression analysis. Mol. Biol. Cell 11:4309-4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oshima, Y. 1997. The phosphatase system in Saccharomyces cerevisiae. Genes Genet. Syst. 72:323-334. [DOI] [PubMed] [Google Scholar]
- 32.Oshima, Y., N. Ogawa, and S. Harashima. 1996. Regulation of phosphatase synthesis in Saccharomyces cerevisiae—a review. Gene 179:171-177. [DOI] [PubMed] [Google Scholar]
- 33.Oyajobi, B. O., R. G. Russell, and A. M. Caswell. 1994. Modulation of ecto-nucleoside triphosphate pyrophosphatase activity of human osteoblast-like bone cells by 1 alpha,25-dihydroxyvitamin D3, 24R,25-dihydroxyvitamin D3, parathyroid hormone, and dexamethasone. J. Bone Miner. Res. 9:1259-1266. [DOI] [PubMed] [Google Scholar]
- 34.Pattison-Granberg, J., and B. L. Persson. 2000. Regulation of cation-coupled high-affinity phosphate uptake in the yeast Saccharomyces cerevisiae. J. Bacteriol. 182:5017-5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Persson, B. L., J. O. Lagerstedt, J. R. Pratt, J. Pattison-Granberg, K. Lundh, S. Shokrollahzadeh, and F. Lundh. 2003. Regulation of phosphate acquisition in Saccharomyces cerevisiae. Curr. Genet. 43:225-244. [DOI] [PubMed] [Google Scholar]
- 36.Rodrigues, C. O., P. M. Dutra, F. S. Barros, T. Souto-Padron, J. R. Meyer-Fernandes, and A. H. Lopes. 1999. Platelet-activating factor induction of secreted phosphatase activity in Trypanosoma cruzi. Biochem. Biophys. Res. Commun. 266:36-42. [DOI] [PubMed] [Google Scholar]
- 37.Schrier, S. M., B. I. Florea, G. J. Mulder, J. F. Nagelkerke, and I. J. AP. 2002. Apoptosis induced by extracellular ATP in the mouse neuroblastoma cell line N1E-115: studies on involvement of P2 receptors and adenosine. Biochem. Pharmacol. 63:1119-1126. [DOI] [PubMed] [Google Scholar]
- 38.Serrano, R., A. Ruiz, D. Bernal, J. R. Chambers, and J. Arino. 2002. The transcriptional response to alkaline pH in Saccharomyces cerevisiae: evidence for calcium-mediated signalling. Mol. Microbiol. 46:1319-1333. [DOI] [PubMed] [Google Scholar]
- 39.Sethuraman, A., N. N. Rao, and A. Kornberg. 2001. The endopolyphosphatase gene: essential in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 98:8542-8547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sherman, F. 2002. Getting started with yeast. Methods Enzymol. 350:3-41. [DOI] [PubMed] [Google Scholar]
- 41.Shnyreva, M. G., E. V. Petrova, S. N. Egorov, and A. Hinnen. 1996. Biochemical properties and excretion behavior of repressible acid phosphatases with altered subunit composition. Microbiol. Res. 151:291-300. [DOI] [PubMed] [Google Scholar]
- 42.Stefan, C., W. Stalmans, and M. Bollen. 1996. Threonine autophosphorylation and nucleotidylation of the hepatic membrane protein PC-1. Eur. J. Biochem. 241:338-342. [DOI] [PubMed] [Google Scholar]
- 43.Stracke, M. L., H. C. Krutzsch, E. J. Unsworth, A. Arestad, V. Cioce, E. Schiffmann, and L. A. Liotta. 1992. Identification, purification, and partial sequence analysis of autotaxin, a novel motility-stimulating protein. J. Biol. Chem. 267:2524-2529. [PubMed] [Google Scholar]
- 44.Svaren, J., and W. Horz. 1997. Transcription factors vs nucleosomes: regulation of the PHO5 promoter in yeast. Trends Biochem. Sci. 22:93-97. [DOI] [PubMed] [Google Scholar]
- 45.Tamai, Y., A. Toh-e, and Y. Oshima. 1985. Regulation of inorganic phosphate transport systems in Saccharomyces cerevisiae. J. Bacteriol. 164:964-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Winzeler, E. A., D. D. Shoemaker, A. Astromoff, H. Liang, K. Anderson, B. Andre, R. Bangham, R. Benito, J. D. Boeke, H. Bussey, A. M. Chu, C. Connelly, K. Davis, F. Dietrich, S. W. Dow, M. El Bakkoury, F. Foury, S. H. Friend, E. Gentalen, G. Giaever, J. H. Hegemann, T. Jones, M. Laub, H. Liao, R. W. Davis, et al. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285:901-906. [DOI] [PubMed] [Google Scholar]
- 47.Wykoff, D. D., and E. K. O'Shea. 2001. Phosphate transport and sensing in Saccharomyces cerevisiae. Genetics 159:1491-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yano, Y., Y. Hayashi, K. Sano, H. Nagano, M. Nakaji, Y. Seo, T. Ninomiya, S. Yoon, H. Yokozaki, and M. Kasuga. 2004. Expression and localization of ecto-nucleotide pyrophosphatase/phosphodiesterase I-1 (E-NPP1/PC-1) and -3 (E-NPP3/CD203c/PD-Iβ/B10/gp130RB13-6) in inflammatory and neoplastic bile duct diseases. Cancer Lett. 207:139-147. [DOI] [PubMed] [Google Scholar]