Abstract
Glycosylphosphatidylinositols (GPIs) are attached to the C termini of some glycosylated secretory proteins, serving as membrane anchors for many of those on the cell surface. Biosynthesis of GPIs is initiated by the transfer of N-acetylglucosamine (GlcNAc) from UDP-GlcNAc to phosphatidylinositol. This reaction is carried out at the endoplasmic reticulum (ER) by an enzyme complex called GPI-N-acetylglucosaminyltransferase (GPI-GlcNAc transferase). The human enzyme has six known subunits, at least four of which, GPI1, PIG-A, PIG-C, and PIG-H, have functional homologs in the budding yeast Saccharomyces cerevisiae. The uncharacterized yeast gene YDR437w encodes a protein with some sequence similarity to human PIG-P, a fifth subunit of the GPI-GlcNAc transferase. Here we show that Ydr437w is a small but essential subunit of the yeast GPI-GlcNAc transferase, and we designate its gene GPI19. Similar to other mutants in the yeast enzyme, temperature-sensitive gpi19 mutants display cell wall defects and hyperactive Ras phenotypes. The Gpi19 protein associates with the yeast GPI-GlcNAc transferase in vivo, as judged by coimmuneprecipitation with the Gpi2 subunit. Moreover, conditional gpi19 mutants are defective for GPI-GlcNAc transferase activity in vitro. Finally, we present evidence for the topology of Gpi19 within the ER membrane.
Glycosylphosphatidylinositols (GPIs) are complex glycolipids that anchor a variety of glycoproteins to the external surface of the plasma membrane (8, 24, 25, 44). GPI-anchored surface proteins of Leishmania, Plasmodium falciparum, and various trypanosome species protect the cell surface of these parasitic protozoa from the host's immune system (8). In humans, a GPI-anchoring deficiency resulting from somatic mutation of the PIG-A gene causes the acquired hematopoietic stem cell disorder paroxysmal nocturnal hemoglobinuria (2). Many of the clinical features of paroxysmal nocturnal hemoglobinuria (e.g., intracellular hemolysis, peripheral blood cytopenias, and thrombosis) can be explained by the absence of GPI-anchored proteins on the surface of blood cells. In Saccharomyces cerevisiae, the GPI initially present on some surface glycoproteins has been proposed to participate in a transglycosylation reaction in which the glycoprotein becomes cross-linked to cell wall β-glucan (7).
GPIs, which have the conserved core structure protein-CO-NH-CH2-CH2-PO4-6-mannose (Man)-α1,2-Man-α1,6-Man-α1,4-GlcN-α1,6-inositol-PO4-lipid, are preassembled at the endoplasmic reticulum (ER) in a multistep pathway prior to their transfer to target proteins (reviewed in references 8, 20, 25, 28, and 39). Many genes involved in the GPI anchoring pathway have been identified, and in most cases yeast and mammalian GPI assembly proteins display amino acid sequence similarity. Mutations blocking GPI assembly or transfer to proteins are lethal in yeast.
The first step in GPI assembly, the transfer of N-acetylglucosamine (GlcNAc) from UDP-GlcNAc to an acceptor phosphatidylinositol (PI) is catalyzed by a multiprotein complex that resides in the ER membrane. The human GPI-GlcNAc transferase complex is an unusually elaborate glycosyltransferase, possessing at least six subunits (44). Four of these, GPI1, PIG-A, PIG-C, and PIG-H, have structural and functional counterparts in the S. cerevisiae Gpi1, Gpi3, Gpi2, and Gpi15 proteins, respectively (15, 22, 23, 27, 31, 40, 42, 43, 46). Among these proteins, PIG-A/Gpi3 is the UDP-GlcNAc-binding and likely catalytic subunit, based both on its sequence similarity to members of a large family of glycosyltransferases (10, 18, 42) and on cross-linking studies using a photoactivatable analogue of UDP-GlcNAc (21). A potential fifth subunit of the mammalian complex has been identified only as a copurifying 5-kDa protein, which may be the homologue of the recently discovered Eri1 subunit of the S. cerevisiae complex, which is similar in size (37). Loss of function mutations in most subunits of the GPI-GlcNAc transferase completely blocks GPI synthesis. The only known exceptions to this are yeast Eri1 and human and yeast Gpi1; loss of either subunit in yeast results in a temperature-sensitive growth defect (23, 37).
The sixth identified subunit of the human GPI-GlcNAc transferase, PIG-P, shares 20% sequence identity to the product of an uncharacterized S. cerevisiae open reading frame of 140 codons, YDR437w (44). Interestingly, the human PIG-P gene, which resides on chromosome 21, is also known as DSCR5, for Down Syndrome critical region 5 (44). Down Syndrome (DS; trisomy 21) is the most common genetic cause of mental retardation. The PIG-P protein is overexpressed twofold in fetal DS brain tissue, leading to the suggestion that aberrant GPI anchoring interferes with brain development (9). Moreover, PIG-P is evidently the only DSCR gene that is expressed in tongue tissue, prompting the suggestion that its overexpression plays a role in the pathophysiology of tongue malformation in DS patients (4). We demonstrate here that YDR437w encodes an essential subunit of the yeast GPI-GlcNAc transferase, and we henceforth refer to this gene as GPI19 (for glycosylphosphatidylinositol anchor 19) in accordance with the nomenclature of other yeast genes involved in GPI anchor production.
MATERIALS AND METHODS
Strains, growth conditions, and transformations.
The S. cerevisiae strains used in this study are listed in Table 1. Yeast cultures were grown in YEPD (1% Bacto yeast extract, 2% Bacto peptone, 2% glucose). Synthetic minimal (SD) medium (30) supplemented with the appropriate nutrients was used to select for plasmid maintenance and gene replacement. Yeast transformations were carried out by the lithium acetate method (16). Escherichia coli DH5α was used to propagate all plasmids. Escherichia coli cells were cultured in Luria broth medium (1% Bacto tryptone, 0.5% Bacto yeast extract, 1% NaCl) and transformed to carbenicillin resistance by standard methods.
TABLE 1.
S. cerevisiae strains useda
| Strain | Relevant genotype | Source |
|---|---|---|
| 1783 | MATaleu2-3, 112 trp1-1 ura3-52 his4 can1r | I. Herskowitz |
| 1784 | MATα | I. Herskowitz |
| 1788 | MATa/α | I. Herskowitz |
| DL2987 | MATa/α gpi19Δ::LEU2/GPI19 | This study |
| DL3050 | MATagpi19Δ::LEU2 pRS316[GPI19] | This study |
| DL3087 | MATagpi19Δ::LEU2 pRS314[gpi19-2] | This study |
| DL3089 | MATagpi19Δ::LEU2 pRS314[gpi19-5] | This study |
| DL3091 | MATagpi19Δ::LEU2 pRS314[gpi19-4] | This study |
| DL3093 | MATagpi19Δ::LEU2 pRS314[gpi19-1] | This study |
| DL3095 | MATagpi19Δ::LEU2 pRS314[gpi19-6] | This study |
| DL3097 | MATagpi19Δ::LEU2 pRS314[gpi19-3] | This study |
| DL3099 | MATagpi19Δ::LEU2 pRS314[GPI19] | This study |
| DL3102 | MATa/α gpi19Δ::LEU2/gpi19Δ::LEU2 pRS314[gpi19-4] | This study |
| DL3103 | MATa/α gpi19Δ::LEU2/gpi19Δ::LEU2 pRS314[gpi19-5] | This study |
| DL3104 | MATa/α gpi19Δ::LEU2/gpi19Δ::LEU2 pRS314[gpi19-3] | This study |
| DL3105 | MATa/α gpi19Δ::LEU2/gpi19Δ::LEU2 pRS314[gpi19-6] | This study |
| DL3106 | MATa/α gpi19Δ::LEU2/gpi19Δ::LEU2 pRS314[gpi19-2] | This study |
| DL3107 | MATa/α gpi19Δ::LEU2/gpi19Δ::LEU2 pRS314[gpi19-1] | This study |
| DL3149 | MATagpi19Δ::LEU2 pRS314[gpi19-7] | This study |
| DL3151 | MATa/α gpi19Δ::LEU2/gpi19Δ::LEU2 pRS314[gpi19-7] | This study |
All strains are derived from the EG123 background (33).
Materials.
UDP[U-14C]GlcNAc (specific activity, 300 mCi/mmol), GDP[1-14C]mannitosol (specific activity, 55 mCi/mmol), and dolichol phosphate (Dol-P) were from American Radiolabeled Chemicals, St. Louis, MO. Zymolyase 20T was obtained from ICN and Endo-H from New England Biolabs.
Genomic deletion of GPI19, cloning, and plasmid construction.
To delete the genomic copy of GPI19 in the EG123 strain background, a DNA fragment spanning from 436 bp 5′ of the GPI19 start codon to 16 bp 3′ of the start codon was amplified by PCR from genomic DNA isolated from yeast strain 1783. A second fragment was amplified from 18 bp 5′ of the GPI19 stop codon to 541 bp 3′ of the stop codon. The 5′ fragment was amplified with primers that placed an XbaI site at the end within the coding sequence and a BamHI site at the opposite end. The 3′ fragment was amplified with primers that placed an ApaI site adjacent to the stop codon and a BamHI site at the opposite end. These fragments were ligated in a three-molecule reaction to the ApaI and XbaI sites of the integrative plasmid pRS305 (32) to create a unique BamHI site between the fragments. The resulting plasmid, pRS305[gpi19Δ::LEU2] (p1955), was linearized with BamHI and used to transform yeast strain 1788 to leucine prototrophy. The replacement of GPI19 sequences was confirmed by PCR. The heterozygous gpi19Δ::LEU2/GPI19 strain (DL2987) was induced to sporulate for tetrad dissection. Viable spores, which segregated 2:2 with inviable spores, were all leucine auxotrophs, confirming the lethality of the deletion.
To construct a genomic clone of GPI19, its coding and regulatory sequences were amplified by PCR from strain 1783 genomic DNA. A 1.1-kb genomic SspI fragment was cloned into the SmaI site of centromeric plasmid pRS316 (32) to yield pRS316[GPI19] (p1995). GPI19 was subcloned into centromeric plasmid pRS314 (32) using the KpnI and SacI sites within the polylinkers of both plasmids to yield pRS314[GPI19] (p1996).
A functional C-terminally hemagglutinin (HA)-tagged form of Gpi19 was constructed as described below. The GPI19 open reading frame was amplified from p1995 and subcloned into pYeF2 (6) using BamHI and NotI sites engineered into the primers. This construction (pYeF2[GPI19]; p2032) fuses Gpi19 in frame at its C terminus with a single copy of the HA epitope and places it under the inducible control of the GAL1 promoter. This plasmid was used to create HA-tagged Gpi19 fusions to invertase (Suc2) for topology studies. Full-length Gpi19-HA was fused to Suc2 by amplification of GPI19-HA with the GAL1 promoter from p2032 using primers that included a SacI site 5′ to the GAL1 promoter and a SalI site immediately 3′ of the HA coding sequence. This fragment was subcloned into the SacI/XhoI sites of pR90 (p2019) (19) to fuse the HA sequence in frame with SUC2. The resulting plasmid, pGAL1::GPI19-HA-SUC2 (p2040), was used to create a related fusion lacking the C-terminal 90 amino acid residues of Gpi19. The GAL1 promoter and the first 150 nucleotides of GPI19 coding sequence was amplified using the 5′ GAL1 primer described above and a primer internal to the GPI19 coding sequence designed with a NotI site. This fragment was subcloned into the SacI/NotI sites of p2040 so as to fuse in frame the N-terminal 50 amino acid residues of Gpi19 with HA-Suc2 (pGAL1::GPI19CΔ-HA-SUC2; p2077).
Isolation of temperature-sensitive gpi19 mutants.
Error-prone PCR was used to generate random mutations within the GPI19 coding sequence. The GPI19 sequence was amplified from p1995 using Taq polymerase and the T7 and T3 primers to include the polylinker sequences on either side of GPI19. Errors were generated during amplification by two modifications to decrease fidelity of the Taq polymerase. First, 0.5 mM MnCl2 was included in each of four separate reactions. Second, a different deoxynucleotide in each of the four reactions was reduced in concentration from 1 mM to 0.1 mM. The products of the four reactions were combined and subcloned into the KpnI/SacI sites of pRS314 (32). Approximately 1,000 independent plasmid clones were isolated and transformed into yeast strain DL3050 (gpi19Δ::LEU2 pRS316[GPI19]) by selection for tryptophan prototrophy. Approximately 3,000 yeast transformants were double replica plated to 5-fluorootic acid (5-FOA)-containing medium (1) at 23°C and 37°C to evict the URA3-based plasmid bearing wild-type GPI19. Replicate colonies that grew in the presence of 5-FOA at low temperature but not at high temperature were isolated for further characterization. A total of eight such colonies were identified. Plasmids were isolated from these temperature-sensitive strains and retransformed into diploid strain DL2987 (gpi19Δ::LEU2/GPI19) for sporulation. DNA sequence analysis of these plasmids revealed that six different gpi19 alleles had been isolated. Mutation sites are available upon request.
The frameshift at codon 25 found among the mutations in both gpi19-5 and gpi19-6 was created in wild-type GPI19 by PCR-mediated site-directed mutagenesis (12) to generate gpi19-7 and was cloned into pRS314 (p2075). Tyr75 was converted to a stop codon (TAA) in gpi19-7 by the same method, creating gpi19-8, which was cloned into pRS314 (p2076). Mutations were confirmed by DNA sequence analysis of the entire GPI19 gene. All primer sequences are available upon request.
In vitro assays of GPI-GlcNAc transferase and Dol-P-mannose synthase.
For GPI-GlcNAc transferase activity, membranes were prepared and assayed for in vitro synthesis of [14C]GlcNAc-PI using a slight modification of the method of Costello and Orlean (5) as described in Sobering et al. (37). The Dol-P-mannose synthase assay was carried out as described by Orlean et al. (29). Lipids were separated by thin-layer chromatography (TLC) on Kieselgel 60 (Merck) using chloroform:methanol:H2O (65:25:4 by volume), and radiolabeled lipids were visualized by fluorography.
Immunodetection of Gpi19 and Gpi2 in complex.
For detection of the Gpi19-Gpi2 association, yeast strain 1788 was cotransformed to uracil and leucine dual prototrophy with pYeF2[GPI19] (which expresses Gpi19-HA under the control of the GAL1 promoter; p2032) and either pRS425MET25[FLAG-GPI2] (p1874) or pRS425[GPI2] (p1860). Transformants were precultured at 30°C in yeast extract peptone (YEP)-2% raffinose and induced for 4 h in YEP-4% galactose, and extracts were prepared as described previously (36). For immunoprecipitation of FLAG-Gpi2, extract (500 μg) was incubated with anti-FLAG M2 agarose beads (20 μl) or protein A-Sepharose (no antibody control) beads (in a 1:1 slurry; both from Sigma) in a buffer containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 5 mM EDTA (pH 8.0), 5 mM EGTA (pH 8.0), 50 mM KF, 30 mM Na2PPi [pH 7.5], 1 mM Na3VO4, and 0.1% NP-40 at 4°C for 1 h with gentle agitation. Immune complexes were washed three times with the same buffer and solubilized in sodium dodecyl sulfate (SDS) sample buffer (final volume of 100 μl). Samples were loaded without heating (heating caused FLAG-Gpi2 to form aggregates), separated on SDS-polyacrylamide gels (8 to 16% gradient; Bio-Rad), and transferred to polyvinylidene difluoride membranes for immunoblot detection of FLAG-Gpi2 with anti-Flag M2 antibodies (Sigma) and of Gpi19-HA with anti-HA antibodies (12CA5; BabCo).
Other assays.
For endoglycosidase H (Endo H) assays, transformants were grown in YEP plus 2% raffinose, followed by induction of Gpi19 fusions by addition of 2% galactose and further incubation at 30°C for 4 h. Cell lysates were prepared as described previously (17) but were supplemented with a final concentration of 80 mM potassium acetate, pH 5.6. Extracts (30 μg protein) were treated with 1.5 μl Endo H (1 U/200 μl) for 1 h at 37°C prior to separation on an SDS-polyacrylamide gel (7%; Bio-Rad) and transferred to PVDF membranes for immunoblot detection of Gpi19-HA-Suc2. Zymolyase sensitivity was measured as described previously (35).
RESULTS
Conditional mutants in GPI19 behave like mutants in GPI anchor biosynthesis.
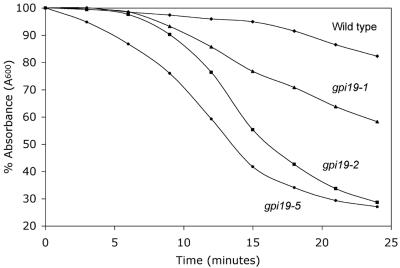
The GPI19 gene is reported in the Saccharomyces genome database (SGD) to be essential for viability. We constructed a gpi19 deletion allele that lacks nearly the entire coding sequence in the EG123 strain background and confirmed the lethality of this mutation (data not shown). As a first step to characterizing the function of GPI19, we created a set of temperature-sensitive gpi19 alleles using error-prone PCR (see Materials and Methods). DNA sequence analysis revealed that we had isolated six independent mutant alleles, each with multiple mutations. Among these, gpi19-1 and gpi19-2 displayed a restrictive temperature of 37°C. By contrast, gpi19-3 through gpi19-6 failed to grow at 34°C. We showed previously that null mutants in the nonessential subunits of the GPI-GlcNAc transferase (Eri1 and Gpi1) display several cell wall related defects (37). First, the growth defects of null mutants in ERI1 and GPI1, which are temperature-sensitive for growth, are suppressed by addition of osmotic support to the medium (e.g., 10% sorbitol). Consistent with this result, we found that all of the gpi19 alleles, with the exception of gpi19-4, were osmotically remedial (data not shown). Second, eri1Δ and gpi1Δ mutants are hypersensitive to cell lysis by treatment with the cell wall lytic enzyme zymolyase (37). Similarly, we found that the gpi19 mutants were hypersensitive to zymolyase treatment (Fig. 1). Finally, the growth defects of eri1Δ and gpi1Δ mutants are suppressed by increased intracellular concentration of UDP-GlcNAc, a substrate for the GPI-GlcNAc transferase (37). This can be accomplished either by overexpression of GFA1, whose product catalyzes the production of glucosamine-6-phosphate, the first committed and rate-limiting step in the production of UDP-GlcNAc (28), or with exogenous glucosamine, which is converted to glucosamine-6-phosphate (3, 45). We found that the least severely impaired alleles of gpi19 (gpi19-1 and gpi19-2) were suppressed for their growth defects at restrictive temperature by either GFA1 overexpression or exogenous glucosamine (Fig. 2 and data not shown).
FIG. 1.
Temperature-sensitive gpi19 mutants are hypersensitive to lysis by treatment with the cell wall digestive enzyme zymolyase. The wild type (1788) and three gpi19 mutants, gpi19-1 (DL3107), gpi19-2 (DL3106), and gpi19-5 (DL3103), were grown in YEPD to mid-log phase at 23°C, washed, and resuspended in water to an initial density of A600 of ∼0.7 prior to treatment with zymolyase 20T (150 μg/ml). Cell lysis was assessed by A600 measurements at the indicated times.
FIG. 2.
Growth defects of conditional gpi19 mutants are suppressed by overexpression of GFA1. The gpi19-2 mutant (DL3106) was transformed with 2μ plasmid pRS202[GFA1] (37), centromeric plasmid pRS316[GPI19] (p1995), or pRS316 (vector). Transformants were streaked onto YEPD and incubated for 3 days at 37°C.
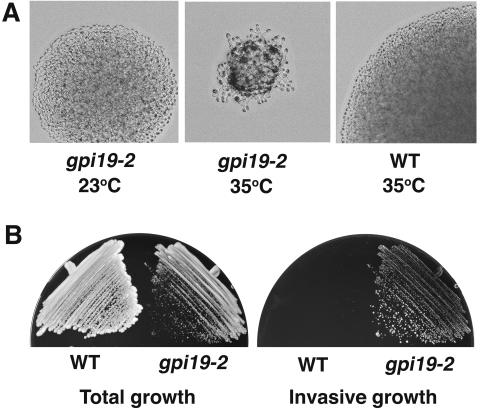
Mutants in GPI anchor biosynthesis also display phenotypes similar to those of hyperactive Ras mutants (36, 37). This is because Ras negatively regulates the activity of the GPI-GlcNAc complex. GPI anchor mutants have been proposed to mimic phenotypically the inhibitory effect of hyperactive Ras on anchor biosynthesis. These phenotypes include filamentous growth and agar invasion at semipermissive temperatures. The gpi19 mutants displayed both weak filamentation phenotypes and invasive growth, which was enhanced by the inclusion of sorbitol in the medium to suppress their growth defects (Fig. 3). These phenotypes collectively support the hypothesis that Gpi19 is important for GPI anchor biosynthesis.
FIG. 3.
Conditional gpi19 mutants display filamentation and agar invasion at semipermissive temperatures. A. A conditional gpi19 mutant displays weak filamentous growth at semipermissive temperature. Single colonies of diploid yeast strains were photographed after growth for 24 h on YEPD plus 10% sorbitol at the indicated temperatures. Strains are the wild type (1788) and gpi19-2 (DL3106). B. A conditional gpi19 mutant displays agar invasive growth. The same strains as in panel A were streaked onto a YEPD plus 10% sorbitol plate and allowed to grow at 35°C for 2 days (total growth). Nonadherent cells were washed from the plate with distilled water, and the plate was incubated for an additional day at 23°C to reveal invasive growth. WT, wild type.
Gpi19 is an essential subunit of the GPI-GlcNAc transferase.
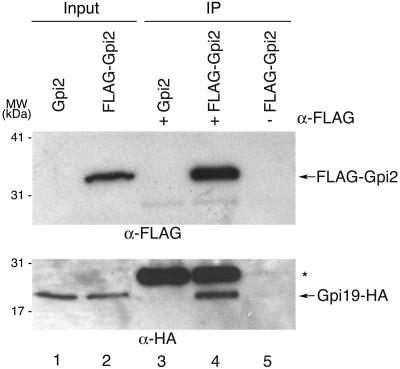
Because Gpi19 is related to mammalian PIG-P (44), and because a large-scale protein localization study revealed that it resides in the endoplasmic reticulum (14), we asked if Gpi19 could be found in association with the GPI-GlcNAc transferase. For this purpose, Gpi19 was fused at its C terminus to an HA epitope (Gpi19-HA) and overexpressed from the GAL1 promoter in yeast cells also overexpressing FLAG-Gpi2 (22, 37), an established GPI-GlcNAc transferase subunit. Immunoprecipitation of FLAG-Gpi2 from extracts made in the presence of nonionic detergent (0.5% NP-40) to disrupt membranes coprecipitated a large fraction of the Gpi19-HA (Fig. 4, lane 4). The observed coprecipitation reflects a true association of Gpi19-HA with FLAG-Gpi2, because Gpi19-HA was not detected in control precipitations either with untagged Gpi2 or in the absence of anti-FLAG antibodies (lanes 3 and 5, respectively). This supports the notion that Gpi19 is a subunit of the GPI-GlcNAc transferase, as revealed previously for mammalian PIG-P (44).
FIG. 4.
Gpi19 associates in vivo with Gpi2. Extracts from a wild-type yeast strain (1788), cotransformed with pYeF2[GPI19] (p2032; expresses Gpi19HA under the inducible control of the GAL1 promoter) and either pRS425MET25 [FLAG-GPI2] (p1874; constitutively expresses FLAG-Gpi2) or pRS425 [GPI2] (p1860; expresses untagged Gpi2), were processed for immunoprecipitation (IP) with α-Flag antibodies. Input protein (30 μg) and a fraction of the IP corresponding to 30 μg of input protein were separated by SDS-polyacrylamide gel electrophoresis and subjected to immunoblot detection of FLAG-Gpi2 (α-FLAG; top panel) or Gpi19-HA (α-HA; lower panel). The asterisk indicates an α-FLAG immunoglobulin G band in the IP lanes. The positions of molecular mass markers (MW) are indicated on the left.
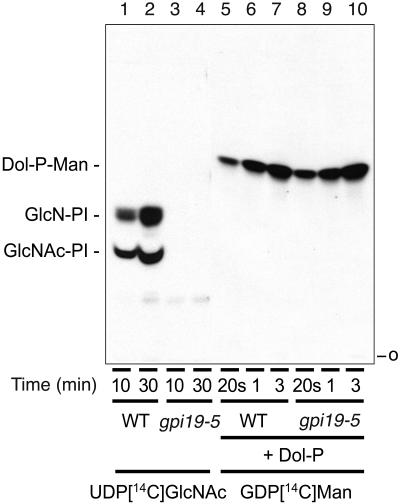
Because Gpi19-HA associates with the GPI-GlcNAc transferase, we asked if its function was required for activity of this enzyme. We examined GPI-GlcNAc transferase activity and the subsequent step (deacetylation of GlcNAc-PI) by thin-layer chromatographic separation of products labeled in vitro upon incubation of mixed membranes from wild-type and gpi19 cells with UDP-[14C]GlcNAc (5). Production of both intermediates (GlcNAc-PI and GlcN-PI) was defective in microsomes derived from the gpi19-5 mutant grown at 25°C (Fig. 5, compare lanes 1 and 2 to lanes 3 and 4), indicating a deficiency in the first step of anchor production, transfer of GlcNAc to an acceptor PI, yielding GlcNAc-PI. Similar results were obtained with microsomes from gpi19-2 and gpi19-3 (data not shown). Microsomes from the conditional gpi1, gpi2, gpi3, and eriΔ mutants are likewise defective in in vitro GPI-GlcNAc transferase activity (22, 23, 37). The activity of a control ER enzyme, Dol-P-Man synthase, was normal in the same microsome preparations (Fig. 5, compare lanes 5 to 7 to lanes 8 to 10). Therefore, we conclude that Gpi19 is essential for GPI-GlcNAc transferase activity.
FIG. 5.
In vitro assay for GPI-GlcNAc transferase activity. Microsomal fractions (100 μg protein) from extracts of a gpi19-5 mutant (DL3089) and its isogenic wild-type strain (DL3099) were incubated with UDP-[14C]GlcNAc at 30°C for the indicated times to measure GPI-GlcNAc transferase activity. The same microsomal fractions (5 μg) were incubated with GDP-[14C]-Man and Dol-P to assay Dol-P-mannose synthase as a control ER enzyme activity (20s, 20-s incubation). The lipids were extracted, separated by TLC, and detected by fluorography. WT, wild type.
Membrane topology of Gpi19.
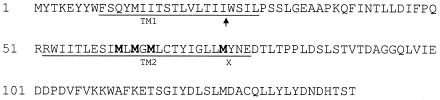
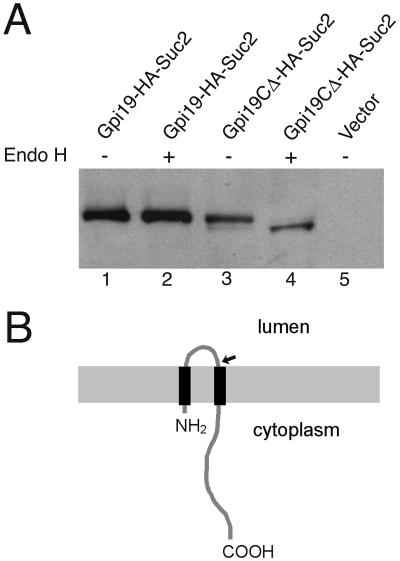
The sequences of Gpi19 and mammalian PIG-P are not especially informative, except for the suggestion that they possess two transmembrane domains (reference 44 and Fig. 6). A membrane topology prediction program (34) suggests that both polypeptide termini of these proteins reside on the cytoplasmic side of the ER membrane. To determine the membrane topology of Gpi19 experimentally, we constructed a pair of GPI19 fusions to SUC2, which encodes the glycoprotein invertase (19). Because invertase is glycosylated only when in the lumen of the ER, its glycosylation state in the context of a fusion to an ER membrane protein indicates the orientation of the transmembrane domain immediately N terminal to it (38). Fusion of HA-Suc2 to the C terminus of full-length Gpi19 did not lead to glycosylation of Suc2 (Fig. 7A, lanes 1 and 2), indicating that the Gpi19 C terminus is cytoplasmic, as predicted. By contrast, fusion of HA-Suc2 to the N-terminal 50 amino acid residues of Gpi19, which includes the first putative transmembrane domain and a short hydrophilic loop (Fig. 6 and 7A, lanes 3 and 4), resulted in glycosylation of Suc2. This indicates that the Gpi19 N terminus is also cytoplasmic and that both transmembrane domains pass through the ER membrane, as predicted.
FIG. 6.
Amino acid sequence of Gpi19. The putative transmembrane domains are underlined. The arrowhead indicates the position of the frameshift mutation in gpi19-5, gpi19-6, and gpi19-7. Potential sites for translational reinitiation are in boldface. The “X” indicates the position of the nonsense codon introduced into gpi19-7 to generate gpi19-8.
FIG. 7.
Membrane topology determination for Gpi19. A. Extracts from wild-type strain 1783, transformed with p2040 (expresses Gpi19-HA-Suc2), p2077 (expresses Gpi19CΔ-HA-Suc2), or pRS314 (vector), were treated with endoglycosidase H to assess the N-glycosylation state. Samples (30 μg protein) were separated by SDS-polyacrylamide gel electrophoresis monitored by immunoblot detection of the HA epitope. B. Cartoon of Gpi19 topology within the ER membrane. The Gpi19CΔ-HA-Suc2 fusion junction is indicated with an arrow.
The N-terminal transmembrane domain of Gpi19 is dispensable.
Sequence analysis of gpi19-5 and gpi19-6 revealed that they possess identical frameshift mutations after codon 24 (Fig. 6) in addition to other alterations that indicated these were independently derived alleles. It seemed unlikely that expression of the N-terminal 24 amino acid residues would be sufficient to complement the lethality of a gpi19Δ mutant. Therefore, we tested the possibility that translation of this mutant was being reinitiated at an internal methionine codon. Reinitiation would occur within a cluster of four methionine residues situated in the second transmembrane domain between Met61 and Met74. We first created the same frameshift mutation after codon 24 in an otherwise wild-type allele by site-directed mutagenesis and confirmed that this allele (gpi19-7) confers temperature-sensitive growth to the gpi19Δ mutant (restrictive temperature of 34°C; data not shown). We further mutated the gpi19-7 allele by converting Tyr75 to a nonsense mutation (Fig. 6). The new allele (gpi19-8) failed to complement the gpi19Δ mutant, confirming that sequences 3′ to the frameshift mutation were important for function of gpi19-7. This result indicates that translation of gpi19-7 is reinitiated between Met61 and Met74 and reveals that the N-terminal half (at least 60 residues) of Gpi19 is dispensable for function at low temperatures.
DISCUSSION
Gpi19 is an essential subunit of the yeast GPI-GlcNAc transferase.
We show in this study that the yeast Gpi19 protein associates with and is required for function of the GPI-GlcNAc transferase, which catalyzes the first step in GPI anchor biosynthesis at the ER. We isolated a set of temperature-sensitive mutants in GPI19, which displayed phenotypic defects characteristic of mutants with GPI deficiencies. These include cell wall weakness, as judged by hypersensitivity to the wall lytic enzyme zymolyase, and filamentation and agar invasion at semipermissive temperatures (36, 37). Additionally, the growth defects of conditional gpi19 mutants at elevated temperature were suppressed by GFA1 overexpression and by exogenous glucosamine, both of which increase the intracellular pool of the GPI-GlcNAc transferase substrate UDP-GlcNAc (3).
The modest sequence similarity of Gpi19 with the human PIG-P subunit of the GPI-GlcNAc transferase suggested that it might be associated with the analogous yeast enzyme complex. We demonstrated by coimmunoprecipitation that Gpi19 associates in vivo with the Gpi2 GPI-GlcNAc transferase subunit. Moreover, temperature-sensitive gpi19 mutants were defective for in vitro GPI-GlcNAc transferase activity, confirming that Gpi19 is essential for this enzymatic step.
We present evidence that Gpi19 passes through the ER membrane twice such that both the N and C termini of Gpi19 are cytoplasmically oriented, consistent with previous predictions (8, 44). However, two of the temperature-sensitive mutants we isolated resulted from frameshift mutations that led to translational reinitiation at a methionine codon in the middle of the GPI19 coding sequence. These results revealed that the N-terminal half of this protein, including the first transmembrane domain and the lumenal loop, is dispensable for function. We interpret this to mean that the second transmembrane domain is sufficient for proper localization and function of Gpi19.
Complexity of the GPI-GlcNAc transferase.
Both the yeast and mammalian GPI-GlcNAc transferases are unusually elaborate enzyme complexes compared to other glycosyltransferases. Although PIG-A/Gpi3 appears to be the catalytic subunit, the roles of other subunits have not been firmly established. PIG-C/Gpi2 may function as a scaffold to anchor the complex to the ER membrane and facilitate interaction among other subunits (8). The nonessential Gpi1 subunit has been speculated to provide a stabilizing role for the GPI-GlcNAc transferase (13). The additional subunits may also provide sites for binding of regulatory proteins. For example, DPM2, a regulator of Dol-P-Man synthase, which is also required for GPI synthesis, associates with and stimulates the mammalian GPI-GlcNAc transferase through interactions with PIG-A, PIG-C, and GPI1 (44). Additionally, the small G-protein Ras2 associates with and inhibits the yeast GPI-GlcNAc transferase (37), although the subunit with which Ras2 associates has not yet been established.
The yeast genome database (www.yeastgenome.org) offers some intriguing connections between subunits of the GPI-GlcNAc transferase and later steps in GPI anchor biosynthesis and attachment. For example, large-scale two-hybrid analysis revealed that Gpi19 associates with an uncharacterized ER-localized protein, Yjr015w (11, 14). Two-hybrid screening also indicates that Yjr015w associates by two hybrid with the Gpi14 GPI-mannosyltransferase (11) and the Gpi16 subunit of the GPI-transamidase complex (41). Similarly, a recent large-scale two-hybrid study of yeast integral membrane proteins revealed interactions between Gpi2 and Gpi16, Gpi7, and Gwt1 (26). Gpi7 is a subunit of the phosphoethanolamine transferase complex proposed to catalyze a late step in GPI synthesis, and Gwt1 is required for the third step in GPI synthesis, inositol acylation. These interactions suggest that various enzymes along the pathway of GPI anchor biosynthesis and attachment are physically connected to the GPI-GlcNAc transferase. Thus, one role of this unusually elaborate complex may be to regulate the activities of enzymes that catalyze subsequent steps in the GPI synthesis pathway.
Acknowledgments
We thank Yuan Lin for construction of the gpi19 mutant library and Ron Stamper for assistance with figures.
This work was supported by NIH grants GM67698 (D.E.L.) and GM46220 (P.O.). NCI training grant 5T32CA09110 supported M.J.R.
REFERENCES
- 1.Boeke, J. D., F. LaCroute, and G. R. Fink. 1984. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 197:345-346. [DOI] [PubMed] [Google Scholar]
- 2.Brodsky, R. A., M. S. Vala, J. P. Barber, M. E. Medof, and R. J. Jones. 1997. Resistance to apoptosis caused by PIG-A gene mutations in paroxysmal nocturnal hemoglobinuria. Proc. Natl. Acad. Sci. USA 94:8756-8760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bulik, D. A., M. Olczak, H. A. Lucero, B. C. Osmond, P. W. Robbins, and C. A. Specht. 2003. Chitin synthesis in Saccharomyces cerevisiae in response to supplementation of growth medium with glucosamine and cell wall stress. Eukaryot. Cell 2:886-890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choi, D. K., Y. Suzuki, S. Yoshimura, T. Togashi, M. Hida, T. D. Taylor, Y. Wang, S. Sugano, M. Hattori, and Y. Sakaki. 2001. Molecular cloning and characterization of a gene expressed in mouse developing tongue, mDscr5 gene, a homolog of human DSCR5 (Down syndrome critical region gene 5). Mamm. Genome 12:347-351. [DOI] [PubMed] [Google Scholar]
- 5.Costello, L. C., and P. Orlean. 1992. Inositol acylation of a potential glycosyl phosphoinositol anchor precursor from yeast requires acyl coenzyme A. J. Biol. Chem. 267:8599-8603. [PubMed] [Google Scholar]
- 6.Cullin, C., and L. Minvielle-Sebastia. 1994. Multipurpose vectors designed for the fast generation of N- or C-terminal epitope-tagged proteins. Yeast 10:105-112. [DOI] [PubMed] [Google Scholar]
- 7.DeNobel, H., and P. N. Lipke. 1994. Is there a role for GPIs in yeast cell wall assembly? Trends Cell Biol. 8:42-45. [DOI] [PubMed] [Google Scholar]
- 8.Eisenhaber, B., S. Maurer-Stroh, M. Novatchkova, G. Schneider, and F. Eisenhaber. 2003. Enzymes and auxiliary factors for GPI lipid anchor biosynthesis and post-translational transfer to proteins. BioEssays 25:367-385. [DOI] [PubMed] [Google Scholar]
- 9.Ferrando-Miguel, R., M. S. Cheon, and G. Lubec. 2004. Protein levels of genes encoded on chromosome 21 in fetal Down Syndrome brain (Part V): overexpression of phosphatidyl-inositol-glycan class P protein (DSCR5). Amino Acids 26:255-261. [DOI] [PubMed] [Google Scholar]
- 10.Geremia, R. A., E. A. Petroni, L. Ielpi, and R. Henrissat. 1996. Towards a classification of glycosyltransferases based on amino acid sequence similarities: prokaryotic α-mannosyltransferases. Biochem. J. 318:133-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hazbun, T. R., L. Malmström, S. Anderson, B. J. Graczyk, B. Fox, et al. 2003. Assigning function to yeast proteins by integration of technologies. Mol. Cell 12:1353-1365. [DOI] [PubMed] [Google Scholar]
- 12.Ho, S.-N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 13.Hong, Y., K. Ohishi, R. Watanabe, Y. Endo, Y. Maeda, and T. Kinoshita. 1999. GPI1 stabilizes an enzyme essential in the first step of glycosylphosphatidylinositol biosynthesis. J. Biol. Chem. 274:18582-18588. [DOI] [PubMed] [Google Scholar]
- 14.Huh, W. K., J. V. Falvo, L. C. Gerk, A. S. Carroll, R. W. Howson, J. S. Weissman, and E. K. O'Shea. 2003. Global analysis of protein localization in budding yeast. Nature 425:686-691. [DOI] [PubMed] [Google Scholar]
- 15.Inoue, N., R. Watanabe, J. Takeda, and T. Kinoshita. 1996. PIG-C, one of the three human genes involved in the first step of glycosylphosphatidylinositol biosynthesis, is a homologue of Saccharomyces cerevisiae GPI2. Biochem. Biophys. Res. Commun. 226:193-199. [DOI] [PubMed] [Google Scholar]
- 16.Ito, H., Y. Fukuda, K. Murata, and A. Kimura. 1983. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153:163-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kamada, Y., U. S. Jung, J. Piotrowski, and D. E. Levin. 1995. The protein kinase C-activated MAP kinase pathway of Saccharomyces cerevisiae mediates a novel aspect of the heat shock response. Genes Dev. 9:1559-1571. [DOI] [PubMed] [Google Scholar]
- 18.Kawagoe, K., J. Takeda, Y. Endo, and T. Kinoshita. 1994. Molecular cloning of murine Pig-a, a gene for GPI-anchor biosynthesis, and demonstration of interspecies conservation of its structure, function, and gene locus. Genomics 23:566-574. [DOI] [PubMed] [Google Scholar]
- 19.Kim, H., Q. Yan, G. von Heijne, G. A. Caputo, and W. J. Lennarz. 2003. Determination of the membrane topology of Ost4p and its subunit interactions in the oligosaccharyltransferase complex of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 100:7460-7464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kinoshita, T., and N. Inoue. 2000. Dissecting and manipulating the pathway for glycosylphosphatidylinositol-anchor biosynthesis. Curr. Opin. Chem. Biol. 4:632-638. [DOI] [PubMed] [Google Scholar]
- 21.Kostova, Z., D. Rancour, A. K. Menon, and P. Orlean. 2000. Photoaffinity labeling with P3-(4-azidoanilido) uridine 5′-triphosphate identifies Gpi3p as the UDPGlcNAc-binding subunit of the enzyme that catalyzes formation of N-acetylglucosaminyl phosphatidylinositol, the first glycolipid intermediate in glycosyl phosphatidylinositol synthesis. Biochem. J. 350:815-822. [PMC free article] [PubMed] [Google Scholar]
- 22.Leidich, S. D., Z. Kostova, R. R. Latek, L. C. Costello, D. A. Drapp, W. Gray, J. S. Fassler, and P. Orlean. 1995. Temperature-sensitive yeast GPI anchoring mutants gpi2 and gpi3 are defective in the synthesis of N-acetylglucosaminyl phosphatidylinositol. J. Biol. Chem. 270:13029-13035. [DOI] [PubMed] [Google Scholar]
- 23.Leidich, S. D., and P. Orlean. 1996. Gpi1, a S. cerevisiae protein that participates in the first step in GPI anchor synthesis. J. Biol. Chem. 271:27829-27837. [DOI] [PubMed] [Google Scholar]
- 24.McConville, M. J., and M. A. J. Ferguson. 1993. The structure, biosynthesis, and function of glycosylated phosphatidylinositols in the parasitic protozoa and higher eukaryotes. Biochem. J. 294:305-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McConville, M. J., and A. K. Menon. 2000. Recent developments in the cell biology and biochemistry of glycosylphosphatidylinositol lipids. Mol. Membr. Biol. 17:1-17. [DOI] [PubMed] [Google Scholar]
- 26.Miller, J. P., R. S. Lo, A. Ben-Hur, C. Desmarias, I. Stagljar, W. S. Noble, and S. Fields. 2005. Large-scale identification of yeast integral membrane protein interactions. Proc. Natl. Acad. Sci. USA 102:12123-12128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miyata, T., J. Takeda, Y. Iida, N. Yamada, N. Inoue, M. Takahashi, K. Maeda, T. Kitani, and T. Kinoshita. 1993. The cloning of PIG-A, a component in the early step of GPI-anchor biosynthesis. Science 259:1318-1320. [DOI] [PubMed] [Google Scholar]
- 28.Orlean, P. 1997. Biogenesis of yeast wall and surface components, p. 229-362. In J. R. Pringle, J. R. Broach, and E. W. Jones (ed.), Molecular and cellular biology of the yeast Saccharomyces cerevisiae Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 29.Orlean, P., C. Albright, and P. W. Robbins. 1988. Cloning and sequencing of the yeast gene for dolichol phosphate mannose synthase, an essential protein. J. Biol. Chem. 263:17499-17507. [PubMed] [Google Scholar]
- 30.Rose, M. D., F. Winston, and P. Hieter. 1990. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 31.Schönbächler, M., A. Horvath, J. Fassler, and H. Riezman. 1995. The yeast SPT14 gene is homologous to the human PIG-A gene and is required for GPI anchor synthesis. EMBO J. 14:1637-1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Siliciano, P. G., and K. Tatchell. 1984. Transcription and regulatory signals at the mating type locus in yeast. Cell 37:969-978. [DOI] [PubMed] [Google Scholar]
- 34.Smith, R. F., B. A. Wiese, M. K. Wojzynski, D. B. Davison, and K. C. Worley. 1996. BCM Search Launcher-an integrated interface to molecular biology data base search and analysis services available on the World Wide Web. Genome Res. 6:454-462. [DOI] [PubMed] [Google Scholar]
- 35.Sobering, A. K., U. S. Jung, K. S. Lee, and D. E. Levin. 2002. Yeast Rpi1 is a putative transcriptional regulator that contributes to preparation for stationary phase. Eukaryot. Cell 1:56-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sobering, A. K., M. R. Romeo, H. A. Vay, and D. E. Levin. 2003. A novel Ras inhibitor, Eri1, engages yeast Ras at the endoplasmic reticulum. Mol. Cell. Biol. 23:4983-4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sobering, A. K., R. Watanabe, M. J. Romeo, B. C. Yan, C. A. Specht, P. Orlean, H. Riezman, and D. E. Levin. 2004. Yeast Ras regulates the complex that catalyzes the first step in GPI-anchor biosynthesis at the ER. Cell 117:637-648. [DOI] [PubMed] [Google Scholar]
- 38.Strahl-Bolsinger, S., and A. Scheinost. 1999. Transmembrane topology of pmt1p, a member of an evolutionarily conserved family of protein O-mannosyltransferases. J. Biol. Chem. 274:9068-9075. [DOI] [PubMed] [Google Scholar]
- 39.Tiede, A., I. Bastisch, J. Schubert, P. Orlean, and R. E. Schmidt. 1999. Biosynthesis of glycosylphosphatidylinositols in mammals and unicellular microbes. Biol. Chem. 380:503-523. [DOI] [PubMed] [Google Scholar]
- 40.Tiede, A., J. Schubert, C. Nischan, I. Jenson, B. Westfall, C. H. Taron, P. Orlean, and R. E. Schmidt. 1998. Human and mouse Gpi1p homologues restore glycosylphosphatidylinositol mambrane anchor biosynthesis in yeast mutants. Biochem. J. 334:609-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Uetz, P., L. Giot, G. Cagney, T. A. Mansfield, R. S. Judson, et al. 2000. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403:623-627. [DOI] [PubMed] [Google Scholar]
- 42.Vossen, J. H., A. F. Ram, and F. M. Klis. 1995. Identification of SPT14/CWH6 as the yeast homolog of hPIG-A, a gene involved in the biosynthesis of GPI anchors. Biochim. Biophys. Acta 1243:549-551. [DOI] [PubMed] [Google Scholar]
- 43.Watanabe, R., N. Inoue, B. Westfall, C. H. Taron, P. Orlean, J. Takeda, and T. Kinoshita. 1998. The first step of glycosylphosphatidylinositol biosynthesis is mediated by a complex of PIG-A, PIG-H, PIG-C, and GPI1. EMBO J. 17:877-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Watanabe, R., Y. Murakami, M. D. Marmor, N. Inuoe, Y. Maeda, J. Hino, K. Kangawa, M. Julius, and T. Kinoshita. 2000. Initial enzyme for glycosylphosphatidylinositol biosynthesis requires PIG-P and is regulated by DPM2. EMBO J. 19:4402-4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Watzele, G., and W. Tanner. 1989. Cloning of the glutamine:fructose-6-phosphate amidotransferase gene from yeast. Pheromonal regulation of its transcription. J. Biol. Chem. 264:8753-8758. [PubMed] [Google Scholar]
- 46.Yan, B. C., B. A. Westfall, and P. Orlean. 2001. Ynl038wp (Gpi15p) is the Saccharomyces cerevisiae homologue of human Pig-Hp and participates in the first step in glycosylphosphatidylinositol assembly. Yeast 18:1383-1389. [DOI] [PubMed] [Google Scholar]