Abstract
L-731,988 inhibits human immunodeficiency virus (HIV) replication through integrase. In this study, approximately 600 nM L-731,988 inhibited the replication of 12 HIV type 1 isolates from multiple clades, including primary isolates and cloned viruses. These data suggest that diketo acids or their derivatives may prove useful on a worldwide basis in treating HIV infection.
The human immunodeficiency virus (HIV) genome encodes three enzymes: protease (PR), reverse transcriptase (RT), and integrase (IN). Agents targeted at RT and PR inhibit viral replication within infected individuals (1, 2, 4). Despite the success of PR and RT inhibitors, recent evidence suggests that the transmission of drug-resistant HIV is increasing and that its rate of transmission can approach 25% (13), making it clear that new therapeutic targets must be identified. IN is an enzyme unique to retroviruses and is required for productive retroviral infection. Given the latter, as well as the clinical effectiveness of other retroviral enzyme inhibitors, IN is an attractive therapeutic target. Recently, the diketo acids were found to curb viral replication through the inhibition of IN (6). Amino acid changes that lead to resistance to diketo acids are located within IN and have been documented previously for HIV type 1 (HIV-1) (6). Analysis of the HIV sequence database shows that changes at 2 of the 3 amino acids conferring resistance to diketo acids are present within the infected population. To investigate the role that natural variation has in the susceptibility of HIV to diketo acids, a series of primary, cloned, and laboratory HIV-1 isolates were examined for susceptibility to L-731,988 (a diketo acid) (Fig. 1) in both cultured cell lines and peripheral blood lymphocytes (PBLs).
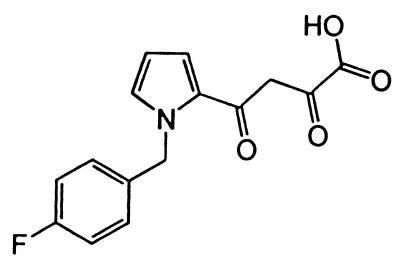
FIG. 1.
Chemical structure of L-731,988.
Laboratory-adapted strains of HIV and the infectious molecular clone HIVNL4-3 can replicate in cultured cell lines, unlike most primary isolates of HIV. Therefore, the susceptibilities to L-731,988 of laboratory-adapted strains (HIVLAI and HIVMVP5180) and HIVNL4-3 clones containing different IN genes (HIVNL4-3:IN7-3, HIVNL4-3:INR104, HIVNL4-3:INR106, and HIVNL4-3:IN92NG003) were determined on cultured MT2 cells by using a vital dye assay (8, 10, 12).
HIVLAI and HIVNL4-3:IN7-3 are clade B, laboratory-adapted HIV strains that are susceptible to diketo acids (6; R. Reinke et al., unpublished data). Cloned isolates were constructed with HIVNL4-3, which contains two unique restriction sites that allow the native IN to be excised and the IN gene of interest to be cloned in frame into the plasmid vector (7). HIVNL4-3:IN7-3 contains the HIVNL4-3 IN gene and was subjected to the same tissue culture and cloning procedures used for the other viruses (7) to control for changes due to culturing of virus and cloning. HIVNL4-3:INR104 and HIV NL4-3:INR106 contain IN genes derived from the North American clinical isolates described previously (9). HIVNL4-3:IN92NG003 contains an IN gene derived from a near-full-length clone of HIV originally isolated in Nigeria (3), and HIVMVP5180 is a group O isolate from Cameroon (5). Nomenclature used throughout indicates the specific IN gene cloned into HIVNL4-3: HIVNL4-3:INR106 contains the IN gene from clinical isolate R106.
HIV-1 primary isolates (HIV92RW016, HIV92RW009, HIV98IN026, HIV93IN101, HIV92THA005, HIV98CN006) were obtained through the National Institutes of Health AIDS Research and Reference Reagent Program and passaged on phytohemagglutinin-stimulated PBLs with interleukin-2; each virus supernatant was collected when cultures exhibited peak staining with polyclonal anti-HIV serum by an immunofluorescence assay. Supernatants were filtered through 0.45-μm-pore-size filters and assayed for exogenous RT activity. Viral supernatants (36,000 cpm of RT activity) were incubated with 0, 0.5, 1.0, and 5.0 μM L-731,988 for 30 min at 37°C. Following incubation, viral supernatants were added to 1.5 × 106 PBLs containing either 0, 0.5, 1.0, or 5.0 μM L-731,988 in a total volume of 2 ml. All infections were performed in triplicate in 24-well plates. Culture supernatants were monitored by indirect immunofluorescence assay and RT assay (11) every 2 days until RT activity within control infections without L-731,988 peaked. The percent inhibition at each drug concentration was calculated on the basis of the control infections. The 50% effective doses (ED50) were calculated for the viruses when RT values for the control infections were at maximum. ED50 were calculated using the CalcuSyn for Windows software package.
All six viruses tested in the MT2 cell assay were susceptible to the inhibitor, exhibiting only minor differences (two- to threefold) in ED50 (Table 1). ED50 for these isolates ranged from 0.5 to 1.6 μM. Similarly, the ED50 of L-731,988 against the primary isolates ranged from 0.6 to 0.9 μM, with the exception of that for HIV92RW009, which increased about sixfold compared to that for HIVLAI (Table 1). Two assays were utilized in this study to examine the susceptibilities of natural variant IN sequences to L-731,988. Despite the differences between the two assays, the data for a control virus, HIVLAI, correlate between the assays (Table 1) (0.51 to 0.65 μM), demonstrating that the antiviral data from both assays are directly comparable.
TABLE 1.
Susceptibilities of HIV-1 isolates to L-731,988
| HIV isolate | Virus cladea | Origin | IN amino acid diver- gence (%)b | Anti- HIV assayc | ED50d (μM) |
|---|---|---|---|---|---|
| LAI | B | United States | MT2 | 0.505 | |
| NL4-3:IN7-3 | B | United States | MT2 | 0.6 | |
| MVP5180 | Group O | Cameroon | 15 | MT2 | 1.59 |
| NL4-3:INR104 | B | United States | 3.5 | MT2 | 0.637 |
| NL4-3:INR106 | B | United States | 2 | MT2 | 1.27 |
| NL4-3:IN92NG003 | A/G | Nigeria | 5.2 | MT2 | 0.685 |
| LAI | B | United States | PBL | 0.654 | |
| 92RW016 | A | Rwanda | 6.6 | PBL | 0.590 |
| 92RW009 | C/A | Rwanda | 5.0 | PBL | 3.14 |
| 98IN026 | C | India | 6.3 | PBL | 0.544 |
| 93IN101 | C | India | 5.2 | PBL | 0.917 |
| 92THA005 | A/E | Thailand | 5.2 | PBL | 0.628 |
| 98CN006 | C | Central African Republic | 5.9 | PBL | 0.923 |
Clades based on gag/env sequences.
Reference sequence is HIVNL4-3. Divergence was calculated based on the number of substitutions out of 288 amino acids.
MT-2 assays were performed at least twice in triplicate (six replicate infections). PBL assays were performed using lymphocytes from one donor in triplicate infections.
ED50 were calculated from the means of values from triplicate infections by using the CalcuSyn for Windows software package.
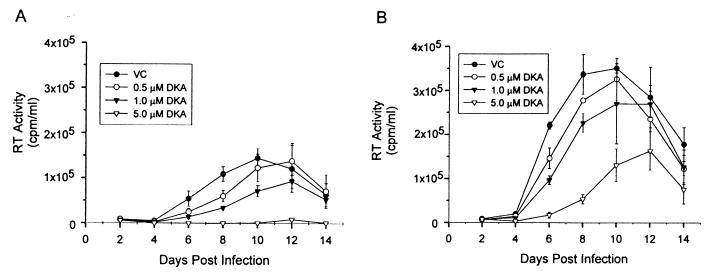
The ability of compounds to inhibit primary isolates of HIV is of paramount importance in their development as drugs. The data presented here show that a group O isolate, group M primary viral isolates representing clades A, B, and C, and a clade A/E recombinant were all susceptible to L-731,988. Compared to the control HIVLAI, most of these viruses exhibited a less-than-twofold difference in susceptibility to L-731,988. In contrast, the isolate HIV92RW009, a C/A recombinant, showed an approximately sixfold decrease in susceptibility to L-731,988. While complete resistance was not observed, this partial-resistance phenotype was detected at all drug concentrations tested, as evidenced by less growth inhibition than with other viruses (Fig. 2). IN sequence analysis of HIV92RW009 failed to reveal amino acid changes matching previously identified resistance mutations for either the diketo acid or the l-chicoric acid class of IN inhibitors (C65, T66, G140, S153, or M154) (6, 7, 14). These data suggest that other amino acid changes within the HIV92RW009 IN may contribute to resistance to diketo acids. Amino acid changes unique to HIV92RW009 IN were a valine-to-alanine change at position 79, a threonine-to-asparagine change at position 124, and a lysine-to-arginine change at position 173. However, the peak viral RT counts for the HIV92RW009 cultures were greater than for other viruses tested. Thus, an increased burst size may contribute to the resistance phenotype.
FIG. 2.
Representative susceptibilities of primary HIV isolates to L-731,988 in PBLs. PBLs were inoculated with HIV in the presence of 0.5, 1.0, or 5 μM L-731,988, and RT activity was measured. (A) Isolate HIV98CN006; (B) isolate HIV92RW009. Abbreviations: VC, virus control; DKA, diketo acid. Each point represents a mean of values from three independent infections, with error bars corresponding to 1 standard deviation.
To date, three mutations have been associated with resistance to the diketo acids: T66I, S153Y, and M154I (6). Two of the viruses tested contain amino acid changes at positions 153 and 154 of IN. Each virus had other mutations that could account for the resistance phenotype, but in particular, HIVMVP5180 contains an S153A change and HIVNL4-3:INR104 has an M154L change. HIVMVP5180 does show an increase in ED50 of roughly threefold, nearly identical to that observed by Hazuda et al. (6) for the S153Y mutation. Although S153 and M154 have been associated with resistance to diketo acids, the changes contained within HIVMVP5180 and HIVNL4-3:INR104 do not dramatically affect their susceptibility to L-731,988, suggesting that mutations occurring at these positions require additional changes, perhaps at position 66 (6), or specific amino acid substitutions for a high-resistance phenotype. Interestingly, S153Y conferred resistance to another diketo acid, L-708,906, but not to L-731,988 (6). It should be noted that if resistance occurs through the synergistic effect of mutations at positions 66, 153, and 154, as suggested by Hazuda et al. (6), the variability at positions 153 and 154 within the natural population may result in a more rapid development of resistance phenotypes. Nevertheless, 15% amino acid divergence for HIVMVP5180 results in only a threefold decrease in susceptibility to L-731,988. Overall, these data suggest that naturally occurring genetic variation within the IN gene is not a major obstacle in the development of the diketo acids for therapeutic use.
Nucleotide sequence accession numbers
The sequences of all IN genes reported herein have been deposited with GenBank. The accession numbers are R104-AF203330, R106-AF203331, 92NG003-U88825, 93IN101-AF500777, 98CN006-AF500778, 98IN026-AF500779, 92RW009-AF500780, 92RW016-AF500781, and 92THA005-AF500782.
Acknowledgments
This work was supported in part by grants from the Public Health Service (AI41360) and the Burroughs-Wellcome Fund (99-2609). W.E.R. is a Burroughs-Wellcome Fund Clinical Scientist in Translational Research. R.R. (grant AI07319) and D.J.L. (grant GM08620) are supported in part by training grants.
We are indebted to Brenda R. McDougall for excellent technical assistance. Manfred G. Reinecke (Texas Christian University, Fort Worth) provided L-731,988.
REFERENCES
- 1.Autran, B., G. Carcelain, T. S. Li, C. Blanc, D. Mathez, R. Tubiana, C. Katlama, P. Debre, and J. Leibowitch. 1997. Positive effects of combined antiretroviral therapy on CD4+ T cell homeostasis and function in advanced HIV disease. Science 277:112-116. [DOI] [PubMed] [Google Scholar]
- 2.Collier, A. C., R. W. Coombs, D. A. Schoenfeld, R. L. Bassett, J. Timpone, A. Baruch, M. Jones, K. Facey, C. Whitacre, V. J. McAuliffe, H. M. Friedman, T. C. Merigan, R. C. Reichman, C. Hooper, and L. Corey. 1996. Treatment of human immunodeficiency virus infection with saquinavir, zidovudine, and zalcitabine. N. Engl. J. Med. 334:1011-1017. [DOI] [PubMed] [Google Scholar]
- 3.Gao, F., D. L. Robertson, C. D. Carruthers, S. G. Morrison, B. Jian, Y. Chen, F. Barré-Sinoussi, M. Girard, A. Srinivasan, A. G. Abimiku, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 1998. A comprehensive panel of near-full-length clones and reference sequences for non-subtype B isolates of human immunodeficiency virus type 1. J. Virol. 72:5680-5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gulick, R. M., J. W. Mellors, D. Havlir, J. J. Eron, C. Gonzalez, D. McMahon, D. D. Richman, F. T. Valentine, L. Jonas, A. Meibohm, E. A. Emini, and J. A. Chodakewitz. 1997. Treatment with indinavir, zidovudine, and lamivudine in adults with human immunodeficiency virus infection and prior antiretroviral therapy. N. Engl. J. Med. 337:734-739. [DOI] [PubMed] [Google Scholar]
- 5.Gürtler, L. G., P. H. Hauser, J. Eberle, A. von Brunn, S. Knapp, L. Zekeng, J. M. Tsague, and L. Kaptue. 1994. A new subtype of human immunodeficiency virus type 1 (MVP-5180) from Cameroon. J. Virol. 68:1581-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hazuda, D. J., P. Pelock, M. Witmer, A. Wolfe, K. Stillmock, J. A. Grobler, A. Espeseth, L. Gabryelski, W. Schleif, C. Blau, and M. D. Miller. 2000. Inhibitors of strand transfer that prevent integration and inhibit HIV-1 replication in cells. Science 287:646-650. [DOI] [PubMed] [Google Scholar]
- 7.King, P. J., and W. E. Robinson, Jr. 1998. Resistance to the anti-human immunodeficiency virus type 1 compound l-chicoric acid results from a single mutation at amino acid 140 of integrase. J. Virol. 72:8420-8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Montefiori, D. C., W. E. Robinson, Jr., S. S. Schuffman, and W. M. Mitchell. 1988. Evaluation of antiviral drugs and neutralizing antibodies to human immunodeficiency virus by a rapid and sensitive microtiter infection assay. J. Clin. Microbiol. 26:231-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reinke, R., N. R. Steffen, and W. E. Robinson, Jr. 2001. Natural selection results in conservation of HIV-1 integrase activity despite sequence variability. AIDS 15:823-830. [DOI] [PubMed] [Google Scholar]
- 10.Robinson, W. E., Jr., M. Cordeiro, S. Abdel-Malek, Q. Jia, S. A. Chow, M. G. Reinecke, and W. M. Mitchell. 1996. Dicaffeoylquinic acid inhibitors of human immunodeficiency virus integrase: inhibition of the core catalytic domain of human immunodeficiency virus integrase. Mol. Pharmacol. 50:846-855. [PubMed] [Google Scholar]
- 11.Robinson, W. E., Jr., D. C. Montefiori, D. H. Gillespie, and W. M. Mitchell. 1989. Complement-mediated, antibody-dependent enhancement of HIV-1 infection in vitro is characterized by increased protein and RNA syntheses and infectious virus release. J. Acquir. Immune Defic. Syndr. 2:33-42. [PubMed] [Google Scholar]
- 12.Robinson, W. E., Jr., M. G. Reinecke, S. Abdel-Malek, Q. Jia, and S. A. Chow. 1996. Inhibitors of HIV-1 replication that inhibit HIV integrase. Proc. Natl. Acad. Sci. USA 93:6326-6331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wegner, S. A., S. K. Brodine, J. R. Mascola, S. A. Tasker, R. A. Shaffer, M. J. Starkey, A. Barile, G. J. Martin, N. Aronson, W. W. Emmons, K. Stephan, S. Bloor, J. Vingerhoets, K. Hertogs, and B. Larder. 2000. Prevalence of genotypic and phenotypic resistance to antiretroviral drugs in a cohort of therapy-naive HIV-1 infected US military personnel. AIDS 14:1009-1015. [DOI] [PubMed] [Google Scholar]
- 14.Zhu, K., M. L. Cordeiro, J. Atienza, W. E. Robinson, Jr., and S. A. Chow. 1999. Irreversible inhibition of human immunodeficiency virus type 1 integrase by dicaffeoylquinic acids. J. Virol. 73:3309-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]