Abstract
HPC2/ELAC2 has been identified as a prostate cancer (CaP) susceptibility gene. Two common missense variants in HPC2/ELAC2 have been identified: a Ser→Leu change at amino acid 217, and an Ala→Thr change at amino acid 541. Tavtigian et al. reported that these variants were associated with CaP in a sample of men drawn from families with hereditary CaP. To confirm this report in a sample unselected for family history, we studied 359 incident CaP case subjects and 266 male control subjects that were frequency matched for age and race and were identified from a large health-system population. Among control subjects, the Thr541 frequency was 2.9%, and the Leu217 frequency was 31.6%, with no significant differences in frequency across racial groups. Thr541 was only observed in men who also carried Leu217. The probability of having CaP was increased in men who carried the Leu217/Thr541 variants (odds ratio = 2.37; 95% CI 1.06–5.29). This risk did not differ significantly by family history or race. Genotypes at HPC2/ELAC2 were estimated to cause 5% of CaP in the general population of inference. These results suggest that common variants at HPC2/ELAC2 are associated with CaP risk in a sample unselected for family history or other factors associated with CaP risk.
There is substantial evidence that prostate cancer (CaP; MIM 176807) has a hereditary component. Linkage of CaP to HPC1 (MIM 601518; Smith et al. 1996, Xu et al. 2000), PCAP (MIM 602759; Berthon et al. 1996), HPCX (MIM 300147; Xu et al. 1998) and CAPB (MIM 603688; Gibbs et al. 1999) has been reported, but these genes have yet to be isolated and may explain a relatively small proportion of disease in the general population. CaP has also been associated with common variants in genes involved in steroid hormone metabolism—including AR (Giovannuci et al. 1997), SRD5A2 (Makridakis et al. 1999; Jaffe et al. 2000), and CYP3A4 (MIM 124010; Rebbeck et al. 1998), vitamin D metabolism (Ingles et al. 1997), and carcinogen metabolism (Rebbeck et al. 1999). The HPC2/ELAC2 gene on chromosome 17p has been identified, by family studies, as being associated with CaP risk (Tavtigian et al. 2000), and at least two common missense variants in HPC2/ELAC2 have been identified. The function of this gene has yet to be elucidated, and the functional significance of the common missense mutations is not known. However, significant cross-species identity of HPC2/ELAC2 exists across a wide range of organisms (Tavtigian et al. 2000). The Ala541Thr missense variant in HPC2/ELAC2 lies adjacent to a highly conserved histidine motif, suggesting that the presence of a Thr541 allele may have deleterious effects on the function of the protein encoded by this gene.
To evaluate whether HPC2/ELAC2 genotypes are associated with nonhereditary CaP risk, a sample of 432 incident CaP case subjects was identified through representative clinics of the University of Pennsylvania Health System (UPHS) between September 1994 and December 1999. The UPHS counts >1 million patient visits per year at the Hospital of the University of Pennsylvania, its ten affiliate hospitals, and ∼130 Clinical Care Associates practices located throughout the metropolitan Philadelphia region. Case status was confirmed by medical records review by use of a standardized abstraction form. Men were excluded from this study if they reported having exposure to finasteride (Proscar) at the time of their CaP diagnosis. Patients who were nonincident case subjects (i.e., those diagnosed >12 mo prior to the date of study ascertainment) or who had a prior diagnosis of cancer at any site also were excluded. After these exclusions, a sample of 359 patients with CaP was available for analysis, although the effective sample size for some analyses was smaller because of missing data.
Six hundred twenty-eight men attending UPHS general medicine clinics were ascertained as potential control subjects. These clinics see a patient population that is demographically similar to those seen in the urologic oncology clinics at UPHS. These men were ascertained concurrently with the individuals who had CaP (i.e., between September 1994 and December 1999). Control subjects were excluded from analysis if they ever had an elevated prostate-specific antigen test (i.e., ⩾4 ng/ml), if they had ever had an abnormal digital rectal examination (DRE), if they had a previous cancer diagnosis, or if they reported having had exposure to finasteride (Proscar) at the time of study ascertainment. After these exclusions were applied, 383 control subjects were available for matching to case subjects. Control subjects were frequency matched to patients with CaP on age (±5 years) and race. After exclusion and matching criteria were applied, a set of 266 control subjects was available for analysis, although the effective sample size for some analyses was smaller because of missing data. Allele-frequency estimates were made in the 383 eligible control subjects (rather than the subset of matched control subjects) to limit the potential effect on allele-frequency estimates that may be induced by matching on case-subject characteristics.
Risk factor, medical history, and CaP diagnostic information was obtained by use of a standardized questionnaire and review of medical records. Information collected included CaP occurrences in first- and second-degree relatives; personal history of benign prostatic hyperplasia and vasectomy; previous cancer diagnoses; demographic information, such as race, educational level, and occupation; and CaP screening history. All study subjects provided informed consent for participation in this research under a protocol approved by the Committee for Studies Involving Human Subjects at the University of Pennsylvania.
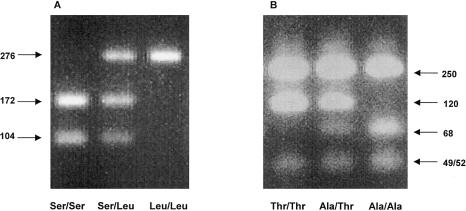
Genomic DNA for the present study was self-collected by each study subject using sterile cheek swabs (Cyto-Pak Cytosoft Brush, Medical Packaging Corporation) and was processed using the protocol of Walker et al. (1999). All PCR reactions were carried out using a master mix that consisted of 4.00 μl of 5× CPCR buffer (0.1M TrisCl, pH 8.8–9.0, 42.52 mM NaCl, 50 mM KCl, 50 mM (NH4)2SO4, 10 mM MgSO4, and 0.5% Triton X-100), 0.4 μl of 10 mM dNTPs, 0.4 μl of each 10 μM primer, and 0.1 μl of Platinum Taq (Life Technologies, Inc.) and ddH2O, for a total reaction volume of 20 μl. Nested PCR reactions were used to amplify the regions containing the polymorphisms of interest. First, primers m5A (5′-CATTCCCATGTATGAACGTCT-3′) and m5P (5′-ATAGTAAGCCCAGGAAGAAGGA-3′) were used to amplify the region containing the Ser217Leu variant. These primers were amplified with a 25-cycle protocol of 95°C for 3 min for 1 cycle; 96°C for 30 s, 55°C for 35 s, and 72°C for 45 s for 25 cycles; followed by an elongation cycle of 72°C for 10 min. These PCR products then were diluted 1:5 and were used in a second PCR reaction at a final dilution of 1:60. The second (nested) set of primers used was m5B (5′-GTTTTCCCAGTCACGACGCATTCCCATGTATGAACGTCT-3′) and m5Q (5′-AGGAAACAGCTATGACCATCTACAAGCATTACAAGGCAGAG-3′). These primers amplified a 276-bp region using the same protocol as described above, but with only 23 cycles rather than 25. Finally, 5 μl of the final 276-bp PCR product was digested overnight with TaqIα (NEB) at 65°C. Genotypes were visualized on a 3% agarose gel (fig. 1A).
Figure 1.
Agarose gel visualization of (A) the Ser217Leu polymorphism and (B) the Ala541Thr polymorphism. PCR product sizes after restriction enzyme digestion with TaqI (for Ser217Leu) or Fnu4HI (for Ala541Thr) are shown.
A second set of nested PCR reactions was used to amplify the Ala541Thr variant. Primers m15A (5′-CCAGCCTTTGTGTAAGTCTAC-3′) and m15P (5′-TCTGGGCAAGTTTGGAAGC-3′) were amplified with a 25-cycle protocol consisting of 95°C for 3 min for 1 cycle; 96°C for 30 s, 55°C for 35 s, and 72°C for 45 s for 25 cycles; followed by an elongation cycle of 72°C for 10 min. The products were then diluted 1:5 and were used in a second PCR reaction at a final dilution of 1:60. The second (nested) set of primers used was m15A (see above) and m15RFLP.R′ (5′-AATTCTTGATAGGAAACAGCTATGACCATCAGCTTTGTGGTCCAGCCCAAC-3′). These primers were used to amplify a 419-bp region amplified using the same protocol as described above, but with only 23 cycles rather than 25. Finally, 20 μl of the final PCR product was then digested for 3 h with Fnu4HI (NEB) at 37°C. Genotypes were visualized on a 3% agarose gel (fig. 1B).
Confirmation of genotype data was undertaken by direct sequencing of 21 individuals identified by RFLP analysis as carrying variant alleles and 8 individuals identified by RFLP analysis as carrying wild-type genotypes. All sequence results were consistent with the RFLP analyses. These analyses were undertaken using an automated ABI377 (ABI Prism), according to manufacturer’s instructions.
In order to evaluate the relationship of Ser217Leu and Ala541Thr variants with the probability of having CaP, we constructed genotype classes that simultaneously considered both missense variants. First, we grouped individuals into four genotype classes, to reflect the joint effects of both missense changes. The following classes were considered: (a) Ser/Ser and Ala/Ala individuals, (b) individuals who carried any Thr541 variant on an Ser/Ser background (no genotypes of this class were observed), (c) individuals who carried any Leu217 variant on an Ala/Ala background, and (d) individuals who carried any Thr541 variant, which were always observed on a background of a Leu217 allele. In addition, we compared individuals who were Ala541 homozygotes with individuals who carried any Thr541 allele. These genotype classes allowed us to compare the joint effects of both missense changes, and to evaluate the potential increase in the deleterious effect of the presence of both variants, relative to that of only one variant.
Genotype-disease associations were undertaken using unconditional logistic regression. Analyses considered the effect of genotype adjusted for potential confounders, including age (at time of diagnosis, in case subjects, and at time of study ascertainment, in control subjects), family history (defined as one or more first-degree relatives and/or two or more second-degree relatives), and race (coded as a discrete variable with three levels: white, black, or other). Subset analyses in whites only and in men with no family history of CaP were also undertaken. All P values were based on two-sided hypothesis tests.
Table 1 presents a description of the sample population. The sample consisted of men aged 41–84 years, ∼85% of whom were white. The racial distribution of our case-subject sample roughly reflects the UPHS prostate cancer–patient population overall. Because of the matching algorithm used here, there was no difference in the distribution of age or race between case subjects and control subjects. Consistently with most previous reports, mean serum PSA levels were significantly higher in case subjects than in control subjects, and case subjects were significantly more likely to have a family history of CaP than were control subjects.
Table 1.
Descriptive Characteristics of Case-Control Sample
| Trait | Control Subjects | Case Subjects | Comparison |
| Mean age in years (SD) | 60.7 (10.0)a | 61.3 (6.8)b | χ2=3.46; df 1; P=.063c |
| Race: | |||
| White | 215 (85.3%) | 306 (86.4%) | FET P=.611 |
| Black | 33 (11.4%) | 37 (10.5%) | |
| Other | 7 (3.3%) | 11 (3.1%) | |
| Mean serum PSA in ng/ml (SD)d | .80 (1.00) | 8.33 (7.18) | χ2=400.06; df 1; P<.0001c |
| Family history of CaP: | |||
| No | 236 (91.5%) | 275 (79.7%) | FET P<.0001 |
| Yes | 22 (8.5%) | 70 (20.3%) |
Range 41–84 years.
Range 41–80 years.
Kruskal-Wallis χ2 approximation.
Serum PSA was measured at time of diagnosis, in case subjects, and at most recent clinic visit, in control subjects.
We estimated the Thr541 allele frequency in 383 control subjects to be 2.9% and the Leu217 allele frequency to be 31.6%. We observed Thr541 alleles only in the presence of Leu217 alleles, consistent with the disequilibrium between these alleles observed by Tavtigian et al. (2000). Among whites, these frequencies were 3.2% and 32.2%, respectively. Among blacks, these frequencies were 0.8% and 28.4%, respectively. There were no statistically significant differences in the Ser217Leu (Fisher's Exact Test [FET] P=.531) or the Ala541Thr (FET P value = .237) allele frequencies by race. Genotype frequencies did not deviate from Hardy-Weinberg proportions in the sample as a whole, in whites, or in blacks.
Genotype frequencies did not differ by decade of birth in our sample for the combined Ser217Leu/Ala541Thr genotype for either case subjects (χ2=2.478, df 4, P=.649) or control subjects (χ2=2.346, df 5, P=.800). In contrast to our results, Tavtigian et al. (2000) reported a moderate effect of birth cohort on the frequency of the Ser217Leu and Ala541Thr variant frequencies in their population. Specifically, the variant allele frequencies appeared to differ in men born before and after 1920. Although ∼20% of the sample of studied by Tavtigian et al. (2000) was born before 1920, <3% of our study subjects were born before 1920. Therefore, our sample may be inadequate to detect the relatively small birth cohort effect reported by Tavtigian and colleagues, if it exists in our population. Additional studies that include a larger proportion of men from earlier birth cohorts are required to evaluate a potential birth cohort effect on genotype frequencies.
Leu217/Thr541 genotypes were significantly more common among case subjects than control subjects (table 2). Even after adjusting for age, race, and family history of CaP, Leu217/Thr541 genotypes predicted CaP risk (odds ratio [OR] = 2.37; 95% CI 1.06–5.29). This result is similar to that reported by Tavtigian et al. (2000) in their comparison of CaP case subjects and “divergent” (i.e., nonfamilial) control subjects. Although the proportions of family history–positive subjects and nonwhites were too small to allow formal tests of interaction, the effect of this genotype did not appear to differ substantially by family-history status or by race. In a subset analysis (table 2), point estimates of CaP risk associated with Leu217/Thr541 genotypes were not substantially different in whites only or family history–negative individuals only, compared with the total sample.
Table 2.
Case-Control Comparisons of HPC2/ELAC2 Genotypes
| Ser217LeuGenotype | Ala541ThrGenotype | Control Subjects | Case Subjects | Total SampleOR (95% CI)a | WhitesOR (95% CI)a | No FamilyHistory of CaPOR (95% CI)a |
| Ser/Ser | Ala/Ala | 121 (46.9%) | 172 (47.9%) | 1.0b | 1.0b | 1.0b |
| Ser/Ser | Any Thr | 0 | 0 | … | … | … |
| Any Leu | Ala/Ala | 128 (49.6%) | 160 (44.6%) | 0.89 (0.63–1.25) | 0.99 (0.69–1.43)c | 0.90 (0.63–1.30)c |
| Any Leu | Any Thr | 9 (3.5%) | 27 (7.5%) | 2.37 (1.06–5.29) | 2.23 (1.00–4.98)c | 2.36 (1.01–5.56)c |
| Any | Ala/Ala | 249 (96.5%) | 332 (92.5%) | 1.0b | 1.0b | 1.0b |
| Any | Any Thr | 9 (3.5%) | 27 (7.5%) | 2.24 (1.02–4.90) | 2.21 (1.01–4.85)c | 2.25 (0.97–5.19)c |
OR adjusted by multiple logistic regression for age, race, and family history of CaP.
Reference group.
Breslow-Day test for homogeneity of OR indicates no differences among strata with P>.5.
Despite the report of Tavtigian et al. (2000) that suggests that mutations in HPC2/ELAC2 explain some hereditary prostate cancer, there are a number of reasons why the present study did not identify differences in the relationship between genotype and disease by family history. First, the sample size may be too small to provide sufficient statistical power to undertake analyses stratified on family history, since only 8% of control subjects and 20% of case subjects reported a family history of prostate cancer. The present sample also consisted of very few individuals from families in which prostate cancer might be considered “hereditary” in nature. Only nine (1.4%) subjects in the present sample reported three or more first- or second-degree relatives with prostate cancer. Second, our family-history information was obtained only by self-report of the case or control subjects, and no validation of family history was undertaken. Unlike most genetic-linkage studies, our study may have relatively more misclassification on prostate cancer family history, and this misclassification may affect our ability to detect effects of genotype by family history. Third, it is possible that, although common HPC2/ELAC2 polymorphisms confer a greater-than-twofold increase an individual’s prostate cancer risk, this effect may not be strong enough to confer a significant family history of CaP in the present study sample. Therefore, we conclude that only limited information about the role of common missense variants at HPC2/ELAC2 and family history of CaP can be learned from the present study.
Additional analyses were undertaken to evaluate the relationship of Leu217/Thr541 genotypes with characteristics of prostate tumors. Leu217/Thr541 genotypes were not associated with an earlier age at diagnosis, the existence of benign prostatic hypertrophy (BPH), Gleason grade, PSA at diagnosis, or extracapsular extension (table 3). PSA values at the time of diagnosis tended to be higher in men who carried one or more variant genotypes. However, this effect was not statistically significant according to a Kruskal-Wallis χ2 test, presumably because of the relatively small sample size and the high degree of variability in PSA. When divided at the sample median PSA value, 16.8% of men with higher PSA values carried the Leu217 or Thr541 genotype compared with 8.7% of men with lower PSA values (FET P=0.026). When broken into tertiles, 21.1% of CaP case subjects with the highest tertile of PSA carried the Leu217 or Thr541 genotype compared with 10.1% in the two lowest tertiles (FET P=.014). These analyses are highly preliminary, but suggest that additional studies of the role of HPC2/ELAC2 genotype and disease severity should be pursued.
Table 3.
Case-Case Associations with HPC2/ELAC2 Genotype
| Variable | Ser/SerAla/Ala | Any LeuAla/Ala | Any LeuAny Thr | Test Statisticand P Value | Ala/Ala | Any Thr | Test Statisticand P Value |
| Age at diagnosis (years) | 61.8 | 60.6 | 61.1 | χ22=1.86; P=.395 | 61.2 | 61.1 | χ21=0.06; P=.808 |
| Median (mean) serum PSA at diagnosis (ng/dl) | 6.0 (7.6) | 6.4 (8.4) | 7.3 (11.3) | χ22=2.89; P=.236a | 6.1 (7.9) | 7.3 (11.3) | χ21=2.34; P=.126a |
| Gleason grade: | |||||||
| <6 | 6 (4.2%) | 6 (4.6%) | 0 (0%) | 12 (4.4%) | 0 (0%) | ||
| 6–8 | 133 (93.0%) | 121 (92.4%) | 23 (95.8%) | FET P = .905 | 254 (92.7%) | 23 (95.8%) | FET P = .509 |
| >8 | 4 (2.8%) | 4 (3.1%) | 1 (4.2%) | 8 (2.9%) | 1 (4.2%) | ||
| BPH: | |||||||
| Yes | 29 (18.4%) | 33 (22.3%) | 6 (23.1%) | FET P = .652 | 62 (17.2%) | 6 (23.1%) | FET P = .800 |
| No | 129 (81.7%) | 115 (77.7%) | 20 (76.9%) | 244 (79.7%) | 20 (76.9%) | ||
| Extracapsular extension: | |||||||
| Yes | 28 (22.6%) | 24 (21.4%) | 5 (26.3%) | FET P = .895 | 52 (22.0%) | 5 (26.3%) | FET P = .774 |
| No | 96 (77.4%) | 88 (78.6%) | 14 (73.7%) | 184 (78.0%) | 14 (73.7%) |
Kruskal-Wallis χ2 approximation.
In contrast to genes that confer relatively high disease risk, we report odds ratio effects associated with HPC2/ELAC2 that are relatively small. However, a relatively large proportion of CaP may be explained by genotypes at HPC2/ELAC2, because Leu217/Thr541 genotypes are relatively common. If we assume a Leu217/Thr541 carrier frequency in the U.S. population to be p=.04, and the effect of this allele is OR = 2.4 (table 2), the percent of CaP risk caused by this genotype can be estimated by use of the formula 100%×p(OR-1)/[p(OR-1)+1] (Lillienfeld and Lillienfeld 1980). The proportion of CaP in the referent population that would be explained by the common missense genotypes is 5.3%. This proportion does not consider other variants (e.g., frameshift mutations) identified by Tavtigian et al. (2000), but it does suggest that these common missense changes may explain a relatively large proportion of prostate cancer case subjects in the population.
We also considered a number of potential study limitations. Our correction for race as a confounder in multiple logistic-regression models (table 2) and our subset analysis among white subjects minimizes the potential for population stratification as an explanation for the present findings (Wacholder et al. 2000). It is unlikely that genotyping errors could account for an elevated proportion of HPC2/ELAC2 in control subjects relative to case subjects, as most variant genotypes and some nonvariant genotypes were confirmed by direct sequencing. Therefore, it is unlikely that genotyping errors induced significant bias in our results. Misclassification of case or control status could also have influenced the inferences of this study. All case subjects were confirmed by review of medical records, and the vast majority are being followed actively for their disease by the clinicians involved in this study. Thus, it is highly unlikely that case subjects have been misclassified. No control subject reported ever being told he had a diagnosis of CaP, and no control subjects had elevated serum PSA or abnormal DRE. Review of medical records confirmed that no control subject was diagnosed with CaP. However, control subjects could have had clinically undetected disease, and this misclassification might bias the reported OR. If we assume that 10% of control subjects had a clinically undetected disease, the unadjusted OR associated with the Leu217/Thr541 variant would be 2.07 versus 2.11, when the actual observed data in table 2 are used. Since the control-subject sample had a clinically normal DRE and serum PSA, it is highly unlikely that as many as 10% of control subjects were misclassified, and it is unlikely that the OR estimates are substantially biased because of control-subject misclassification.
Although the present results suggest that HPC2/ELAC2 is associated with CaP risk, additional confirmatory epidemiological and basic science studies will be required to better understand the association reported here. Nonetheless, this report suggests that HPC2/ELAC2 is involved in the etiology of CaP in men ascertained without respect to family history or other determinants of CaP risk.
Acknowledgments
The authors wish to thank Sean Tavtigian, for sharing relevant sequence and genotype data for HPC2/ELAC2, and Diana Iliev, for providing technical assistance with the genotyping assays. This study was supported by grants from the Public Health Service (ES08031 and CA85074 [to T.R.R.]) and the University of Pennsylvania Cancer Center. The authors wish to thank Drs. M. D. Cirigliano, G. W. Crooks, D. Farhadi, S. J. Gluckman, D. Goldmann, W. Greer, C. Guerra, D. A. Horowitz, J. Jaeger, M. Rusk, P. Szaparay, and V. Weil for their valuable assistance in ascertaining study subjects.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- Genome Database, http://www.gdb.org/ (for HPC2/ELAC2)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for CaP [MIM 176807], HPC1 [MIM 601518], PCAP [MIM 602759], HPCX [MIM 300147], CAPB [MIM 603688], and CYP3A4 [MIM 124010])
References
- Berthon P, Valeri A, Cohen-Akenine A, Drelon E, Paiss T, Wohr G, Latil A, Millasseau P, Mellah I, Cohen N, Blanche H, Bellane-Chantelot C, Demenais F, Teillac P, Le Duc A, de Petriconi R, Hautmann R, Chumakov I, Bachner L, Maitland NJ, Lidereau R, Vogel W, Fournier G, Mangin P, Cussenot O (1998) Predisposing gene for early onset prostate cancer, localized on chromosome 1q42.2-43. Am J Hum Genet 62:1416–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs M, Stanford JL, McIndoe RA, Jarvik GP, Kolb S, Goode EL, Chakrabarti L, Schuster EF, Buckley VA, Miller EL, Brandzel S, Li S, Hood L, Ostrander EA (1999) Evidence for a rare prostate cancer susceptibility locus at chromosome 1p36. Am J Hum Genet 64:776–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannuci E, Stampfer MJ, Krithivas K, Brown M, Dahl D, Brufsky A (1997) The CAG repeat within the androgen receptor gene and its relationship to prostate cancer. Proc Natl Acad Sci USA 94:3320–3323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingles S, Ross RK, Yu MC, Irvine RA, La Pera G, Haile RW, Henderson B (1997) Association of prostate cancer risk with genetic polymorphisms in vitamin D receptor and androgen receptor. J Natl Cancer Inst 89:166–170 [DOI] [PubMed] [Google Scholar]
- Jaffe JM, Walker AH, MacBride S, Peschel R, Tomaszewski J, Van Arsdalen K, Wein AJ, Malkowicz SB, Rebbeck TR (2000) Association of SRD5A2 genotype and pathologic characteristics of prostate tumors. Cancer Res 60:1626–1630 [PubMed] [Google Scholar]
- Lillienfeld AM, Lillienfeld DE (1980) Foundations of epidemiology, second edition. Oxford University Press, New York [Google Scholar]
- Makridakis N, Ross RK, Pike MC, Crocitto LE, Kolonel LN, Pearce CL, Henderson BE, Reichardt JK (1999) Association of a missense substitution in the SRD5A2 gene with prostate cancer in African American and Hispanic men in Los Angeles, USA. Lancet 354:975–978 [DOI] [PubMed] [Google Scholar]
- Rebbeck TR, Jaffe JM, Walker AH, Wein AJ, Malkowicz SB (1998) Modification of clinical characteristics of prostate cancers by CYP3A4 genotype. J Natl Cancer Inst 90:1225–1229 [DOI] [PubMed] [Google Scholar]
- Rebbeck TR, Jaffe JM, Walker AH, White DL, Wein AJ, Malkowicz SB (1999) Glutathione-S-transferase-μ (GSTM1) and -θ (GSTT1) genotypes in the etiology of prostate cancer. Cancer Epidemiol Biomarkers Prev 8:283–287 [PubMed] [Google Scholar]
- Smith T, Freije D, Carpten JD, Gronberg H, Xu J, Isaacs SD, Brownstein MJ, Bova GS, Guo H, Bujnovszky P, Nusskern DR, Damber JE, Bergh A, Emanuelsson M, Kallioniemi OP, Walker-Daniels J, Bailey-Wilson JE, Beaty TH, Meyers DA, Walsh PC, Collins FS, Trent JM, Isaacs WB (1996) Major susceptibility locus for prostate cancer on chromosome 1suggested by a genome-wide search. Science 274:1371–1374 [DOI] [PubMed] [Google Scholar]
- Tavtigian SV, Simard J, Labrie F Skolnick MH, Neuhausen SL, Rommens J, Cannon Albright LA (2000) A strong candidate prostate cancer predisposition gene at chromosome 17p. Am J Hum Genet Suppl 67:7 [Google Scholar]
- Wacholder S, Rothman N, Caporaso N (2000) Population stratification in epidemiologic studies of common genetic variants and cancer: quantification of bias. J Natl Cancer Inst 92:1151–1158 [DOI] [PubMed] [Google Scholar]
- Walker AH, White DL, Jaffe JM, Kanetsky PA, Rebbeck TR (1999) Collection of genomic DNA by buccal swabs for PCR-based biomarker assays. Environ Health Perspect 107:3–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J (2000) Combined analysis of hereditary prostate cancer linkage to 1q24-25: results from 772 hereditary prostate cancer families from the International Consortium for Prostate Cancer Genetics. Am J Hum Genet 66:945–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Meyers D, Freije D, Isaacs S, Wiley K, Nusskern D, Ewing C, Wilkens E, Bujnovszky P, Bova GS, Walsh P, Isaacs W, Schleutker J, Matikainen M, Tammela T, Visakorpi T, Kallioniemi OP, Berry R, Schaid D, French A, McDonnell S, Schroeder J, Blute M, Thibodeau S, Trent J (1998) Evidence for a prostate cancer susceptibility locus on the X chromosome. Nat Genet 20:175–179 [DOI] [PubMed] [Google Scholar]