Abstract
Imprinted genes represent a curious defiance of normal Mendelian genetics. Mammals inherit two complete sets of chromosomes, one from the mother and one from the father, and most autosomal genes will be expressed from both the maternal and the paternal alleles. Imprinted genes, however, are expressed from only one chromosome, in a parent-of-origin–dependent manner. Because silent and active promoters are present in a single nucleus, the differences in activity cannot be explained by transcription-factor abundance. Thus, transcription of imprinted genes represents a clear situation in which epigenetic mechanisms restrict gene expression and, therefore, offers a model for understanding the role of DNA modifications and chromatin structure in maintaining appropriate patterns of expression. Furthermore, because of their parent-of-origin–restricted expression, phenotypes determined by imprinted genes are susceptible not only to genetic alterations in the genes but also to disruptions in the epigenetic programs controlling regulation. Imprinted genes are often associated with human diseases, including disorders affecting cell growth, development, and behavior.
Introduction
The nonequivalence of maternally and paternally contributed genomes was first identified in elegant nuclear-transfer studies (McGrath and Solter 1984; Surani et al. 1984). Subsequently, uniparental disomies (UPDs), in which either single chromosomes or parts thereof are inherited solely through the maternal or the paternal germlines, have been studied extensively in mice, to identify regions of the genome that carry imprinted genes (Cattanach 1986). For example, paternal UPD of the distal end of mouse chromosome 7 results in early embryonic lethality, a phenotype that can be explained by either loss of a maternal-specific transcript or the double dose of a paternal-specific transcript in these animals (Ferguson-Smith et al. 1991). Likewise, human geneticists have identified, by association of uniparental inheritance of these regions with specific diseases, chromosomal regions likely to carry imprinted genes; for example, paternal UPD of human 11p15.5 (syntenic with mouse distal 7) is associated with Beckwith-Wiedemann syndrome (BWS [MIM 130650]) (Henry et al. 1991; Weksberg et al. 1993b). Other diseases clearly associated with imprinted genes include Prader-Willi (PWS [MIM 176270]), Angelman (AS [MIM 105830]), and Russell-Silver syndromes (MIM 180860) and Albright hereditary osteodystrophy (MIM 103580). Imprinted genes contribute to language development and social affiliation (Skuse et al. 1997) and probably to other complex behavioral phenotypes, including alcohol preference, schizophrenia, and bipolar affective disorder, in humans (Nicholls 2000). Finally, disruption in the monoallelic expression of imprinted genes may be the most common mutation associated with cancer (Feinberg 2000). An excellent compilation and description of studies demonstrating parent-of-origin effects has been recently published (Morison and Reeve 1998).
Several approaches have been used to isolate imprinted genes. These include positional cloning and candidate-gene testing to find genes responsible for the UPD phenotypes in mice and humans (e.g., see Barlow et al. 1991; Lee et al. 1997), as well as genomewide scans that depend on the differential expression or epigenetic modification of maternal and paternal alleles of imprinted genes (e.g., see Hatada et al. 1993; Kaneko-Ishino et al. 1995; Piras et al. 2000). It has become clear that imprinted genes are not randomly distributed throughout the genome but, rather, are often concentrated in discrete clusters. Thus, the identification of one imprinted gene has often led to the rapid determination that nearby genes are also imprinted. Finally, allele-specific expression of several genes has been discovered serendipitously during analysis of their loss-of-function phenotypes in mouse studies. The paternal-specific expression of IGF2 (MIM 147470), the first endogenous gene to be identified as imprinted, was discovered in this way (DeChiara et al. 1991). The frequency with which imprinted genes have been fortuitously identified in knockout studies suggests that ∼0.1%–1% of all mammalian genes are imprinted (Barlow 1995). To date, ∼3 dozen imprinted genes have been identified in mice and humans.
This review will focus on recent experiments investigating the molecular basis for monoallelic expression of genes within two imprinted gene clusters: human 11p15.5/mouse distal 7 (associated with BWS and with Wilms tumor [MIM 194070]) and human 15q11-q13/central mouse chromosome 7 (associated with PWS and AS). Taken together, these studies indicate that mechanisms for parent-of-origin–specific gene expression are likely to vary from gene to gene, since the cell exploits cis-acting sequences and transcription factors already involved in determining the cell type–specific patterns of expression. Thus, dissection of imprinting pathways contributes to a general understanding of mechanisms for controlling the expression of nonimprinted genes in the mammalian cell.
Allele-Specific Expression: A Developmental Process
Imprinting can be considered a multistep developmental process. First, the chromosome must be marked as to its parental origin. Presumably, this occurs either during gametogenesis or in the zygote, prior to fusion of the two gametes, while the maternal and paternal chromosomes are still physically separate. Second, a parent-of-origin mark must be stably maintained as the cells divide and differentiate. The imprint or mark might remain identical to the original mark on the gametic chromosomes or may be a secondary derivative of that mark. Third, a parent-of-origin mark must be recognized by the transcriptional machinery, so as to result in monoallelic expression. Finally, and specific to germ cells, the mark must be erased and reset. A failure at any of these steps would result in a loss of imprinting (LOI) mutation. As described below, there is evidence for mutations in each of these steps in animal and human disease models.
Establishing the Mark: Imprinting and DNA Methylation
CpG methylation has received great attention as an excellent candidate for the genomic-imprinting mark, on the basis of two very useful properties of the DNA methyltransferase 1 (DNMT1) enzyme. First, DNMT1 has been demonstrated to associate with DNA-replication forks. Second, it shows a strong substrate preference for hemimethylated DNA. Given the semiconservative replication of DNA, these two features of the enzyme indicate that DNA, once methylated, will tend to stay methylated, thus providing a mechanism for the stable maintenance of an imprint during cell division and differentiation (Bestor and Verdine 1994).
Using CpG-sensitive restriction enzymes or bisulfite sequencing, researchers in many labs have identified parent-of-origin–specific differences in CpG methylation in almost all imprinted genes that have been examined. However, only for three genes—H19 (MIM 103280), IGF2R (MIM 147280), and Snrpn (MIM 182279)—does this methylation fit the strictest criteria of a genomic imprint—that is, the differences are present in gametes and are maintained throughout development (Stoger et al. 1993; Tremblay et al. 1995; Shemer et al. 1997). Furthermore, the functional significance of the methylated sequences for each of these genes is supported by mutational analyses that demonstrate the essential role of these elements in imprinted expression of linked genes (Wutz et al. 1997; Thorvaldsen et al. 1998; Yang et al. 1998).
A role for methylation is further supported by the demonstration that mice deficient in Dnmt1-gene function show a loss of imprinting at almost all loci tested (Li et al. 1993; Shemer et al. 1997; Caspary et al. 1998). The exact phenotype is gene specific, consistent with the association of methylation with both silent and expressed loci; for example, the H19 gene, which is normally methylated on the silent paternal allele, becomes biallelically expressed after loss of Dnmt1 function. In contrast, methylation of Igf2R and Igf2 is normally on the active allele, and Dnmt1-deficient mice fail to express these genes from either chromosome. These experiments do not distinguish between a role for Dnmt1 function in establishing the gametic imprint, maintaining the mark in somatic cells, or altering gene expression in response to the real gametic imprint.
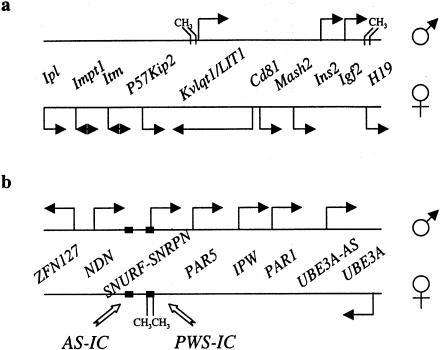
Monoallelic expression of at least one gene, Mash2, proceeds even in the absence of Dnmt1 activity (Caspary et al. 1998; Tanaka et al. 1999). This gene is part of the human 11p15.5/mouse distal 7 cluster (fig. 1a), where monoallelic expression of the H19, Igf2, Kvlqt1, and P57Kip2 genes is dependent on methylation (Li et al. 1993; Caspary et al. 1998). Perhaps these results mean only that CpG sequences crucial for imprinting of Mash2 are less sensitive to the loss of Dnmt1 function, given compensation by other methyltransferases still present in the mouse. However, organisms that lack methyltransferase enzyme activity can still maintain stable states of gene activation and repression, including parent-of-origin–specific effects (e.g., see Dalgaard and Klar 1999; Wolffe and Matzke 1999). Thus, DNA methylation is not necessarily crucial in genomic imprinting.
Figure 1 .
Clusters of imprinted genes on mouse distal 7 (syntenic with human 11p15.5) (a) and human 15q11-q13 (syntenic with mouse central 7) (b). Transcripts are depicted by arrows, with the direction indicated if known. Regions showing parent-of-origin–dependent DNA methylation are shown as CH3. The thickened segments of the lines represent the AS-IC and PWS-IC elements, as indicated. These sites are required in cis for normal imprinting on human 15q11-13.
Molecular bases by which methylation can alter transcription patterns are becoming increasingly clear (Ng and Bird 1999). Methylation can block expression directly, by interfering with the binding of transcriptional activator complexes, or it may act indirectly, by recruiting the factors that induce repressive chromatin structures. Methylation is also associated with active alleles, at several loci. In this case, the methylation is expected to interfere with recruitment of a transcriptional repressor. The exact role for methylation in monoallelic expression both in setting the genomic imprint and in altering the patterns of gene expression is best characterized for the mouse H19/Igf2 locus.
Multiple Roles for Methylation: Imprinting at the Mouse H19/Igf2 Locus
Understanding the molecular basis for the paternal specific expression of Igf2 is of long-standing interest in the field of imprinting. Mouse Igf2 was the first endogenous gene whose parent-of-origin–specific expression was recognized. In addition, overexpression of IGF2 has been a favored mechanism for the etiology of both BWS and Wilms tumor. Overexpression of Igf2 can occur through paternal UPD, through rearrangements on the maternal chromosome, or through LOI mutations, in which expression of the normally silent maternal Igf2 allele is noted (Ping et al. 1989; Weksberg et al. 1993a; Elliot and Maher 1994; Joyce et al. 1997).
Igf2 is part of a cluster of imprinted genes whose organization is well conserved in mice and humans (fig. 1a). Igf2 and its neighbor, the maternal-specific H19 gene, are coregulated. They share enhancers, at least for expression in several endodermal tissues (Leighton et al. 1995b) and in skeletal muscle (Kaffer et al. 2000). These enhancers all lie 3′ of the H19 gene and, thus, downstream of the Igf2 and the H19 promoters (fig. 2a).
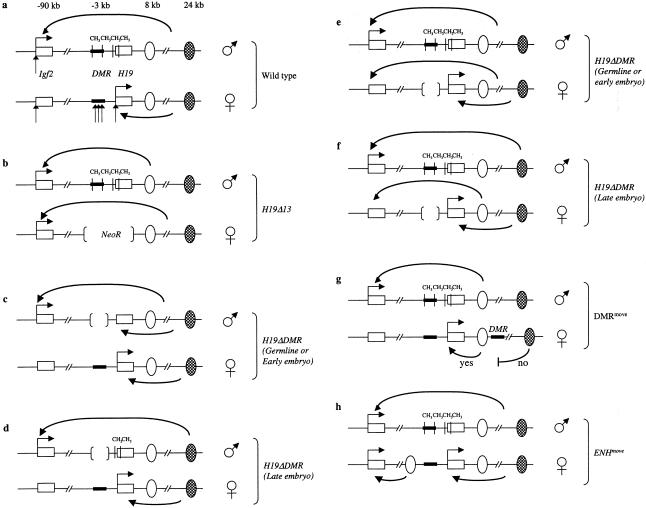
Figure 2 .
Effect that mutations in the H19DMR region have on expression of H19 and Igf2. a, Wild-type expression. Most wild-type cells express only the maternal H19 allele and only the paternal Igf2 allele. Expression of both genes is driven by shared enhancer elements. Endodermal enhancers (unblackened ovals) and skeletal muscle enhancers (blackened ovals) are ∼8 and ∼24 kb upstream of the H19 promoter, which is ∼90 kb upstream of the Igf2 promoter. Paternal chromosome–specific CpG methylation is noted in upstream sequences called “H19DMR” (thickened segments of lines). Differential methylation of this region is observed in sperm and is maintained during the global methylation changes observed during early embryogenesis. After implantation, the hypermethylation (CH3) spreads to include the H19 promoter and gene body. The maternal chromosome shows hypersensitivity to nuclease digestion (vertical arrows) in the DMR region and at the H19 promoter, whereas the Igf2 promoter appears to be equally sensitive to digestion on both chromosomes. b, Mechanistically linked imprinting of H19 and Igf2. The H19D13 allele replaces the H19 gene, including its promoter and H19DMR, with the NeoR gene. The shared endodermal and mesodermal enhancers are unaffected by this mutation, which, on maternal inheritance, results in biallelic expression of Igf2. c, H19DMR, necessary to silence paternal expression of H19. H19DDMR removes sequences that are differentially methylated on the paternal chromosome. When inherited through the paternal germline or removed from the paternal chromosome early during embryogenesis, H19 expression becomes biallelic. d, Silencing of the paternal H19 allele, mediated by epigenetic changes driven by H19DMR but not directly dependent on DMR function. When DMR sequences are removed from the paternal chromosome only late during embryogenesis, H19 expression remains monoallelic. e, H19DMR, necessary to silence expression of Igf2 from the maternal allele. When inherited through the maternal germline or removed from the maternal chromosome early during embryogenesis, Igf2 expression becomes biallelic. f, Silencing of the maternal Igf2 allele, directly dependent on the action of H19DMR. Even when the DMR is removed from the maternal chromosome only late during embryogenesis, Igf2 expression becomes biallelic. g, H19DMR and transcriptional insulator function. In DMRmove, H19DMR was inserted between the H19/Igf2 endodermal and mesodermal enhancers. After maternal inheritance of the DMRmove chromosome, H19 expression in skeletal muscle, but not in the liver, was lost. h, Biallelic expression of Igf2, allowed by moving of the endodermal enhancer elements closer to Igf2, where they are no longer separated from the Igf2 promoter by the DMR. “Early embryo” (c and e) indicates that deletions were generated in preimplantation embryos. “Late embryo” (d and f) describes deletions generated in differentiated muscle cells.
Likewise, the reciprocal imprinting of the Igf2/H19 gene pair is also mechanistically linked. LOI mutations associated with Wilms tumor generally show biallelic expression of IGF2, coupled with loss of expression of H19 from the maternal chromosome (Steenman et al. 1994; Reik et al. 1995; Catchpoole et al. 1997). In mice, a deletion of the H19 gene and 10 kb of upstream sequence that leaves the shared enhancers intact causes biallelic expression of Igf2 (Leighton et al. 1995a; fig. 2b). Together, these results suggest that silencing of paternal Igf2 might ultimately be dependent on the activity of the H19 gene, and, thus, attention has been focused on mechanisms of imprinting at that locus.
Genetic analysis of imprinting of the H19 gene itself has been greatly facilitated by the finding that relatively small transgenes carrying the H19 gene, its promoter, its enhancer elements, and several kilobases of upstream sequences can mimic the maternal-specific expression of the endogenous locus (Bartolomei et al. 1993). Deletion analyses of these transgenes demonstrated that promoter and enhancer sequences required for tissue-specific and temporally appropriate expression of H19 are not sufficient for making the transgene maternal-chromosome specific. Rather, sequences encompassing at least approximately 2 to 4/7 kb upstream of the H19 promoter are necessary to mark the H19 transgene as paternal in origin (Pfeifer et al. 1996; Elson and Bartolomei 1997; Ainscough et al. 1998; Kaffer et al. 2000) and to induce maternal-specific expression of the transgene.
Experiments to define differences in the paternal and maternal chromosomes underline the importance of these upstream sequences. These sequences are methylated in sperm but not in oocytes, and the paternal chromosome remains differentially hypermethylated throughout embryogenesis (Tremblay et al. 1995). Thus, this region, H19DMR (differentially methylated region), appears to carry a methylation imprint. During development, the hypermethylation on the paternal allele expands from H19DMR to include the H19 promoter and the gene body (fig. 2a). More recently, several labs have independently identified nuclease-hypersensitive sites in H19DMR that are specific to the maternal chromosome (Hark and Tilghman 1998; Szabo et al. 1998; Khosla et al. 1999).
The maternal H19 promoter also becomes nuclease hypersensitive, consistent with the binding of transcription factors and activation of expression of the maternal H19 allele. (Bartolomei et al. 1993; Ferguson-Smith et al. 1993). Strikingly, no allelic differences in nuclease hypersensitivity at the Igf2 promoter have been reported, which suggests that both the maternal and paternal Igf2 promoters might be equally ready for activation but that some other factor prevents enhancement on the maternal chromosome (Sasaki et al. 1992).
Mice inheriting a deletion of the H19DMR through the father show loss of imprinting at H19—that is, these mice are biallelic for expression of H19 (Thorvaldsen et al. 1998; fig. 2c). Thus, H19DMR acts to silence the paternal H19 promoter. To address the mechanism of silencing, H19DMR was flanked with Cre lox sites and was deleted in a temporally regulated manner, by use of cell type–specific Cre recombinase transgenes (Srivastava et al. 2000). It was therefore possible to generate mice that had inherited a wild-type paternal chromosome in which H19DMR was deleted either in early embryos or in differentiated cells (fig. 2c and d). When H19DMR was deleted from the paternal chromosome in differentiated cells, the paternal H19 allele remained silent. However, when a wild-type chromosome is inherited through the sperm but H19DMR is deleted early during embryogenesis, repression of the paternal allele is lost. Thus, H19DMR’s role in monoallelic expression is temporary, and its silencing activity is not required for direct interaction with the transcriptional machinery. Bisulfite-sequencing experiments demonstrate that H19DMR is required during early embryogenesis, to direct methylation of the H19 promoter. These changes are then stably maintained independent of DMR and probably directly repress the paternal allele. These results are consistent with earlier experiments, which showed that the repression of the paternal H19 is actually a process that proceeds as the embryo develops (Jinno et al. 1995; Szabo and Mann 1995; Svensson et al. 1998).
In sum, genetic experiments demonstrate that H19DMR is necessary to maintain the H19 imprint during early development and to convert the imprint into a signal that actually represses the transcriptional machinery. Molecular studies support the notion that DMR carries the actual imprint, but genetic studies to date have not been able to address this issue.
H19DMR is also required for monoallelic expression of Igf2 (Thorvaldsen et al. 1998; fig. 2e). Its deletion on the maternal chromosome results in inappropriate activation of the maternal Igf2 allele. Thus, H19DMR is also a silencer for Igf2. Strikingly, the mechanism for silencing of Igf2 is distinct from the mechanism used to silence paternal H19 (Srivastava et al. 2000; fig. 2e–f). Deletion of DMR in a cell type–specific manner demonstrates that its presence is continually necessary to silence the maternal Igf2 promoter, indicating that maternal H19DMR and/or bound proteins interact directly with the transcriptional machinery, to block expression of maternal Igf2. One model to explain H19DMR’s action in suppressing maternal Igf2 expression posits that it functions as a transcriptional insulator. On the maternal chromosome, the insulator prevents activation of the Igf2 promoter by the distal enhancers. On the paternal chromosome, the methylation imprint inactivates the insulator, allowing activation of the paternal Igf2 allele.
The hallmark of a transcriptional insulator is its ability to prevent activation of a promoter, in a strictly position-dependent manner. An insulator prevents transcription only when juxtaposed between a promoter and the enhancers on which that promoter depends for its activation. The ability of H19DMR sequences to act as a transcriptional insulator has been demonstrated in vivo by use of transgenes inserted at heterologous positions in the chromosome (Hark et al. 2000) and by manipulation of the H19/Igf2 locus itself (Kaffer et al. 2000; fig. 2g). In the latter experiment, DMR was removed to a position downstream of the H19 gene, between the endodermal and skeletal muscle enhancer elements. This construct, DMRmove, thus places the putative insulator between the H19 promoter and the muscle enhancers and thereby mimics the topology of the Igf2 locus (fig. 2g). With maternal inheritance, activation of H19 in skeletal muscle but not in liver was blocked, consistent with the presence of insulator activity on the H19DMR insert. With paternal inheritance, the relocated H19DMR is methylated, and its insulator function abrogated (C. R. Kaffer and K. Pfeifer, unpublished observations).
The ability of H19DMR sequences to act as insulators has also been demonstrated in vitro by use of integrated (Bell and Felsenfeld 2000; Hark et al. 2000; Kaffer et al. 2000) and episomal (Kanduri et al. 2000) minigene constructs. The ability of transfected DNA constructs to mimic expression patterns and chromatin conformations of the maternal locus is good evidence that imprinting at H19/Igf2 is solely a paternal marking of the chromosome.
The molecular basis for paternal-specific inactivation of the insulator is provided by biochemical analyses that show that binding of the CCCTC-binding factor (CTCF) protein to DMR sequences is inhibited by CpG methylation of the protein’s recognition sites (Bell and Felsenfeld 2000; Hark et al. 2000). CTCF is a transcription factor that has been demonstrated elsewhere to play a role in insulator activity at the chicken β-globin locus (Bell et al. 1999). That CTCF actually plays a role in the cell is strongly supported by in vivo footprinting results that demonstrate maternal chromosome–specific CTCF (Szabo et al. 2000).
Together, these results present a conceptually simple model for imprinting at H19/Igf2. The maternal chromosome represents the default or unimprinted state. The H19 promoter is active, as is a transcriptional insulator that prevents activation of the Igf2 promoter by the shared enhancer elements. The paternal chromosome is marked or imprinted at DMR, and this imprint has two functions. First, it directs further epigenetic modification of the H19 promoter, which blocks its activity. Second, the methylation prevents binding of CTCF protein and thus prevents activation of the insulator element, thereby permitting enhancer-mediated activation of the Igf2 promoter. Experiments that introduced chromosome 11 into mouse cells demonstrated that these mechanisms for transcriptional regulation are likely to be conserved in humans (Gabriel et al. 1998b).
LOI Mutations at Human IGF2
Biallelic expression of IGF2 without deletion or rearrangement on either chromosome is frequently associated with BWS and with Wilms tumor. In some cases, this LOI at IGF2 is associated with normal, maternal-specific expression of H19 (Brown et al. 1996; Joyce et al. 1997). In other cases, LOI at IGF2 is associated with inappropriate silencing and hypermethylation of both H19 alleles (Steenman et al. 1994; Reik et al. 1995; Catchpoole et al. 1997). How can these LOI phenotypes be viewed, in light of the mouse studies just described?
The first LOI phenotype—biallelic IGF2/normal H19 expression—does present in the mouse, in which the maternal chromosome is inherited in its wild-type form but in which an H19DMR deletion occurs late during embryonic development (fig. 2f). Thus, by analogy with the mouse, these patients with LOI may be better described as having loss of insulation rather than imprinting. The interesting issue is the molecular basis for loss of insulation in these patients. In mice, this phenotype is caused by a deletion of the insulator element. In most patients, no chromosomal abnormalities or genetic alterations are observed. Intriguingly, however, LOI can be associated with chromosomal inversion/deletions in the BWSCR1 region, which spans the KVLQT1 (KCNQ1) gene, several hundred kilobases upstream of IGF2 (Mannens et al. 1994; Hoovers et al. 1995; fig. 1a).
Mouse studies predict that the maternal Igf2 promoter is ready for activation but is silent because of the lack of activation by enhancer elements. If this is also the case for human IGF2, there are only two formal explanations for the ability of a mutation to permit activation of the maternal IGF2. In the first case, insulator function of H19DMR is unaffected, and the normal IGF2 enhancers remain unable to activate the maternal IGF2 promoter. Then the effect of BWS mutations must be to somehow “create” a new enhancer element for IGF2. To explain molecular data, this new enhancer would need to activate IGF2 in the appropriate tissues and at levels like that of the endogenous enhancer. Such a mutation was, in fact, engineered in the mouse by removal of the normal endodermal enhancers to a position midway between H19 and Igf2 and, thus, on the proximal side of the Igf2 promoter, relative to the H19DMR insulator (Webber et al. 1998; fig. 2h). The alternative—and, I think, more likely—scenario is that the H19DMR insulator function is abolished in these patients with LOI. This loss of insulation then allows expression of maternal IGF2 to be driven by the normal enhancers. In this scenario, LOI occurs via an epigenetic or genetic alteration in sequences required to act with the H19DMR to organize the maternal IGF2 into a transcriptionally silent domain. Perhaps the organization of the locus into domains of expressed and unexpressed genes uses paired insulator/boundary domains like the scs/scs′ system described in Drosophila (Udvardy 1985; Kellum and Schedl 1991, 1992). Further directed mutagenesis in mice, mimicking the translocation chromosomes of patients with BWS, will help elucidate the true nature of these mutations.
The second type of LOI mutation in BWS shows biallelic IGF2 associated with loss of expression of the maternal H19 gene (Brown et al. 1996; Joyce et al. 1997). By analogy with the mouse, this phenotype would be more usefully described as gain of imprinting, because the patient has, in effect, two imprinted chromosomes. No such phenotype is observed in mice, and it is hard to imagine how to generate such a phenotype via genetic alterations. Bestor and Tycko (1996) have described an epigenetic mechanism that could create de novo methylation during DNA synthesis. When imprinted domains pair during mitosis (LaSalle and LaLande 1996), strand exchange that occurs infrequently between methylated and nonmethylated sequences will present hemimethylated sites to the DNMT1 enzyme. DNMT1 maintenance activity thus is used by the organism to create de novo methylation, given an already methylated template. Such a phenomenon has been described in the fungus Ascobulus (Colot et al. 1996).
A Second Imprint on 11p15.5?
One clear finding of both mouse and human studies is that imprinting at the H19/IGF2 locus can be mechanistically separated from imprinting of other genes in the cluster. First, the effect of deletion of the H19 gene and H19DMR is restricted to the H19, Igf2, and Ins2 loci; the deletion has no effect on imprinting of Mash2, Kvlqt1, or p57Kip2 (Leighton et al. 1995a; Caspary et al. 1998). Second, the effects of LOI at IGF2 are, in most cases, restricted to that gene and to H19 (Reik et al. 1995). Finally, genes in the distal 7 cluster are differentially affected by Dnmt1 loss-of-function mutations. Together, these findings suggest that there are probably additional cis-acting imprinting centers (ICs) at human 11p15.5/mouse distal 7. The disruption of the KVLQT1 locus in multiple cases of BWS suggests that KVLQT1 may harbor these elements (Lee et al. 1997).
Two labs have identified a region near KVLQT1 exon 10, KvDMR, that is hypermethylated specifically on the maternal chromosome (Lee et al. 1999; Mitsuya et al. 1999; Smilinich et al. 1999). Preliminary evidence supports the notion that this methylation is a true imprint, in that it appears to be oocyte specific and not acquired developmentally in response to allele-specific transcription. The functional significance of KvDMR is not yet clear. LOI at the site is associated with a number of cases of BWS, including cases that also show LOI at IGF2 but, more generally, cases that are normal for IGF2 function. Lee et al. (1999) speculate that KvDMR might be a regulatory site working analogously to the H19DMR, whose methylation permits expression of the downstream KVLQT1 and p57KIP2 promoters.
If independent mechanisms determine imprinting for discrete clusters of genes within the 11p15.5/distal 7 supercluster, it remains puzzling why the clusters are then grouped in the first place. One explanation is that the region contains sequences that make the whole region permissive for imprinting but that individual IC centers are further necessary to implement the imprint. This description does not account for the ability of relatively small H19 transgenes to direct their own imprinting at a wide range of chromosomal integration sites. Imprinting of transgenes has not been demonstrated for other genes in the cluster.
Imprinting at a Distance: 15q11-q13
A second well-characterized cluster of imprinted genes is located on human chromosome 15q11-q13 (central mouse 7) (fig. 1b). PWS and AS are two clinically distinct neurobehavioral disorders that are each most commonly caused by an identical 4-Mbp deletion of this region. However, the deletion is always paternal in origin for PWS but maternal in origin for AS (for reviews, see Jiang et al. 1998; Nicholls et al. 1998). The maternally expressed UBE3A gene maps toward the telomeric end of the deletion, and loss of UBE3A is likely to explain AS (Nakao et al. 1994; Rougeulle et al. 1997; Vu and Hoffman 1997). At least six paternal-specific transcripts—ZNF127, NDN, SNURF/SNRPN, PAR5, IPW, and PAR1—have been identified, and all map to the centromeric end of the deletion (Ozcelik et al. 1992; Sutcliffe et al. 1994; Wevrick et al. 1994; MacDonald and Wevrick 1997; Sutcliffe et al. 1997; Gray et al. 1999; Jong et al. 1999; fig. 1b). More recently, eight novel imprinted transcripts have been mapped to the locus, but their regulation by the IC mutations (see below) has not yet been tested (Lee and Wevrick 2000). The mapped imprinted genes span >2 Mb. A region in the middle of the cluster that encompasses the SNURF/SNRPN promoter is hypermethylated on the maternal chromosome at all stages of development and is therefore a good candidate for a gametic imprint (Sutcliffe et al. 1994; Glenn et al. 1996). This imprint is conserved in the mouse (Shemer et al. 1997; Gabriel et al. 1998a).
One genetically interesting class of patients with PWS have biparental inheritance of chromosome 15, but both chromosomes behave maternally—that is, are hypermethylated at the SNURF/SNRPN locus and express only UBE3A (Reis et al. 1994; Buiting et al. 1995; Saitoh et al. 1996). In rare patients with PWS who have microdeletions causing this imprinting defect, the PWS imprinting center (PWS-IC) has been mapped to a 4.3-kbp region spanning the SNURF/SNRPN promoter and coincident with the methylation imprint (Ohta et al. 1999a; fig. 1b). These PWS-IC mutations can be transmitted silently over multiple generations, with the phenotype apparent only when the imprint needs to be reset from maternal to paternal. Thus, it has been suggested that, at this locus in humans and in mice, PWS-IC controls switching in the male germline (Yang et al. 1998). However, recent experiments demonstrate that PWS-IC, in both humans and mice, is required for maintainance of the paternal identity of the chromosome during early embryogenesis (Bielinska et al. 2000).
At a superficial level, PWS-IC thus resembles H19DMR. Methylation of either element results in silencing of linked genes—either SNURF/SNRPN and other paternal genes at 15q11-q13 or H19 at 11p15.5. In both cases, they are certainly required during early embryogenesis, and probably also in germ cells, to mark the silent chromosome. Of course, PWS-IC is able to silence transcription of genes that are orders of magnitude distant, relative to the effects of H19DMR. The mechanism by which the silent state is transmitted over such long distances is not understood. The overlap of the IC with SNURF/SNRPN is intriguing, and it is not clear whether this is coincidence or whether expression of that promoter and/or the RNA product itself are somehow required to keep the paternal chromosome transcriptionally active. However, at least some insertional mutations at the SNURF/SNRPN locus do not disrupt imprinting (Yang et al. 1998; Tsai et al. 1999).
The ability of the imprint to activate maternal UBE3A is perhaps explained by the recent discovery of a paternally expressed antisense transcript overlapping UBE3A but proximal to it, relative to PWS-IC (Rougeulle et al. 1998; fig. 1b). The authors who reported this discovery proposed that UBE3A imprinting might be the indirect effect of paternal-specific expression of the antisense transcript. In keeping with this hypothesis, UBE3A is biallelic in all tissues in which the antisense tissue is not expressed, but it is monoallelic in the brain, where the antisense transcript is expressed.
The previous discussion of imprinting at 15q11-q13 is oversimplified, however, because it has not accounted for the presence of an AS-IC in addition to PWS-IC. This 1.15-kb element is ∼40 kb upstream of the SNURF/SNRPN promoter (fig. 1b) (Ohta et al. 1999b) and is defined by the microdeletions that cause maternally inherited chromosomes to show a paternal phenotype—that is, hypomethylation at the SNRPN promoter, expression of paternal transcripts, and loss of maternal-specific transcription of UBE3A. This situation appears to be very different from the H19/Igf2 system, in which it is presumed that the chromosome is imprinted in only one germline and passes through the other unmarked.
Several models describe mechanisms for gamete-specific imprinting events at 15q11-q13 (Buiting et al. 1995; Dittrich et al. 1996; Burger et al. 1997; Ohta et al. 1999a). One unifying model proposes that the role of both IC elements is to regulate expression of paternal-specific genes (Brannan and Bartolomei 1999). PWS-IC is a cis-regulatory element required for activation of the paternal expression pattern, possibly via its role in initiating or maintaining transcription of SNURF/SNRPN. Thus, loss of PWS-IC prevents activation of the paternal program. In the female germline, the imprint, which is dependent on some transcript or sequence from AS-IC, is established at the SNURF/SNRPN promoter. This imprint prevents SNURF/SNRPN transcription and, thereby, halts the rest of the paternal program. As predicted by this model, deletion of PWS-IC is epistatic to deletion of AS-IC.
Summary
Although imprinting remains largely mysterious, recent experiments have made considerable progress in elucidating some of the mechanisms for monoallelic expression of imprinted genes. These results indicate that, although DNA methylation plays a crucial role, its direct effect on transcription will vary from locus to locus. At the H19/Igf2 locus, methylation represses transcription of the paternal H19 allele by directly blocking the activation of the H19 promoter but activates expression of Igf2 by its simultaneous inactivation of a transcriptional insulator. On human 15q11-q13, methylation appears to directly block activation of the maternal SNURF/SNRPN promoter, perhaps analogous to its effect on paternal H19. However, it activates UBE3A expression, possibly indirectly, by blocking expression of an antisense RNA. These examples of imprinting effects on promoter activation, insulator function, and long-range chromatin structure suggest that imprinting has evolved in the mammal by using conventional mechanisms of transcriptional regulation. Thus, through clever engineering, the cell is able to use methylation imprints as both positive and negative signals. Dissecting monoallelic expression pathways will therefore contribute toward an understanding of normal gene regulation and of the molecular basis for diseases associated with disregulation at imprinted loci.
Electronic-Database Information
The URL and accession numbers for data in this article are as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim (for BWS [MIM 130650], PWS [MIM 176270], AS [MIM 105830], Russell Silver syndrome [180860], Albright hereditary osteodystrophy [MIM 103580], Wilms tumor [MIM 194070], Igf2 [MIM 147470], H19 [MIM 103280], Snrpn [MIM 182279], and Igf2R [MIM 147280])
References
- Ainscough JF, John RM, Surani MA (1998) Mechanism of imprinting on mouse distal chromosome 7. Genet Res 72:237–245 [DOI] [PubMed] [Google Scholar]
- Barlow DP (1995) Gametic imprinting in mammals. Science 270:1610–1613 [DOI] [PubMed] [Google Scholar]
- Barlow DP, Stoger R, Herrmann BG, Saito K, Schweifer N (1991) The mouse insulin-like type-2 receptor is imprinted and closely linked to the Tme locus. Nature 349:84–87 [DOI] [PubMed] [Google Scholar]
- Bartolomei MS, Webber AL, Brunkow ME, Tilghman SM (1993) Epigenetic mechanisms underlying the imprinting of the mouse H19 gene. Genes Dev 7:1663–1673 [DOI] [PubMed] [Google Scholar]
- Bell AC, Felsenfeld G (2000) Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405:482–485 [DOI] [PubMed] [Google Scholar]
- Bell AC, West AG, Felsenfeld G (1999) The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell 98:387–396 [DOI] [PubMed] [Google Scholar]
- Bestor TH, Tycko B (1996) Creation of genomic methylation patterns. Nat Genet 12:363–367 [DOI] [PubMed] [Google Scholar]
- Bestor TH, Verdine GL (1994) DNA methyltransferases. Curr Opin Cell Biol 6:380–389 [DOI] [PubMed] [Google Scholar]
- Bielinska B, Blaydes SM, Buiting K, Yang T, Krajewska-Walasek M, Horsthemke B, Brannan CI (2000) De novo deletions of SNRPN exon 1 in early human and mouse embryos result in a paternal to maternal imprint switch. Nat Genet 25:74–78 [DOI] [PubMed] [Google Scholar]
- Brannan CI, Bartolomei MS (1999) Mechanisms of genomic imprinting. Curr Opin Genet Dev 9:164–170 [DOI] [PubMed] [Google Scholar]
- Brown KW, Villar AJ, Bickmore W, Clayton-Smith J, Catchpoole D, Maher ER, Reik W (1996) Imprinting mutation in the Beckwith-Wiedemann syndrome leads to biallelic IGF2 expression through an H19-independent pathway. Hum Mol Genet 5:2027–2032 [DOI] [PubMed] [Google Scholar]
- Buiting K, Saitoh S, Gross S, Dittrich B, Schwartz S, Nicholls RD, Horsthemke B (1995) Inherited microdeletions in the Angelman and Prader-Willi syndromes define an imprinting centre on human chromosome 15. Nat Genet 9:395–400 [DOI] [PubMed] [Google Scholar]
- Burger J, Buiting K, Dittrich B, Gross S, Lich C, Sperling K, Horsthemke B, Reis A (1997) Different mechanisms and recurrence risks of imprinting defects in Angelman syndrome. Am J Hum Genet 61:88–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspary T, Cleary MA, Baker CC, Guan XJ, Tilghman SM (1998) Multiple mechanisms regulate imprinting of the distal chromosome 7 gene cluster. Mol Cell Biol 18:3466–3474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catchpoole D, Lam WW, Valler D, Temple IK, Joyce JA, Reik W, Schofeld PN, Maher ER (1997) Epigenetic modification and uniparental inheritance of H19 in Beckwith-Wiedemann syndrome. J Med Genet 34:353–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattanach BM (1986) Parental origin effects in mice. J Embryol Exp Morph 97(Suppl):137–150 [PubMed]
- Colot V, Maloisel L, Rossignol JL (1996) Interchromosomal transfer of epigenetic states in Ascobulus: transfer of DNA methylation is mechanistically related to homologous recombination. Cell 86:855–864 [DOI] [PubMed] [Google Scholar]
- Dalgaard JZ, Klar AJ (1999) Orientation of DNA replication establishes mating-type switching pattern in S. pombe. Nature 400:181–184 [DOI] [PubMed] [Google Scholar]
- DeChiara TM, Robertson EJ, Efstratiadis A (1991) Parental imprinting of the mouse insulin-like growth factor II gene. Cell 64:849–859 [DOI] [PubMed] [Google Scholar]
- Dittrich B, Buiting K, Korn B, Rickard S, Buxton J, Saitoh S, Nicholls RD, Poustka A, Winterpacht A, Zabel B, Horsthemke B (1996) Imprint switching on human chromosome 15 may involve alternative transcripts of the SNRPN gene. Nat Genet 14:163–170 [DOI] [PubMed] [Google Scholar]
- Elliott M, Maher ER (1994) Beckwith-Wiedemann syndrome. J Med Genet 31:560–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elson DA, Bartolomei MS (1997) A 5′ differentially methylated sequence and the 3′-flanking region are necessary for H19 transgene imprinting. Mol Cell Biol 17:309–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg AP (2000) DNA methylation, genomic imprinting and cancer. Curr Top Microbiol Immunol 249:87–99 [DOI] [PubMed] [Google Scholar]
- Ferguson-Smith AC, Cattanach BM, Barton SC, Beechey CV, Surani MA (1991) Embryological and molecular investigations of parental imprinting on mouse chromosome 7. Nature 351:667–670 [DOI] [PubMed] [Google Scholar]
- Ferguson-Smith AC, Sasaki H, Cattanach BM, Surani MA (1993) Parental-origin-specific epigenetic modifications of the mouse H19 gene. Nature 362:751–755 [DOI] [PubMed] [Google Scholar]
- Gabriel JM, Gray TA, Stubbs L, Saitoh S, Ohta T, Nicholls RD (1998a) Structure and function correlations at the imprinted mouse Snrpn locus. Mamm Genome 9:788–793 [DOI] [PubMed]
- Gabriel JM, Higgins MJ, Gebuhr TC, Shows TB, Saitoh S, Nicholls RD (1998b) A model system to study genomic imprinting of human genes. Proc Natl Acad Sci USA 95:14857–14862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glenn CC, Saitoh S, Jong MT, Filbrandt MM, Surti U, Driscoll DJ, Nicholls RD (1996) Gene structure, DNA methylation, and imprinted expression of the human SNRPN gene. Am J Hum Genet 58:335–346 [PMC free article] [PubMed] [Google Scholar]
- Gray TA, Saitoh S, Nicholls RD (1999) An imprinted, mammalian bicistronic transcript encodes two independent proteins. Proc Natl Acad Sci USA 96:5616–5621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM (2000) CTCF mediates methylation-sensitive enhancer blocking activity at the H19/Igf2 locus. Nature 405:486–489 [DOI] [PubMed] [Google Scholar]
- Hark AH, Tilghman SM (1998) Chromatin conformation of the H19 epigenetic mark. Hum Mol Genet 7:1979–1985 [DOI] [PubMed]
- Hatada I, Sugama T, Mukai T (1993) A new imprinted gene cloned by a methylation-sensitive genome scanning method. Nucleic Acids Res 21:5577–5582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry I, Bonaiti-Pellie C, Chehensse V, Beldjord C, Schwartz C, Utermann G, Junien C (1991) Uniparental disomy in a genetic cancer-predisposing syndrome. Nature 351:665–667 [DOI] [PubMed] [Google Scholar]
- Hoovers HMN, Kalikin LM, Johnson LA, Alders M, Redeker B, Law DJ, Bliek J, Steenman M, Benedict M, Wiegant J, Lengauer C, Taillon-Miller P, Schlessinger D, Edwards MC, Elledge SJ, Ivens A, Westerveld A, Little P, Mannens M, Feinberg AP (1995) Multiple genetic loci within 11p15 defined by Beckwith-Wiedemann syndrome rearrangement breakpoints with subchromosomal transferable fragments. Proc Natl Acad Sci USA 92:12456–12460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Tsai TF, Bressler J, Beaudet AL (1998) Imprinting in Angelman and Prader-Willi syndromes. Curr Opin Genet Dev 8:334–342 [DOI] [PubMed] [Google Scholar]
- Jinno Y, Ikeda Y, Yun K, Maw M, Masuzaki H, Fukada H, Inuzuka K, Fujishita A, Ohtani Y, Okimoto T, Ishimaru T, Niikawa N (1995) Establishment of functional imprinting of the H19 gene in human developing placentae. Nat Genet 10:318–324 [DOI] [PubMed] [Google Scholar]
- Jong MT, Gray TA, Ji Y, Glenn CC, Saitoh S, Driscoll DJ, Nicholls RD (1999) A novel imprinted gene, encoding a RING zinc-finger protein, and overlapping antisense transcript in the Prader-Willi syndrome critical region. Hum Mol Genet 8:783–793 [DOI] [PubMed] [Google Scholar]
- Joyce JA, Lam WK, Catchpoole DJ, Jenks P, Reik W, Maher ER, Schofield PN (1997) Imprinting of IGF2 and H19: lack of reciprocity in sporadic Beckwith-Wiedemann syndrome. Hum Mol Genet 9:1543–1548 [DOI] [PubMed]
- Kaffer CR, Srivastava M, Park KY, Ives E, Hsieh S, Batlle J, Grinberg A, Huang SP, Pfeifer K (2000) A transcriptional insulator at the imprinted H19/Igf2 locus. Genes Dev 14:1908–1919 [PMC free article] [PubMed] [Google Scholar]
- Kanduri C, Holmgren C, Pilartz M, Franklin G, Kanduri M, Liu L, Ginjala V, Ulleras E, Mattsson R, Ohlsson R (2000) The 5′-flanking of the murine H19 gene in an unusual chromatin conformation unidirectionally blocks enhancer-promoter communication. Curr Biol 10:449–457 [DOI] [PubMed] [Google Scholar]
- Kaneko-Ishino T, Kuroiwa Y, Miyoshi N, Kohda T, Suzuki R, Yokoyama M, Viville S, Barton SC, Ishino F, Surani MA (1995) Peg1/Mest imprinted gene on chromosome 6 identified by cDNA subtraction hybridization. Nat Genet 11:52–59 [DOI] [PubMed] [Google Scholar]
- Kellum R, Schedl P (1991) A position-effect assay for boundaries of higher order chromosomal domains. Cell 64:941–950 [DOI] [PubMed] [Google Scholar]
- ——— (1992) A group of scs elements function as domain boundaries in an enhancer-blocking assay. Mol Cell Biol 12:2424–2431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khosla S, Aitchison A, Gregory R, Allen ND, Feil R (1999) Parental allele-specific chromatin configuration in a boundary-imprinting-control element upstream of the mouse H19. Mol Cell Biol 19:2556–2566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaSalle JM, LaLande M (1996) Homologous association of oppositely imprinted chromosomal domains. Science 272:725–728 [DOI] [PubMed] [Google Scholar]
- Lee MP, DeRaun MR, Mitsuya K, Galonek HL, Brandenburg S, Oshimura M, Feinberg AP (1999) Loss of imprinting of a paternally expressed transcript, with antisense orientation to KvLQT1 occurs frequently in Beckwith-Wiedemann syndrome and is independent of insulin-like growth factor II imprinting. Proc Natl Acad Sci USA 96:5203–5208 [DOI] [PMC free article] [PubMed]
- Lee MP, Hu RJ, Johnson LA, Feinberg AP (1997) Human KVLQT1 gene shows tissue-specific imprinting and encompasses Beckwith-Wiedemann syndrome chromosomal rearrangements. Nat Genet 15:181–185 [DOI] [PubMed] [Google Scholar]
- Lee S, Wevrick R (2000) Identification of novel imprinted transcripts in the Prader-Willi syndrome and Angelman syndrome deletion region: further evidence for regional imprinting control. Am J Hum Genet 66:848–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leighton PA, Ingram RS, Eggenschwiler J, Efstratiadis A, Tilghman SM (1995a) Disruption of imprinting caused by deletion of the H19 gene region in mice. Nature 375:34–39 [DOI] [PubMed] [Google Scholar]
- Leighton PA, Saam JR, Ingram RS, Stewart CL, Tilghman SM (1995b) An enhancer deletion affects both H19 and Igf2 expression. Genes Dev 9:2079–2089 [DOI] [PubMed] [Google Scholar]
- Li E, Beard C, Jaenisch R (1993) Role for DNA methylation in genomic imprinting. Nature 366:362–365 [DOI] [PubMed] [Google Scholar]
- MacDonald HR, Wevrick R (1997) The necdin gene is deleted in Prader-Willi syndrome and is imprinted in human and mouse. Hum Mol Genet 6:1873–1878 [DOI] [PubMed] [Google Scholar]
- Mannens M, Hoovers JMN, Redeker E, Verjaal M, Feinberg AP, Little P, Boavida M, Coad N, Steenman M, Bliek J, Niikawa N, Tonoki H, Nakamura Y, de Boer EG, Slater RM, John R, Cowell JK, Junien C, Henry I, Tommerup N, Weksburg R, Pueschel SM, Leschot NJ, Westerveld A (1994) Parental imprinting of human chromosome region 11p15.3-pter involved in the Beckwith-Wiedemann syndrome and various human neoplasia. Eur J Hum Genet 2:3–23 [DOI] [PubMed] [Google Scholar]
- McGrath J, Solter D (1984) Completion of mouse embryogenesis requires both the maternal and paternal genomes. Cell 37:179–183 [DOI] [PubMed] [Google Scholar]
- Mitsuya M, Meguro M, Lee MP, Katoh M, Schulz TC, Kugoh H, Yoshida MA, Niikawa N, Feinberg AP, Oshimura M (1999) LIT1, an imprinted antisense RNA in the human KvLQT1 locus identified by screening for differentially expressed transcripts using monochromosomal hybrids. Hum Mol Genet 8:1209–1217 [DOI] [PubMed] [Google Scholar]
- Morison IM, Reeve AE (1998) A catalogue of imprinted genes and parent-of-origin effects in humans and animals. Hum Mol Genet 7:1599–1609 [DOI] [PubMed] [Google Scholar]
- Nakao M, Sutcliffe JS, Durtschi B, Mutirangura A, Ledbetter DH, Beaudet AL (1994) Imprinting analysis of three genes in the Prader-Willi/Angelman region: SNRPN, E6-associated protein, and PAR-2. Hum Mol Genet 3:309–315 [DOI] [PubMed] [Google Scholar]
- Ng HH, Bird A (1999) DNA methylation and chromatin modification. Curr Opin Genet Dev 9:158–163 [DOI] [PubMed] [Google Scholar]
- Nicholls RD (2000) The impact of genomic imprinting for neurobehavioral and developmental disorders. J Clin Invest 105:413–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls RD, Saitoh S, Horsthemke B (1998) Imprinting in Prader-Willi and Angelman syndrome. Trends Genet 14:194–200 [DOI] [PubMed] [Google Scholar]
- Ohta T, Buiting K, Kokkonen H, McCandless S, Heeger S, Leisti H, Driscoll DJ, Cassidy SB, Horsthemke B, Nicholls RD (1999a) Molecular mechanism of Angelman syndrome in two large families involves an imprinting mutation. Am J Hum Genet 64:385–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta T, Gray TA, Rogan PK, Buiting K, Gabriel JM, Saitoh S, Muralidhar B, Bilienska B, Krajewska-Walasek M, Driscoll DJ, Horsthemke B, Butler MG, Nicholls RD (1999b) Imprinting-mutation mechanisms in Prader-Willi syndrome. Am J Hum Genet 64:397–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozcelik T, Leff SE, Robinson W, Donlon T, Lalande M, Sanjines E, Schinzel A, Francke U (1992) Small nuclear ribonucleoprotein polypeptide N (SNRPN), an expressed gene in the Prader-Willi critical region. Nat Genet 2:265–269 [DOI] [PubMed] [Google Scholar]
- Pfeifer K, Leighton PA, Tilghman SM (1996) The structural H19 gene is required for transgene imprinting. Proc Natl Acad Sci USA 93:13876–13883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ping AJ, Reeve AE, Law DJ, Young MR, Boehnke M, Feinberg AP (1989) Genetic linkage of Beckwith-Wiedemann syndrome to 11p15. Am J Hum Genet 44:720–723 [PMC free article] [PubMed] [Google Scholar]
- Piras G, El Kharroubi A, Kozlov S, Escalante-Alcalde D, Hernandez L, Copeland NG, Gilbert DJ, Jenkins NA, Stewart CL (2000) Zac1 (Lot1), a potential tumor suppresser gene, and the gene for ε-sarcoglycan are maternally imprinted genes: identification by a subtractive screen of novel uniparental fibroblast lines. Mol Cell Biol 20:3308–3315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reik W, Brown KW, Schneid H, Le Bouc Y, Bickmore W, Maher ER (1995) Imprinting mutations in the Beckwith-Wiedemann syndrome suggested by an altered imprinting pattern in the IGF2-H19 domain. Hum Mol Genet 4:2379–2385 [DOI] [PubMed] [Google Scholar]
- Reis A, Dittrich B, Greger V, Buiting K, Lalande M, Gillessen-Kaesback G, Anvret M, Horsthemke B (1994) Imprinting mutations suggested by abnormal DNA methylation patterns in familial Angelman and Prader-Willi syndromes. Am J Hum Genet 54:741–747 [PMC free article] [PubMed] [Google Scholar]
- Rougeulle C, Cardoso D, Fontes M, Colleaux L, Lalande M (1998) An imprinted antisense RNA overlaps UBE3A and a second maternally expressed transcript. Nat Genet 19:15–16 [DOI] [PubMed] [Google Scholar]
- Rougeulle C, Glatt H, Lalande M (1997) The Angelman syndrome candidate gene, UBE3A/E6-AP, is imprinted in brain. Nat Genet 17:14–15 [DOI] [PubMed] [Google Scholar]
- Saitoh S, Buiting K, Rogan PK, Buxton JL, Driscoll DJ, Arnemann J, Konig R, Malcolm S, Horsthemke B, Nicholls RD (1996) Minimal definition of the imprinting center and fixation of a chromosome 15q11-q13 epigenotype by imprinting mutations. Proc Natl Acad Sci USA 93:7811–7815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki H, Jones PA, Chaillet JR, Ferguson-Smith AC, Barton SC, Reik W, Surani MA (1992) Parental imprinting: potentially active chromatin of the repressed maternal allele of the mouse insulin-like growth factor II (Igf2) gene. Genes Dev 6:1843–1856 [DOI] [PubMed] [Google Scholar]
- Shemer R, Birger Y, Riggs AD, Razin A (1997) Structure of the imprinted mouse Snrpn gene and establishment of its parental-specific methylation pattern. Proc Natl Acad Sci USA 94:10267–10272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skuse DH, James RS, Bishop DV, Coppin B, Dalton P, Aamodt-Leeper G, Bacarese-Hamilton M, Creswell C, McGurk R, Jacobs PA (1997) Evidence from Turner's syndrome of an imprinted X-linked locus affecting cognitive function. Nature 387:705–708 [DOI] [PubMed] [Google Scholar]
- Smilinich NJ, Day CD, Fitzpatrick GV, Caldwell GM, Lossie AC, Cooper PR, Smallwood AC, Joyce JA, Schofield PN, Reik W, Nicholls RD, Weksburg R, Driscoll DJ, Maher ER, Shows TB, Higgins MJ (1999) A maternally methylated CpG island in KvLQT1 is associated with an antisense paternal transcript and loss of imprinting in Beckwith-Wiedemann syndrome. Proc Natl Acad Sci USA 96:8064–8069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava M, Hsieh S, Grinberg A, Williams-Simon L, Huang SP, Pfeifer K (2000) H19 and Igf2 monoallelic expression is regulated in two distinct ways by a shared cis acting regulatory region upstream of H19. Genes Dev 14:1186–1195 [PMC free article] [PubMed] [Google Scholar]
- Steenman MJ, Rainier S, Dobry CJ, Grundy P, Horon IL, Feinberg AP (1994) Loss of imprinting at IGF2 is linked to reduced expression and abnormal methylation of H19 in Wilms' tumor. Nat Genet 7:433–439 [DOI] [PubMed] [Google Scholar]
- Stoger R, Kubicka P, Liu C-G, Kafri T, Razin A, Cedar H, Barlow DP (1993) Maternal-specific methylation of the imprinted mouse Igf2r locus identifies the expressed locus as carrying the imprinting signal. Cell 73:61–71 [DOI] [PubMed] [Google Scholar]
- Surani MA, Barton SC, Norris ML (1984) Development of reconstituted eggs suggests imprinting of the genome during gametogenesis. Nature 308:548–550 [DOI] [PubMed] [Google Scholar]
- Sutcliffe JS, Han M, Christian SL, Ledbetter DH (1997) Neuronally-expressed necdin gene: an imprinted candidate gene in the Prader-Willi syndrome. Lancet 350:1520–1521 [DOI] [PubMed] [Google Scholar]
- Sutcliffe JS, Nakao M, Christian S, Orstavik KH, Tommerup N, Ledbetter DH, Beaudet AL (1994) Deletions of a differentially methylated CpG island at the SNRPN gene define a putative imprinting control region. Nat Genet 8:52–58 [DOI] [PubMed] [Google Scholar]
- Svensson K, Mattsson R, James TC, Wentzel P, Pilartz M, MacLaughlin J, Miller SJ, Olsson T, Eriksson UJ, Ohlsson R (1998) The paternal allele of the H19 gene is progressively silenced during early mouse development: the acetylation status of histones may be involved in the generation of variegated expression patterns. Development 125:61–69 [DOI] [PubMed] [Google Scholar]
- Szabo PE, Mann JR (1995) Allele-specific expression and total expression levels of imprinted genes during early mouse development: implications for imprinting mechanisms. Genes Dev 9:3097–3108 [DOI] [PubMed] [Google Scholar]
- Szabo PE, Pfeifer GP, Mann JR (1998) Characterization of novel parent-specific epigenetic modifications upstream of the imprinted mouse H19 gene. Mol Cell Biol 18:6767–6776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo PE, Tang SH, Rentsendorj A, Pfeifer GP, Mann JR (2000) Maternal-specific footprints at putative CTCF sites in the H19 imprinting control region give evidence for insulator function. Curr Biol 10:607–610 [DOI] [PubMed] [Google Scholar]
- Tanaka M, Puchyr M, Gertsenstein M, Harpal K, Jaenisch R, Rossant J, Nagy A (1999) Parental origin–specific expression of Mash2 is established at the time of implantation with its imprinting mechanism highly resistant to genome-wide demethylation. Mech Dev 87:129–142 [DOI] [PubMed] [Google Scholar]
- Thorvaldsen JL, Duran KL, Bartolomei MS (1998) Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and Igf2. Genes Dev 12:3693–3702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay KD, Saam JR, Ingram RS, Tilghman SM, Bartolomei MS (1995) A paternal-specific methylation imprint marks the alleles of the mouse H19 gene. Nat Genet 9:407–413 [DOI] [PubMed] [Google Scholar]
- Tsai T-F, Jiang YH, Bressler J, Armstrong D, Beaudet AL (1999) Paternal deletion from Snrpn to Ube3a in the mouse causes hypotonia, growth retardation and partial lethality and provides good evidence for a gene contributing to Prader-Willi syndrome. Hum Mol Genet 8:1357–1364 [DOI] [PubMed] [Google Scholar]
- Udvardy A, Maine E, Schedl P (1985) The 87A7 chromomere: identification of novel chromatin structures flanking the heat shock locus that may define the boundaries of higher order domains. J Mol Biol 185:341–358 [DOI] [PubMed] [Google Scholar]
- Vu TH, Hoffman AR (1997) Imprinting of the Angelman syndrome gene, UBE3A, is restricted to brain. Nat Genet 17:12–13 [DOI] [PubMed] [Google Scholar]
- Webber A, Ingram RS, Levorse J, Tilghman SM (1998) Location of enhancers is essential for imprinting of H19 and Igf2. Nature 391:711–715 [DOI] [PubMed] [Google Scholar]
- Weksberg R, Shen DR, Fei, Song QL, Squire J (1993a) Disruption of insulin-like growth factor imprinting in Beckwith-Wiedemann syndrome. Nat Genet 5:143–150 [DOI] [PubMed] [Google Scholar]
- Weksberg R, Teshima I, Williams BR, Greenberg CR, Pueschel SM, Chernos JE, Fowlow SB, Hoyme E, Anderson IJ, Whiteman DA (1993b) Molecular characterization of cytogenetic alterations associated with the Beckwith-Wiedemann syndrome (BWS) phenotype refines the localization and suggests the gene for BWS is imprinted. Hum Mol Genet 2:549–556 [DOI] [PubMed] [Google Scholar]
- Wevrick R, Kerns JA, Francke U (1994) Identification of a novel paternally expressed gene in the Prader-Willi syndrome region. Hum Mol Genet 3:1877–1882 [DOI] [PubMed] [Google Scholar]
- Wolffe AP, Matzke MA (1999) Epigenetics: regulation through repression. Science 286:481–486 [DOI] [PubMed] [Google Scholar]
- Wutz A, Smrzka OW, Schweifer N, Schellander K, Wagner EF, Barlow (1997) Imprinted expression of the Igf2r gene depends on an intronic CpG island. Nature 389:745–749 [DOI] [PubMed] [Google Scholar]
- Yang T, Adamson TE, Resnick JL, Leff S, Wevrick R, Francke U, Jenkins NA, Copeland NG, Brannan CI (1998) A mouse model for Prader-Willi syndrome imprinting-centre mutations. Nat Genet 19:25–31 [DOI] [PubMed] [Google Scholar]