Abstract
Lysosomal free sialic acid–storage diseases include the allelic disorders Salla disease (SD) and infantile sialic acid–storage disease (ISSD). The defective gene, SLC17A5, coding for the lysosomal free sialic acid transporter was recently isolated by positional cloning. In the present study, we have identified a large number of mutations in SLC17A5 in patients presenting with either Salla disease or the ISSD phenotype. We also report for the first time the exon-intron boundaries of SLC17A5. All Finnish patients with SD (n=80) had a missense mutation changing a highly conserved arginine to cysteine (R39C); 91% of them were homozygotes for this old founder mutation. The compound-heterozygote patients, with the founder mutation in only one allele, presented with a more severe phenotype than did the homozygote patients. The same R39C mutation was also found both in most of the Swedish patients with SD and in a heterozygous form in five patients from central Europe who presented with an unusually severe (intermediate) SD phenotype. Ten different mutations, including deletions, insertions, and missense and nonsense mutations, were identified in patients with the most severe ISSD phenotype, most of whom were compound heterozygotes. Our results indicate some genotype-phenotype correlation in free sialic acid–storage diseases, suggesting that the phenotype associated with the homozygote R39C mutation is milder than that associated with other mutations.
Introduction
Lysosomal free sialic acid–storage diseases (SASD [MIM 269920]) include the allelic disorders Salla disease (SD), enriched in the Finnish population, and infantile sialic acid–storage disease (ISSD [Aula and Gahl, in press]). The clinical symptoms of SD consist of moderate to severe psychomotor retardation and ataxia presenting usually at age <1 year. The progression of symptoms in SD is slow; the oldest patient died at age 72 years. The clinical picture of ISSD is much more severe. The affected newborns exhibit dysmorphic features, an enlarged liver and spleen, and a failure to thrive immediately after birth. These patients have a shortened life span, usually <2 years. Progressive cerebellar atrophy and dysmyelination have been documented by magnetic-resonance imaging in patients with SD (Haataja et al. 1994; Varho et al. 1999). In addition to the main phenotypes of SD and ISSD, rare intermediate phenotypes have also been reported (Fois et al. 1987; Schleutker et al. 1995b).
As a result of defective egress of free sialic acid from lysosomes, patients with SD excrete large amounts of free sialic acid in their urine and store it excessively, at 10–30 times the normal rates, in several types of tissues and cultured fibroblasts (Renlund et al. 1986; Mancini et al. 1989). In patients with ISSD, the urinary excretion and intracellular storage is even higher, up to 100 times the normal rate. Electron-microscopic and histochemical studies have shown that excessive sialic acid accumulates in the lysosomes (Aula et al. 1979). We have recently identified the gene, SLC17A5, responsible for the lysosomal-membrane sialic acid transport and have found mutations in Finnish patients with either SD or ISSD (Verheijen et al. 1999). The gene is located on chromosome 6q and has an open reading frame (ORF) of 1485 bp. It encodes sialin, a protein of 495 amino acids, which is predicted to be an integral lysosomal membrane protein with 12 transmembrane (TM) domains. In the present study, we analyzed, for mutations in the SLC17A5 gene, a total of 100 Finnish and non-Finnish patients with SD and 10 patients with ISSD. We initially screened the patients’ DNA for the identified Finnish mutation (R39C) and then sequenced the entire coding region and the exon-intron boundaries in those patients who did not have the Finnish founder mutation, SallaFIN, in both alleles. A wide spectrum of different DNA alterations was identified in disease alleles. The characterization of mutations in SLC17A5, when compared with the clinical findings, suggests a correlation between the molecular defect and the phenotype of the patients.
Patients and Methods
Patients and Clinical Summary
A clinical summary of the sialic acid–storage disorders is presented in table 1. All diagnoses were based on typical clinical findings, histology, and biochemical assays of sialic acid. Most of the Finnish patients with SD (n=73) presented the classical form of the disease. On a clinical basis, seven patients (K.N., L.J., I.P., K.M., M.M., L.Mt., and L.Mo.) had a more severe type of SD. The onset of symptoms was earlier than that in typical SD; the patients were spastic, were unable to walk, and had seizures and severe growth retardation. The free sialic acid levels in their urine were slightly higher than those in typical patients with SD. The first signs of the disease were detected at age <6 mo.
Table 1.
Patients and Summary of Clinical Findings in SASD
| Disease | Clinical Characterization | Age at Onset | Free Sialic Acidin Urinea | No. of Cases in Study |
| SD | Hypotonia and ataxia, ocular squint, delayed development, few words, able to walk; IQ 20–30; life span near normal | 6–12 mo | 12 fold (5–36) | 73 Finnish, 15 Swedish,b 2 Englishc |
| Intermediate “severe Salla” | Severe hypotonia, later ataxia, spasticity; growth delay; no organomegaly; seizures; shortened life span | 1–6 mo | 15–34 fold | 7 Finnish, 1 Dutch,d 1 Italian,e 1 Germanb |
| ISSD | Intrauterine hydrops, neonatal ascites, enlarged liver; dysmorphic features; failure to thrive; early death (age <2 years) | Intrauterine | 102 fold (18–245) | 10 Whitef |
The clinical findings and course of the disease were, for Swedish patients (n=15), all compatible with the findings in the Finnish patients with SD. Also, two patients in one British family (Le.C. and Le.L.) had the Finnish type of the disease. Three patients—one Italian (P.E.), one Dutch (B.E.), and one German (G.A.)—had a severe form of SD, the intermediate type, according to the clinical data. Ten patients—four British (C.G., C.J., Mc.I., and S.L.), one Italian (A.B.), one Yugoslavian (A.Z.), two French (P.M. and D.R.), and two French-Canadian (J.G. and R.D.)—were classified as having the severe, early-onset, fatal ISSD.
Allele-Specific PCR to Detect the Finnish Mutation
We performed allele-specific PCR (Wu et al. 1989) using either 5′-CAGTGTGCTGCTCTGCTC-3′ (wild type) or 5′-CAGTGTGCTGCTCTGCTT-3′ (mutant) as a 5′ primer and 5′-AGGCCAAAATTGCTAAGTTGT-3′ as a 3′ primer. Cycling conditions were 11 min at 95°C, 30 s at 95°C, 30 s at 57°C, and 1 min at 72°C, for 35 cycles, with thermal cycler PTC-225 (MJ Research) and AmpliTaq Gold (Roche).
cDNA Sequencing
Fibroblast cell lines were established from patients with either SD or ISSD who have been described elsewhere. Fibroblasts were cultured in DMEM (Gibco BRL, Life Technologies) supplemented with 10% fetal calf serum and antibiotics. Total RNA was isolated from fibroblasts by use of the RNeasy Midi Kit (Qiagen), according to the manufacturer’s instructions. The first-strand cDNA was subsequently generated by a standard protocol using AMV-RT enzyme (NEB). cDNA was amplified by PCR with a PTC-225 thermal cycler (MJ Research) and Dynazyme (Finnzymes), under the appropriate conditions. PCR products were analyzed on 1% agarose gel and subsequently were sequenced using the ABI Prism Big Dye Terminator Cycle Sequencing kit (Applied Biosystems) and an ABI 377 (Applied Biosystems) fluorescent sequencer. We sequenced the complete ORF, using four different primer pairs: P1, forward 5′-GTGGCGAGTACACCTGCTCA-3′ and reverse 5′-CTTCTAGTGCTCTGAGTACAA-3′; P2, forward 5′-TGGAGGATATGTTGCCAGCA-3′ and reverse 5′-GTGTGCAACTACGATAGCCC-3′; P3, forward 5′-ATCAGCTTTCTTCACAGAAGTCA-3′ and reverse 5′-CAATATCCAGATGGTTGATGC-3′; and P4, forward 5′-GTAGCTGCTGGCTTCATTGG-3′ and reverse 5′-ATACATTAATAGAGGCAGGATTA-3′.
Genomic DNA Sequencing
We amplified exons 2–11 from genomic DNA, using intronic primers flanking the exons, as shown in table 2. All specific primer sets were designed in the intronic sequences flanking each exon, at a distance of ⩾40 bp from the exon-intron boundary. The PCR reactions were carried out in a final volume of 50 μl in a standard reaction condition, with Dynazyme DNA polymerase (Finnzymes). Exons were amplified for 30–35 cycles at either 94°C for 1 min or 53°C–64°C (depending on the primer pair) for 1 min and 72°C for 1 min, followed by a 10-min extension step at 72°C. PCR products were visualized on 1% agarose gel and subsequently were sequenced using an ABI 377 fluorescent sequencer (Applied Biosystems).
Table 2.
Primers for Amplification and Sequencing of SLC17A5 Exons
| Exon and PCR and Sequencing Primers | AmplifiedProduct(bp) | AnnealingTemperature(°C) |
| 2: | ||
| Forward, 5′-AGGAGTTCAAGACCAGCCTAAGCAACATG-3′ | 370 | 54 |
| Reverse, 5′-TACACAAACAAAGTATAGTTTCTGTAGGATGA-3′ | ||
| 3: | ||
| Forward, 5′-AATCTCATTTATATGTACTTACATGCCAGT-3′ | 415 | 53 |
| Reverse, 5′-GGTTCATATTCATACCAAAGAATGACATC-3′ | ||
| 4: | ||
| Forward, 5′-CATGGAACCTGATTCGTTTGATTCTTATC-3′ | 310 | 58 |
| Reverse, 5′-AACATTGCATCGTTCTGGTATGCAGGCC-3′ | ||
| 5: | ||
| Forward, 5′-CCCATCCTCTGTAAGCAGTAGACTTGTC-3′ | 300 | 58 |
| Reverse, 5′-CAGGAAGTGTTTCAGAACTGTGGTTCAG-3′ | ||
| 6: | ||
| Forward, 5′-TCAAGACATGTAAAAATTGTTGTTGGTGC-3′ | 345 | 54 |
| Reverse, 5′-CTACTTTGTCTAGAGCATGCACAATAGG-3′ | ||
| 7: | ||
| Forward, 5′-GAATTCTGGATAGAGTTTTAGCTGATACC-3′ | 420 | 53 |
| Reverse, 5′-AGGACAGCAGAGTAAAATGGAAGATACAG-3′ | ||
| 8: | ||
| Forward, 5′-TTCAAGTGGTGGTCATTGGCTGTGATGA-3′ | 345 | 58 |
| Reverse, 5′-GTGATTAGAGCGAGGTGTTCTGTGTTGAG-3′ | ||
| 9: | ||
| Forward, 5′-GTCTTCCAGGCAATATAGTGTTTCTACAG-3′ | 390 | 56 |
| Reverse, 5′-AAGTGCTGGGATTACAGGTGTGAGTCACC-3′ | ||
| 10: | ||
| Forward, 5′AGGCTGCAGTGAGCTGAGATCATCCCAC-3′ | 380 | 55 |
| Reverse, 5′-ATACTTTCCCATCCATTAAGGCATTTAGC-3′ | ||
| 11: | ||
| Forward, 5′-ATGTGGCTGTTTTAACACTCTTGTGACAC-3′ | 330 | 64 |
| Reverse, 5′-ATAGAATGCTTACACAATACAGAAGGCAC-3′ |
Northern Hybridization
Total RNAs were extracted from respective fibroblast cultures, with a Qiagen RNeasy Midi Kit used according to the manufacturer’s protocol. An RNA filter was made using standard protocols. A 32P-labeled 140-bp PCR fragment amplified from SLC17A5 cDNA was used in the hybridization, which was performed at 68°C for 1 h with ExpressHyb solution (Clontech Laboratories). The filter was washed as recommended by the manufacturer. To evaluate the loading of the samples, the filter was subsequently rehybridized with 32P-labeled β-actin (Clontech).
Genomic Structure of SLC17A5
We searched the human genome database for genomic clones, using the BLASTN algorithm (Altschul et al. 1990) and the SLC17A5 cDNA sequence. We found one Homo sapiens clone (HSJ397H23) that contained 22 unordered pieces of genomic DNA covering the SLC17A5 cDNA. This sequence was not finished and could contain errors. We assembled the right order of the pieces and their orientation. This sequence contained 10 complete exons, covering most of the ORF of SLC17A5. The missing 5′ exon was determined by sequencing a genomic clone (PAC 202M22) covering the SLC17A5 gene. We chose as the sequencing primer a sequence derived from the next intron. Intron primers flanking all intron-exon boundaries were chosen (table 3), and all exons were amplified by PCR using genomic DNA isolated from cultured human fibroblasts. All amplified exons and boundaries were sequenced using an Applied Biosystems ABI 377 fluorescent sequencer. Using this approach, we determined all 11 exons covering the whole ORF. Exon 11 covered also the 3′ UTR. Because of the missing 5′ sequence, the exact length of exon 1 could not be determined. For genomic amplification of the first exon, an exon 1 primer was used together with a reverse primer from the intron boundary of the first intron: forward, 5′-CGGCATGCGTAGACCGACG-3′; reverse, 5′-CTCGCGCCTGAGCAGCTC-3′.
Table 3.
Sequence of the Exon-Intron Boundaries of the SLC17A5 Gene
| Exon | Positions | 5′ Splice Donor | 3′ Splice Acceptor |
| 1 | ⩽94 | GGGCCGAAGCCG-gtaagcggcggg | caatctttatttcag-CTCCAGTGTGC |
| 2 | 95–291 | CATAATCAAACG-gtaggtgctatt | ttctgatttttgtag-GGTAAGAAGTAC |
| 3 | 292–525 | GGACTAGGAGAG-gtaattttcttt | gttcccttctctcag-GGTGTTACATTT |
| 4 | 526–613 | TTTCATATGCAG-gtgagataacaa | tatgtgttgttctag-GAGCACAGCTTG |
| 5 | 614–700 | TCTACTTTTTTG-gtaagtttaaaa | ttctattttaag-GTACTATTGGAATA |
| 6 | 701–819 | TTAAGAAATCAG-gtatggactttg | tgcctggttcttcag-CTTTCTTCACAG |
| 7 | 820–978 | AATGTTCAAGAG-gtaagaaaaaat | tcttattccttccag-AATGGGTTTTTA |
| 8 | 979–1111 | TAGCCTTATAG-gtaagtgaagcaa | ctttttaactccag-GAATGATTGGGA |
| 9 | 1112–1259 | TATTGCTCCTTC-gtgagtactaat | aaaatgtttctacag-GTATGCTGGTAT |
| 10 | 1260–1350 | CTGACCCCTGAT-gtaagtagataa | tgtcttcctttatag-AACACTGTTGGA |
| 11 | ⩾1351 |
Results
SallaFIN
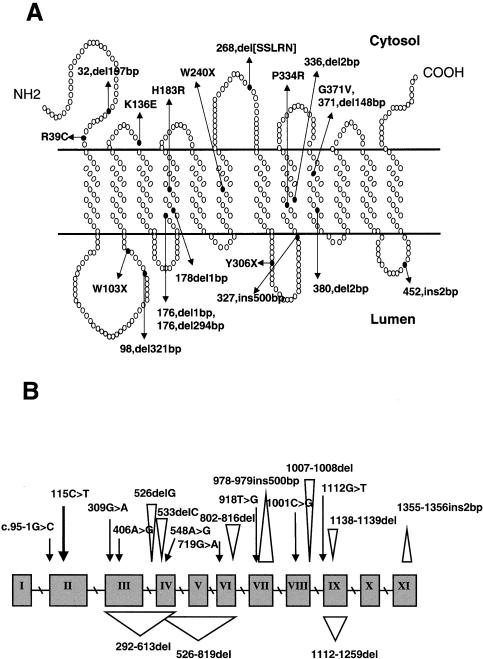
Allele-specific PCR was used to monitor for the 115C→T mutation in the leukocyte DNA from 80 Finnish patients with SD. Altogether, 73 patients proved to be homozygous for this nucleotide change. The mutation changes a highly conserved arginine (R39) into cysteine, leading to an alteration just before the first TM domain of the predicted polypeptide chain (fig. 1A). Seven patients were found to be compound heterozygotes, having the SallaFIN mutation in only one allele and another mutation in the other allele. Thus, the SallaFIN mutation was found in 95% of Finnish SD chromosomes, indicating, again, a founder effect in the Finnish population, a finding that is characteristic to diseases belonging to the Finnish disease heritage (Peltonen et al. 1999). Of 15 Swedish patients with SD, 12 were homozygous for the SallaFIN mutation, 2 were heterozygous, and 1 did not have this mutation in either allele. Somewhat surprisingly, we found the SallaFIN mutation in a heterozygous form in five non-Scandinavian patients with SD. These patients are of British, Dutch, Italian, and German origin and have no known relationship to Finland, nor are they known to be related to each other (table 4).
Figure 1 .
A, Sialin mutations in patients with free SASD shown in the predicted model of sialin in the lysosomal membrane are denoted in terms of amino acid position. Only the start points of deletions and insertions are indicated. SSLRN = in-frame deletion of Ser, Ser, Leu, Arg, and Asn. B, SLC17A5 mutations in patients with free SASD. Point mutations are marked with an arrow (↓), deletion points with an upside-down triangle (▿), and insertion points with a triangle (▵).
Table 4.
Mutations in Non-Scandinavian Patients with SASD
|
Allele 1 |
Allele 2 |
|||
| Phenotype | NucleotideChange | PredictedPolypeptideConsequence | NucleotideChange | PredictedPolypeptideConsequence |
| Severe SD (intermediate): | ||||
| Le.L. | 115C→T | R39C | 1138–1139del a | Frameshift, 7 novel amino acids, premature stop |
| Le.C. | 115C→T | R39C | 1138–1139del a | Frameshift, 7 novel amino acids, premature stop |
| G.A. | 115C→T | R39C | 292–611del a | Deletion of exons 3 and 4 |
| B.E. | 115C→T | R39C | ? | ? |
| P.E. | 115C→T | R39C | ? | ? |
| ISSD: | ||||
| S.L. | 802–816del | In-frame deletion of 5 amino acids in the cytosolic loop between TM domains 6 and 7 | 802–816del | In-frame deletion of 5 amino acids in the cytosolic loop between TM domains 6 and 7 |
| C.J. | 802–816del | In-frame deletion of 5 amino acids in the cytosolic loop between TM domains 6 and 7 | 1112G→T a | G371V |
| Mc.I.b | 802–816del | In-frame deletion of 5 amino acids in the cytosolic loop between TM domains 6 and 7 | 1355–1356insAAa | Frameshift, read-through of stop codon |
| J.G.b | 802–816del | In-frame deletion of 5 amino acids in the cytosolic loop between TM domains 6 and 7 | ? | ? |
| R.D.b | 802–816del | In-frame deletion of 5 amino acids in the cytosolic loop between TM domains 6 and 7 | ? | ? |
| C.G. | 802–816del | In-frame deletion of 5 amino acids in the cytosolic loop between TM domains 6 and 7 | ? | ? |
| P.M. | 526delG | 37 novel amino acids, premature stop | 918T→Ga | Y306X |
| D.R.b | 533delC | Frameshift, 33 novel amino acids, premature stop | 1112–1259del | Deletion of exon 9 |
| A.Z.b | 548A→G | H183R | 1001C→G | P334R |
| A.B.b | 978–979ins500bp | Premature stop | 978–979ins500bp | Premature stop |
Mutation not detectable in cDNA.
Reported by Verheijen et al. (1999).
Carrier Frequency
In 226 control DNA samples from all regions of Finland, we applied allele-specific PCR to screen for the R39C mutation and found one carrier. This suggests a populationwide carrier frequency of ∼1/200. We also screened for this mutation in a study sample of 107 individuals originating from the northeastern region of Finland, where SD is known to be more prevalent. Two carriers were identified, suggesting a higher regional frequency of the disease allele, ∼1%, which is well in accordance with previous estimates (Aula et al. 1986).
Mutations in Finnish Compound-Heterozygote Patients with SD
To identify the mutations in the other alleles of the Finnish compound-heterozygote patients with SD, we first sequenced the coding region, using, as a template, reverse-transcribed RNA from patients’ fibroblasts. Two patients (K.N. and L.J.) were found to share a 294-bp (nucleotide positions 526–819) deletion in their other allele. This deletion is predicted to result in an in-frame deletion of 98 amino acids (fig. 1A). The deleted region encompasses exons 4–6, also including the same five amino acids (SSLRN) deleted in six severely affected patients with ISSD (see below). One patient (I.P.) had a missense mutation, 406G→A, in the other allele, which changes a lysine into glutamic acid (K136E) in the cytosolic loop just before the third TM domain. In five patients (K.M., L.Mt., L.Mo., M.M., and V.A.), cDNA sequencing did not reveal the mutation of the other allele. In these cases, we sequenced genomic DNA, including the exon-intron boundaries and part of the introns, to identify all disease mutations. We were able to find the second mutation in three additional Finnish patients (table 5). Two sisters (L.Mt. and L.Mo.) and one other patient (M.M.) have a 2-bp deletion (1007–1008del) in exon 8. This results in a frameshift and, consequently, in the formation of 13 novel amino acids followed by a premature stop codon. In total, we identified three different mutations, in addition to SallaFIN, in Finnish patients with SD. In two compound-heterozygote cases, the mutation of the other allele remains unknown.
Table 5.
Mutations in the Scandinavian Patients with SD
|
Allele 1 |
Allele 2 |
|||
| Phenotype | NucleotideChange | PredictedPolypeptideConsequence | NucleotideChange | PredictedPolypeptideConsequence |
| Classical SD: | ||||
| Swedish (n = 12) | 115C→T | R39C | 115C→T | R39C |
| Finnish (n = 73) | 115C→T | R39C | 115C→T | R39C |
| V.A. | 115C→T | R39C | ? | ? |
| Severe SD (intermediate): | ||||
| Finnish: | ||||
| K.N. | 115C→T | R39C | 526–819del | Deletion of exons 4–6 |
| L.J. | 115C→T | R39C | 526–819del | Deletion of exons 4–6 |
| I.P. | 115C→T | R39C | 406A→G | K136E |
| K.M. | 115C→T | R39C | ? | ? |
| M.M. | 115C→T | R39C | 1007–1008dela | 13 Novel amino acids, premature stop |
| L.mt. | 115C→T | R39C | 1007–1008dela | 13 Novel amino acids, premature stop |
| L.mo. | 115C→T | R39C | 1007–1008dela | 13 Novel amino acids, premature stop |
| Swedish: | ||||
| K.H. | 95–291del, 197 bp (cDNA), ∼95−1G→C (genDNA)a | Removes splice site, deletion of exon 2 | 1007–1008dela | 13 Novel amino acids, premature stop |
| J.T. | 115C→T | R39C | 309G→Aa | W103X |
| A.S. | 115C→T | R39C | 719G→Aa | W240X |
Mutation not detectable in cDNA.
Mutations in Non-Finnish Compound-Heterozygote Patients with SD
The mutations in the other alleles of the two Swedish heterozygotes could not be detected in their fibroblast cDNA. By genomic sequencing, we identified two novel mutations (table 5). One patient (J.T.) was found to have a nonsense mutation (309G→A) in exon 3, predicted to result in only a short (102 amino acids) translation product from this allele. In the other patient (A.S.), we identified also a nonsense mutation (719G→A) in exon 6. An intronic mutation (∼95−1G→C) was found in one patient (K.H.) who does not have the R39C mutation in either allele. This nucleotide substitution occurs one base upstream of the first base of exon 2 and, consequently, results in exon skipping, since cDNA sequencing revealed a deletion of 197 bp covering the whole of exon 2. The other allele of this patient carried the same 2-bp deletion (1007–1008del) as was seen in three Finnish compound heterozygotes.
As mentioned above, five foreign patients were found, on the basis of allele-specific PCR results, to carry the SallaFIN mutation resulting in R39C substitution in one allele. No mutations in the other allele could be established by cDNA sequencing. The sequencing of genomic DNA revealed the mutation in the other allele in the German patient (G.A.), who had a genomic deletion encompassing exons 3 and 4. The English sisters were found to have a 2-bp deletion (1138–1139del2bp) that results in a frameshift and a premature stop codon. The mutation of the other allele in the Dutch (B.E.) and in the Italian (P.E.) patients has yet to be identified. Altogether, we identified five novel mutations in the non-Finnish compound-heterozygote patients with SD.
Mutations in Patients with ISSD
None of the patients with ISSD had the SallaFIN mutation. Using the initial cDNA and further genomic DNA sequencing, we were able to find 10 different mutations, including missense mutations, deletions, and insertions (table 4). Surprisingly, we found an identical 15-bp deletion (802–816del15bp) in six seemingly unrelated patients(C.G., C.J., J.G., Mc.I., R.D., and S.L.) of Canadian, English, and French origin. One of these patients (S.L.) is a child of consanguineous parents and is homozygous for the mutation, whereas all the others are heterozygotes. This mutation is predicted to lead to a deletion of five amino acids (SSLRN) in the cytosolic loop between TM domains 6 and 7. The mutation in the other allele of these heterozygotes has been identified in two patients. In one case (Mc.I.), a 2-bp insertion (1355–1356ins2bp) in exon 11 was found. This causes a frameshift and is predicted to lead to formation of novel amino acids and to read through of the normally used stop codon. Another patient (C.J.) had a missense mutation (1112G→T) of the first base of exon 9, changing a conserved glycine into valine. One French patient (P.M.) was shown to have a heterozygous 1-bp deletion (526delG) that results in a frameshift—formation of 37 novel amino acids followed by a premature stop. In the other allele, a nonsense mutation (918T→G) was found in exon 7. A homozygous 500-bp insertion (978ins500bp) leading to a truncated polypeptide was found in an Italian patient, and two different missense mutations (H183R and P334R) were detected in a Yugoslavian patient. Altogether, we have identified 10 different mutations in our study sample of 10 patients with ISSD.
Northern Analysis
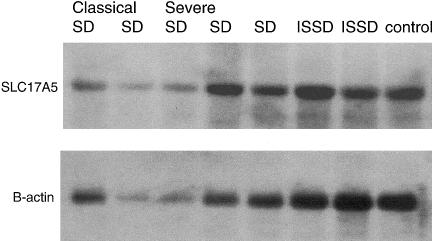
We hybridized a SLC17A5-specific cDNA probe to a northern filter with total RNA from patients and one control. Steady-state mRNA from fibroblasts of two classical, R39C-homozygote, and three severely affected compound-heterozygote patients with SD (K.N., R39C/526–819del; L.Mt., R39C/1007–1008del; and I.P., R39C/406A→G), as well as from two patients with ISSD (S.L., 802–816del/802–816del; and Mc.I., 802–816del/1355–1356ins) were analyzed. On the basis of the mutations in these patients, we anticipated a difference in the expression levels of SLC17A5 RNA but could not detect significant differences, in the steady-state mRNA levels, between the patients and the control (fig. 2).
Figure 2 .
Expression of SLC17A5 in patients with classical SD, severe SD, and ISSD. A northern blot made from total RNAs was hybridized with a 32P-labeled 140-bp PCR probe amplified from SLC17A5 cDNA and subsequently rehybridized with 32P-labeled β-actin, to evaluate the loading of the samples.
Discussion
We analyzed all known Finnish patients with SD and found the same missense mutation (115C→T/R39C) in 95% of all disease chromosomes. This missense mutation, SallaFIN, which changes an arginine into cysteine, is clearly a founder mutation that, because of the population history and geographic isolation of Finland, enriched in the Finnish population (Nevanlinna 1972). The presence of a founder mutation already had been anticipated, on the basis of haplotype analyses that revealed a common haplotype in 95% of the disease chromosomes in a 60-kb area flanking the disease locus (Schleutker et al. 1995b). Our results regarding SD are in line with the results of earlier observations of several other monogenic disorders in Finland. One major founder mutation has been established systematically for all the diseases belonging to the Finnish disease heritage (Peltonen 1997; Peltonen et al. 1999).
We investigated the carrier frequency of the SallaFIN mutation in Finland—both nationwide and in the northeastern region—where there is a higher prevalence of the disease. The results show that the estimated carrier frequency is four times higher in the Salla-Kuusamo area (1.87%), compared with that in the overall population (0.44%). This is well in accordance with previous estimates based on prevalence of the disease (Aula et al. 1986).
Finding the SallaFIN in Swedish families with SD that are not known to be related to Finnish pedigrees is not surprising in light of the admixture of these populations during the centuries in which they shared a common history. However, the presence of the SallaFIN mutation in central and southern European families is unexpected and provides interesting speculations about the age, origin, and migration of this disease mutation. On the basis of the long genetic interval (10 cM) with significant linkage disequilibrium, the SallaFIN mutation can be concluded to have been introduced into the Finnish population relatively recently (∼20 generations ago [Schleutker et al. 1995a]). Where this mutation originally came from is not known, but its presence in central and southern European SD alleles suggests that this mutation arrived in Finland with immigrants from Europe.
One major founder mutation covering 95% of disease alleles provides excellent diagnostic possibilities in Finland. A simple gene test for the R39C mutation has been developed using minisequencing technology (Syvanen et al. 1992), and this test can also be used for carrier detection and prenatal diagnosis of SD. Even carrier screening in selected populations may become feasible, either for the SD mutation alone or in combination with other prevalent mutations in the population.
The mutations identified in the SLC17A5 gene show a wide spectrum of disease-causing DNA variations. We identified missense and nonsense mutations, splice-site mutations, and insertions and deletions. On the basis of the distribution of the mutations, no mutational hotspot was suggested, although some clustering of mutations is seen in exons 4 and 8 (fig. 1B). Some of the nonsense mutations can be predicted to result in a short polypeptide product from this allele, whereas some deletions result in the formation of a few novel amino acids and a longer but, most probably, nonfunctional protein product. In northern analyses, we could not detect any differences in the total steady-state mRNA levels between patients with classical SD (R39C homozygotes), those with the severe form of SD (compound heterozygotes), and those with ISSD (fig. 2). However, 8 of 19 mutations could not be identified in the cDNA produced from the mRNA of these cell lines, suggesting a significantly lower steady-state transcript level for these mutant alleles. These alleles represented nonsense mutations, deletions, and an insertion.
The phenotypic variation seen in SD seems, to some extent, to correlate with the presence of the R39C mutation. All those patients with SD who are homozygous for the SallaFIN mutation can be classified as having the classical, slowly progressing SD phenotype. They show the typical symptoms of SD: early-onset psychomotor retardation and ataxia, hypotonia, and elevated excretion of free sialic acid in their urine (up to 30 times the normal values; table 1). With one exception (V.A.), the compound-heterozygote patients are more severely affected than are the R39C homozygotes. The amount of free sialic acid in urine is usually higher in the heterozygotes than in the homozygotes (table 1), and their phenotype is more severe. Interestingly, the non-Finnish patients who have the SallaFIN mutation also have a milder phenotype than do those who have altogether different mutations. Many compound-heterozygote patients have been classified as having the phenotype intermediate between the classical SD and ISSD phenotypes (Schleutker et al. 1995b). The most severe phenotype of the disease, ISSD, is found in individuals in whom neither of the mutated alleles carries the SallaFIN mutation. The presence of the R39C mutation in the sialin polypeptide thus seems to preserve more of the protein’s normal function, compared with other mutations found in compound-heterozygote patients. If the lysosomal sialic acid transporter, like several membrane-transporter proteins, functions as a multimer on the lysosomal membrane, the R39C mutation does not seem to greatly disturb either the primary folding or the potential multimerization of the protein. The other mutations may distort the secondary structure of the protein and hinder the membrane targeting or the formation of an active dimer or multimer. Although some mutant alleles seem to result in very low transcript levels, interference of the polypeptides is suggested, on the basis of the more severe clinical phenotype of the majority of compound-heterozygote patients with the SallaFIN mutation in their other allele. Our ongoing studies of the expression of the normal and mutant SLC17A5 alleles will shed further light on the structure and function of the sialin protein.
Acknowledgments
We thank the patients and families and the following physicians for contributing to this work: A. Fensom, M. Cantz, E. Lemyre, and I. Maire. Paula Hakala is thanked for her excellent technical assistance. This study was supported financially by the Academy of Finland; the Pediatric Research Foundation (Ulla Hjelt Fund); the Rinnekoti Research Foundation, Espoo, Finland; and the Research and Science Foundation of Farmos.
Electronic-Database Information
The accession number and URLs for data in this article are as follows:
- BLAST, http://www.ncbi.nlm.nih.gov/BLAST
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/omim (for SASD [MIM 269920]) [PubMed]
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410 [DOI] [PubMed] [Google Scholar]
- Aula P, Autio S, Raivio KO, Rapola J, Thoden CJ, Koskela SL, Yamashina I (1979) “Salla disease”: a new lysosomal storage disorder. Arch Neurol 36:88–94 [DOI] [PubMed] [Google Scholar]
- Aula P, Gahl W. Disorders of free sialic acid storage. In: Scriver CR BA, Sly WS, Valle D (eds) The metabolic and molecular basis of inherited disease. McGraw-Hill, New York (in press) [Google Scholar]
- Aula P, Renlund M, Raivio KO, Koskela SL (1986) Screening of inherited oligosaccharidurias among mentally retarded patients in northern Finland. J Ment Defic Res 30:365–368 [DOI] [PubMed] [Google Scholar]
- Fois A, Balestri P, Farnetani MA, Mancini GM, Borgogni P, Margollicci MA, Molinelli M, Alessandrini C, Gerli R (1987) Free sialic acid storage disease: a new Italian case. Eur J Pediatr 146:195–198 [DOI] [PubMed] [Google Scholar]
- Haataja L, Parkkola R, Sonninen P, Vanhanen SL, Schleutker J, Aarimaa T, Turpeinen U, Renlund M, Aula P (1994) Phenotypic variation and magnetic resonance imaging (MRI) in Salla disease, a free sialic acid storage disorder. Neuropediatrics 25:238–244 [DOI] [PubMed] [Google Scholar]
- Mancini GM, de Jonge HR, Galjaard H, Verheijen FW (1989) Characterization of a proton-driven carrier for sialic acid in the lysosomal membrane: evidence for a group-specific transport system for acidic monosaccharides. J Biol Chem 264:15247–15254 [PubMed] [Google Scholar]
- Mancini GM, Hu P, Verheijen FW, van Diggelen OP, Janse HC, Kleijer WJ, Beemer FA, Jennekens FG (1992) Salla disease variant in a Dutch patient: potential value of polymorphonuclear leucocytes for heterozygote detection. Eur J Pediatr 151:590–595 [DOI] [PubMed] [Google Scholar]
- Nevanlinna HR (1972) The Finnish population structure: a genetic and genealogical study. Hereditas 71:195–236 [DOI] [PubMed] [Google Scholar]
- Peltonen L (1997) Molecular background of the Finnish disease heritage. Ann Med 29:553–556 [DOI] [PubMed] [Google Scholar]
- Peltonen L, Jalanko A, Varilo T (1999) Molecular genetics of the Finnish disease heritage. Hum Mol Genet 8:1913–1923 [DOI] [PubMed] [Google Scholar]
- Renlund M, Tietze F, Gahl WA (1986) Defective sialic acid egress from isolated fibroblast lysosomes of patients with Salla disease. Science 232:759–762 [DOI] [PubMed] [Google Scholar]
- Robinson RO, Fensom AH, Lake BD (1997) Salla disease—rare or underdiagnosed? Dev Med Child Neurol 39:153–157 [DOI] [PubMed] [Google Scholar]
- Schleutker J, Laine AP, Haataja L, Renlund M, Weissenbach J, Aula P, Peltonen L (1995a) Linkage disequilibrium utilized to establish a refined genetic position of the Salla disease locus on 6q14-q15. Genomics 27:286–292 [DOI] [PubMed] [Google Scholar]
- Schleutker J, Leppanen P, Mansson JE, Erikson A, Weissenbach J, Peltonen L, Aula P (1995b) Lysosomal free sialic acid storage disorders with different phenotypic presentations—infantile-form sialic acid storage disease and Salla disease—represent allelic disorders on 6q14-15. Am J Hum Genet 57:893–901 [PMC free article] [PubMed] [Google Scholar]
- Syvanen AC, Ikonen E, Manninen T, Bengtstrom M, Soderlund H, Aula P, Peltonen L (1992) Convenient and quantitative determination of the frequency of a mutant allele using solid-phase minisequencing: application to aspartylglucosaminuria in Finland. Genomics 12:590–595 [DOI] [PubMed] [Google Scholar]
- Varho T, Komu M, Sonninen P, Holopainen I, Nyman S, Manner T, Sillanpaa M, Aula P, Lundbom N (1999) A new metabolite contributing to N-acetyl signal in 1H MRS of the brain in Salla disease. Neurology 52:1668–1672 [DOI] [PubMed] [Google Scholar]
- Verheijen FW, Verbeek E, Aula N, Beerens CE, Havelaar AC, Joosse M, Peltonen L, Aula P, Galjaard H, van der Spek PJ, Mancini GM (1999) A new gene, encoding an anion transporter, is mutated in sialic acid storage diseases. Nat Genet 23:462–465 [DOI] [PubMed] [Google Scholar]
- Wu DY, Ugozzoli L, Pal BK, Wallace RB (1989) Allele-specific enzymatic amplification of beta-globin genomic DNA for diagnosis of sickle cell anemia. Proc Natl Acad Sci USA 86:2757–2760 [DOI] [PMC free article] [PubMed] [Google Scholar]