Abstract
The photoreceptor cell–specific ATP-binding cassette transporter gene (ABCA4; previously denoted “ABCR”) is mutated in most patients with autosomal recessive (AR) Stargardt disease (STGD1) or fundus flavimaculatus (FFM). In addition, a few cases with AR retinitis pigmentosa (RP) and AR cone-rod dystrophy (CRD) have been found to have ABCA4 mutations. To evaluate the importance of the ABCA4 gene as a cause of AR CRD, we selected 5 patients with AR CRD and 15 patients with isolated CRD, all from Germany and The Netherlands . Single-strand conformation–polymorphism analysis and sequencing revealed 19 ABCA4 mutations in 13 (65%) of 20 patients. In six patients, mutations were identified in both ABCA4 alleles; in seven patients, mutations were detected in one allele. One complex ABCA4 allele (L541P;A1038V) was found exclusively in German patients with CRD; one patient carried this complex allele homozygously, and five others were compound heterozygous. These findings suggest that mutations in the ABCA4 gene are the major cause of AR CRD. A primary role of the ABCA4 gene in STGD1/FFM and AR CRD, together with the gene's involvement in an as-yet-unknown proportion of cases with AR RP, strengthens the idea that mutations in the ABCA4 gene could be the most frequent cause of inherited retinal dystrophy in humans.
Inherited chorioretinal dystrophies show a high degree of clinical and genetic heterogeneity (RetNet Web site). Clinical classification is relatively straightforward for those diseases that display typical fundoscopic and electrophysiological abnormalities—for example, gyrate atrophy (MIM 258870), choroideremia (MIM 303100), Stargardt disease (STGD1 [MIM 248200]), and Best vitelliform macular dystrophy (MIM 153700). Linkage studies in each of these diseases have suggested the involvement of a single locus, thereby facilitating the positional cloning of the underlying genes (Valle and Simell 1989; Cremers et al. 1990; Allikmets et al. 1997b; Petrukhin et al. 1998). In other inherited chorioretinal diseases, however, the clinical and genetic classification is much more difficult, because the phenotypes are less distinctive. Consequently, the identification of the genes involved in this group of retinal disorders often relies on a “brute force” approach, in which promising candidate genes are screened for mutations in a large cohort of patients with chorioretinal dystrophies (Dryja 1997 and references therein). Traditionally, retinitis pigmentosa (RP [MIM 268000]) is considered a rod-cone dystrophy, with earlier and more serious involvement of the rod photoreceptors as compared with cone photoreceptors. Conversely, there is a group of patients in whom the cone degeneration is more prominent than the degeneration of rods. In this cone-rod pattern of retinal dystrophy, or cone-rod dystrophy (CRD [MIM 120970, MIM 601777, MIM 600624, MIM 603649, MIM 604116, MIM 600977]), the photopic cone b-wave amplitude is reduced more than the scotopic rod b-wave amplitude in the electroretinogram (ERG) (Szlyk et al. 1993). At the end stages of both rod-cone and cone-rod dystrophy, the ophthalmologic characteristics tend to coalesce, in part because the chorioretinal degenerations follow a common apoptotic pathway (Chang et al. 1993).
Recent studies have suggested that the photoreceptor-specific ATP-binding cassette transporter ABCR—recently renamed “ABCA4” (MIM 601691)—works as an N-retinylidene-phosphatidylethanolamine flippase on the disk membrane of both cone and rod photoreceptor cells (Sun et al. 1999; Weng et al. 1999; Molday et al. 2000). We and others have shown the involvement of the ABCA4 gene in STGD1/FFM and in a few families and patients with autosomal recessive (AR) RP (RP19 [MIM 601718]) and AR CRD (CORD3 [MIM 604116]) (Allikmets et al. 1997b; Cremers et al. 1998; Martinez-Mir et al. 1998; Lewis et al. 1999; Maugeri et al. 1999; Papaioannou et al. 2000). Moreover, heterozygous ABCA4 mutations were found in 16% of patients with age-related macular degeneration (AMD [MIM 153800]) (Allikmets et al. 1997a). Although the significance of this finding is under debate (Stone et al. 1998), at least two ABCA4 mutations (G1961E and D2177N) have been shown, in a large multicenter study, to be statistically more frequent in patients with AMD compared with controls (Allikmets and The International ABCR Screening Consortium 2000).
These studies suggest a model in which the most severe retinal disorder associated with ABCA4 mutations, RP, is caused by two ABCA4 null alleles, whereas CRD and STGD1 are due to combinations of ABCA4 mutations yielding residual ABCA4 function (Cremers et al. 1998; Martinez-Mir et al. 1998; van Driel et al. 1998; Allikmets 1999; Lewis et al. 1999; Maugeri et al. 1999; Shroyer et al. 1999; Lewis and Lupski 2000). On the basis of this model and the grouping of ABCA4 mutations in different classes of severity, we estimated that ABCA4 mutations could be responsible for CRD in 1 in 50,000 individuals (Maugeri et al. 1999). Since AR CRD represents a relatively rare disorder, we hypothesized that ABCA4 could be an important cause of AR CRD. In this study, we tested this hypothesis by evaluating the ABCA4 gene in patients with AR CRD.
The ABCA4 genotype-phenotype–correlation model (Cremers et al. 1998; van Driel et al. 1998; Maugeri et al. 1999) suggests that retinal dystrophies form a phenotypic continuum. In this study, we selected a group of patients whose condition clearly is clinically distinct from STGD1 and classic RP. Clinical investigations were performed in two different centers: the Department of Ophthalmology at University Medical Centre in Nijmegen and the Augenklinik in Heidelberg. At the time of diagnosis, the ophthalmologists were not aware of the results of the mutation analysis.
Rod-cone and cone-rod patterns of retinal degeneration were diagnosed predominantly on the basis of the ERG patterns. Patients were diagnosed as having CRD when the b-wave of the photopic ERG (cone response) was more severely reduced than the b-wave of the scotopic ERG. In addition, these patients demonstrated the following features: decreased visual acuity, impaired color vision, central or paracentral scotomas, and fundoscopic evidence of maculopathy. Patients with either STGD1 or fundus flavimaculatus were excluded from this study. Yellow-white, irregularly shaped flecks in the posterior pole and macular atrophy in the later stages of the disease were considered characteristic features of these types of inherited macular degeneration. Furthermore, these patients often demonstrate blocking of the normal background fluorescence, on fluorescein angiography, and normal or nearly normal full-field ERG results. Patients with RP show scotopic rod amplitudes more severely reduced than their photopic cone amplitudes and typically develop night blindness and progressive impairment of the midperipheral visual field in adolescence. Fundoscopic abnormalities include narrowed retinal vessels, bone-spicule pigmentation, and waxy pallor of the optic disk. Patients with these RP characteristics were also excluded from this study.
Twenty unrelated patients were included in this study. In five families, two or more siblings were affected, suggesting AR inheritance. In each of the remaining 15 families, only one patient with CRD was identified. Thirteen of these patients were considered typical CRD cases. Representative fundus photographs are shown in figure 1a and b. In seven patients, either the ophthalmologic examination was not complete, or some clinical findings commonly not associated with CRD were observed, although the ERG recordings showed a cone-rod type of retinal degeneration. Three patients (9250, 9369, and 9661) demonstrated narrow retinal vessels and bone spicules in the midperiphery in the later stages of their disease (fig. 1d). One patient (9378) showed typical patchy areas of chorioretinal atrophy in the midperiphery (fig. 1c). In another patient (8547), there was only minimal progression of the retinal dystrophy. Visual fields were not available in patient 11630. His ERG showed virtually extinguished cone responses in the presence of slightly subnormal rod responses. Finally, in patient 13163, the retinal dystrophy was in a final stage, with nonrecordable ERG patterns. These seven patients were therefore classified as having “atypical” CRD.
Figure 1.
Fundus appearance in four patients with CRD. a, Patient 11872, with perifoveal atrophy of the retinal pigment epithelium (RPE) (bull's eye) and atrophy of the RPE outside the macula. b, Patient 9633, with pericentral atrophy of the RPE. Arteries are slightly attenuated. c, Patient 9378, with central hypo- and hyperpigmentation, patchy atrophy of the RPE temporal to the macula, normal vessels, and normal optic disk. d, Patient 9369, with atrophy of the RPE around the optic disk, bone spicules along arteries and venules in the midperiphery, and attenuated arterioles. The periphery of the retina is normal. Patients 9378 (c) and 9369 (d) were classified as atypical.
Using the single-strand conformation polymorphism (SSCP) and direct-sequencing techniques (Allikmets et al. 1997b; Gerber et al. 1998; Maugeri et al. 1999), we searched for mutations in the 50 exons and flanking intron sequences of the ABCA4 gene. Nineteen ABCA4 mutations were identified in 13 of 20 patients (table 1). In six patients, mutations were found in both ABCA4 alleles; in seven patients, one ABCA4 mutation was found. ABCA4 mutations were identified in 4 (80%) of 5 patients with AR CRD and in 9 (60%) of 15 patients with isolated CRD. Thirteen mutant ABCA4 alleles were found in 9 of 13 patients who showed the “classic” symptoms of CRD. Six mutant ABCA4 alleles were identified in four of seven patients classified as having atypical CRD.
Table 1.
ABCA4 Mutations in Patients with CRD
|
ABCA4 Allele 1 |
ABCA4 Allele 2 |
||||
| Patient | Inheritance | Nucleotide Changes | Effects | Nucleotide Changes | Effects |
| 9250a | Isolated | 1622T→C;3113C→T | L541P;A1038Vb | 194G→A | G65Eb |
| 9303 | AR | 1622T→C;3113C→T | L541P;A1038Vb | ||
| 9336 | Isolated | 6658C→T | Q2220X | ||
| 9369a | AR | 6601–6602delAG | Frameshift | ||
| 9370 | Isolated | 1622T→C;3113C→T | L541P;A1038Vb | ||
| 9371 | Isolated | 1622T→C;3113C→T | L541P;A1038Vb | 1622T→C;3113C→T | L541P;A1038Vb |
| 9378a | Isolated | 768G→T | 5′ Splice mutationb | ||
| 9553 | AR | 2588G→C | ΔG863/G863Ab | IVS35del-2→+2del4 | 3′ Splice mutation |
| 9633 | Isolated | 1622T→C;3113C→T | L541P;A1038Vb | 4469G→A | C1490Yb |
| 9650 | Isolated | 3364G→A | E1122Kb | ||
| 9887 | Isolated | 4793C→A | A1598D | 6329G→A | W2110X |
| 11872 | Isolated | 634C→T | R212Cb | ||
| 13163a | AR | 1622T→C;3113C→T | L541P;A1038Vb | IVS36+1G→A | 5′ Splice mutationb |
Among 13 different mutations, 8 had been identified previously in patients with STGD1 and, with the exception of the frequent 2588G→C founder mutation (Maugeri et al. 1999), were not present in ⩾100 healthy control individuals (Fishman et al. 1999; Lewis et al. 1999; Maugeri et al. 1999). Five mutations were novel. One is a missense mutation (A1598D) that was not found in 100 control individuals. Four are thought to represent null mutations, either because they introduce a stop codon (W2110X and Q2220X), result in a frameshift (6601–6602delAG), or disrupt a splice site (IVS35-2→+2del4). A complex L541P;A1038V allele was found in 7 of 19 mutant alleles. Surprisingly, this complex ABCA4 allele was exclusively found in 6 of 14 German patients with CRD. One patient carried this complex allele homozygously, three patients were compound heterozygotes, and two patients were heterozygous (table 1 and fig. 2).
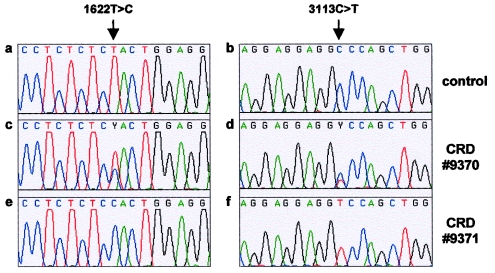
Figure 2.
DNA sequences in patients carrying the complex ABCA4 allele L541P;A1038V. a–b, Normal nucleotide sequences of exon 12 (a) and exon 21 (b) in a control individual. c–d, Heterozygous 1622T→C (c) and 3113C→T (d) nucleotide changes in patient 9370. e–f, Homozygous 1622T→C (e) and 3113C→T (f) nucleotide changes in patient 9371.
In this study, we identified ABCA4 mutations in 65% of the patients investigated, with no significant difference between patients that we classified as having typical versus atypical CRD (69% vs. 57%). In total, ABCA4 mutations have been identified in 80% of the patients with AR CRD and in 60% of those with isolated CRD. Assuming AR inheritance for all patients included in this study, we identified ABCA4 mutations in 19 (47.5%) of the 40 alleles. In patients with STGD1, a disease that is thought to be genetically homogeneous, ABCA4 mutation analysis revealed mutations in ∼60% of the alleles (Lewis et al. 1999; Maugeri et al. 1999). Apparently, a proportion (∼30%) of the ABCA4 mutations currently are missed because of the limitations of the SSCP analysis technique and because many splice mutations could be located in the noncoding sequence. Hence, it is possible that some of the seven CRD patients in whom we did not find an ABCA4 mutation do in fact carry ABCA4 mutations. Moreover, some of the six cases of isolated CRD without ABCA4 mutations might be caused by non–ABCA4-associated de novo autosomal dominant gene defects. These data suggest that, in >70% of patients with AR CRD, the disease is due to pathological ABCA4 mutations. We conclude that mutations in the ABCA4 gene are the major cause for AR CRD.
A prominent pathological role of ABCA4 in AR CRD underscores the “ABCA4 genotype-phenotype model” (fig. 3), in which we predicted AR CRD to occur whenever the ABCA4 activity is severely but not completely compromised—for example, by a combination of “moderate” and “severe” ABCA4 mutations (Cremers et al. 1998; van Driel et al. 1998; Maugeri et al. 1999). This model was based in part on the observation that two ABCA4 null mutations were invariably found in families with atypical AR RP (Cremers et al. 1998; Martinez-Mir et al. 1998) and in part on genotype-phenotype comparison in a family with RP and CRD (Cremers et al. 1998) and in patients with STDG1 (Allikmets et al. 1997b; Lewis et al. 1999; Maugeri et al. 1999). A frequent mild ABCA4 founder mutation (2588G→C) was found in 37% of European patients with STGD1 (Maugeri et al. 1999) and was never found homozygously in this or other groups of patients (Allikmets et al. 1997b; Lewis et al. 1999; Maugeri et al. 1999). In most of the compound-heterozygous patients with STGD1, the 2588G→C mutation was accompanied by severe ABCA4 mutations (Maugeri et al. 1999).
Figure 3.
Genotype-phenotype–correlation model and estimated incidence of ABCA4 mutations in patients with retinal dystrophies (adapted from work by van Driel et al. [1998]). The model shows the inverse correlation between the ABCA4 residual activity and the severity of the retinal dystrophy. The most severe phenotype, RP, is caused by two null/severe ABCA4 mutations. If the ABCA4 activity is partially retained, patients will develop either CRD, as the result of a severe mutation inherited together with a moderate mutation, or STGD1, as the result of a combination of a severe and a mild ABCA4 mutation.
Although we consider the genotype-phenotype model an oversimplification of the diverse functional effects that mutations can have on the presumed N-retinylidene-phosphatidylethanolamine flippase activity of ABCA4 (Sun et al. 1999; Weng et al. 1999) and on ABCA4-associated pathology, it allows us to evaluate the severity of novel mutations and of combinations thereof and to test the model's validity in patients with ABCA4-associated pathology other than STGD1. On the basis of the model, patients with CRD should not carry two null ABCA4 alleles. Although the number of patients with CRD in which both ABCA4 mutations are found is still low and the consequences of individual mutations have not been functionally tested, this indeed seems to be the case.
According to the model (fig. 3), the mild 2588G→C mutation, in combination with an ABCA4 mutation on the other allele, should cause STGD1 and not CRD or RP. Nevertheless, one patient with CRD (9553) was a compound heterozygote for the 2588G→C mutation and a severe splice-site mutation. As reported elsewhere in patients with STGD1 (Lewis et al. 1999; Rozet et al. 1999; Shroyer et al. 2000), it is possible that patient 9553 carries an allele with the 2588G→C mutation and another, as-yet-undetected mutation. Moreover, the paucity of the 2588G→C mutation in patients with CRD is in contrast to its frequent occurrence in Western European patients with STGD1 (1/20 cases with CRD vs. 15/40 cases with STGD1 [Maugeri et al. 1999]; P<.01, one-sided Fisher's exact test), strengthening our hypothesis that it only contributes to STGD1 pathology. Seven alleles in six German patients with CRD were found to carry two missense mutations (L541P;A1038V). The frequency of this allele is surprisingly high in our group of patients with CRD, comparable to the frequency of the 2588G→C ABCA4 founder mutation in European patients with STGD1 (7/40 alleles in patients with CRD vs. 15/80 alleles in patients with STGD1) (Maugeri et al. 1999). The ABCA4 allele carrying only the A1038V mutation is one of the most frequent alleles in patients with STGD1 in the United States (15/150) (Lewis et al. 1999). However, among a total of 357 non-German patients with STGD1, only 4 (1.1%) were shown to carry the L541P;A1038V allele (Rozet et al. 1998; Fishman et al. 1999; Lewis et al. 1999; Maugeri et al. 1999; Papaioannou et al. 2000; Simonelli et al. 2000). In contrast, the complex allele was found in 22 (14.2%) of 155 German patients with STGD1 (Maugeri et al. 1999; B. H. Weber, personal communication), indicating that this complex allele most likely is not specifically involved in CRD.
In conclusion, we have found that, despite the high clinical variability observed among patients with CRD, mutations in the ABCA4 gene are responsible for the majority of the cases. These findings confirm our previous hypothesis on the importance of ABCA4 involvement in CRD (Maugeri et al. 1999), which is surprising, in view of the high degree of genetic heterogeneity seen in other retinal dystrophies.
Future studies will concentrate on the ABCA4 analysis of patients with AR RP, which, on the basis of our genotype-phenotype model and previous estimates (Maugeri et al. 1999), should carry ABCA4 mutations in ∼8% of the cases. Since other known AR RP genes are mutated in ⩽5% of the patients (Dryja et al. 1997, 1999; Rivolta et al. 2000), mutations in ABCA4 may well be the most frequent cause of nonsyndromic retinal dystrophy in humans.
Acknowledgments
The authors thank Mrs. B. van den Helm and S. D. van der Velde-Visser for expert technical assistance. This research was supported by the British Retinitis Pigmentosa Society, the Foundation Fighting Blindness, the Rotterdamse Vereniging Blindenbelangen, the Algemene Nederlandse Vereniging ter Voorkoming van Blindheid, Stichting Blindenhulp, Stichting De Drie Lichten, the Landelijke Stichting voor Blinden en Slechtzienden, the Gelderse Blindenvereniging, and the Stichting voor Ooglijders.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim (for gyrate atrophy [MIM 258870], choroideremia [MIM 303100], STGD1 [MIM 248200], Best vitelliform macular dystrophy [MIM 153700], RP [MIM 268000], CRD [MIM 120970, MIM 601777, MIM 600624, MIM 603649, MIM 604116, MIM 600977], ABCA4 [MIM 601691], RP19 [MIM 601718], CORD3 [MIM 604116], and AMD [MIM 153800])
- RetNet, Retinal Information Network, http://www.sph.uth.tmc.edu/Retnet/resource.htm
References
- Allikmets R (1999) Molecular genetics of age-related macular degeneration: current status. Eur J Ophthalmol 9:255–265 [DOI] [PubMed] [Google Scholar]
- Allikmets R, International ABCR Screening Consortium, The (2000) Further evidence for an association of ABCR alleles with age-related macular degeneration. Am J Hum Genet 67:487–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allikmets R, Shroyer NF, Singh N, Seddon JM, Lewis RA, Bernstein PS, Peiffer A, Zabriskie NA, Li Y, Hutchinson A, Dean M, Lupski JR, Leppert M (1997a) Mutation of the Stargardt disease gene (ABCR) in age-related macular degeneration. Science 277:1805–1807 [DOI] [PubMed] [Google Scholar]
- Allikmets R, Singh N, Sun H, Shroyer NF, Hutchinson A, Chidambaram A, Gerrard B, Baird L, Stauffer D, Peiffer A, Rattner A, Smallwood P, Li Y, Anderson KL, Lewis RA, Nathans J, Leppert M, Dean M, Lupski JR (1997b) A photoreceptor cell-specific ATP-binding transporter gene (ABCR) is mutated in recessive Stargardt macular dystrophy. Nat Genet 15:236–246 [DOI] [PubMed] [Google Scholar]
- Chang GQ, Hao Y, Wong F (1993) Apoptosis: final common pathway of photoreceptor death in rd, rds, and rhodopsin mutant mice. Neuron 11:595–605 [DOI] [PubMed] [Google Scholar]
- Cremers FPM, van de Pol TJR, van Driel M, den Hollander AI, van Haren FJJ, Knoers NVAM, Tijmes N, Bergen AAB, Rohrschneider K, Blankenagel A, Pinckers AJLG, Deutman AF, Hoyng CB (1998) Autosomal recessive retinitis pigmentosa and cone-rod dystrophy caused by splice site mutations in the Stargardt's disease gene ABCR. Hum Mol Genet 7:355–362 [DOI] [PubMed] [Google Scholar]
- Cremers FPM, van de Pol TJR, van Kerkhoff EPM, Wieringa B, Ropers H-H (1990) Cloning of a gene that is rearranged in patients with choroideraemia. Nature 347:674–677 [DOI] [PubMed] [Google Scholar]
- Dryja TP (1997) Gene-based approach to human gene-phenotype correlations. Proc Natl Acad Sci USA 94:12117–12121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dryja TP, Rucinski DE, Huang Chen S, Berson EL (1999) Frequency of mutations in the gene encoding the α subunit of rod cGMP-phosphodiesterase in autosomal recessive retinitis pigmentosa. Invest Ophthalmol Vis Sci 40:1859–1865 [PubMed] [Google Scholar]
- Fishman GA, Stone EM, Grover S, Derlacki DJ, Haines HL, Hockey RR (1999) Variation of clinical expression in patients with Stargardt dystrophy and sequence variations in the ABCR gene. Arch Ophthalmol 117:504–510 [DOI] [PubMed] [Google Scholar]
- Gerber S, Rozet JM, van de Pol TJR, Hoyng CB, Munnich A, Blankenagel A, Kaplan J, Cremers FPM (1998) Complete exon-intron structure of the retina specific ATP binding transporter gene (ABCR) allows the identification of novel mutations underlying Stargardt disease. Genomics 48:139–142 [DOI] [PubMed] [Google Scholar]
- Lewis RA, Lupski JR (2000) Macular degeneration: the emerging genetics. Hosp Pract (Off Ed) 35:41–50 [PubMed] [Google Scholar]
- Lewis RA, Shroyer NF, Singh N, Allikmets R, Hutchinson A, Li Y, Lupski JR, Leppert M, Dean M (1999) Genotype/phenotype analysis of a photoreceptor-specific ATP-binding cassette transporter gene, ABCR, in Stargardt disease. Am J Hum Genet 64:422–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Mir A, Paloma E, Allikmets R, Ayuso C, del Rio T, Dean M, Vilageliu L, Gonzàlez-Duarte R, Balcells S (1998) Retinitis pigmentosa caused by a homozygous mutation in the Stargardt disease gene ABCR. Nat Genet 18:11–12 [DOI] [PubMed] [Google Scholar]
- Maugeri A, van Driel MA, van de Pol TJR, Klevering BJ, van Haren FJJ, Tijmes N, Bergen AAB, Rohrschneider K, Blankenagel A, Pinckers AJLG, Dahl N, Brunner HG, Deutman AF, Hoyng CB, Cremers FPM (1999) The 2588G→C mutation in the ABCR gene is a mild frequent founder mutation in the western European population and allows the classification of ABCR mutations in patients with Stargardt disease. Am J Hum Genet 64:1024–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molday LL, Rabin AR, Molday RS (2000) ABCR expression in foveal cone photoreceptors and its role in Stargardt macular dystrophy. Nat Genet 25:257–258 [DOI] [PubMed] [Google Scholar]
- Papaioannou M, Ocaka L, Bessant D, Lois N, Bird A, Payne A, Bhattacharya S (2000) An analysis of ABCR mutations in British patients with recessive retinal dystrophies. Invest Ophthalmol Vis Sci 41:16–19 [PubMed] [Google Scholar]
- Petrukhin K, Koisti MJ, Bakall B, Li W, Xie G, Marknell T, Sandgren O, Forsman K, Holmgren G, Andreasson S, Vujic M, Bergen AAB, McGarty-Dugan V, Figueroa D, Austin CP, Metzker ML, Caskey CT, Wadelius C (1998) Identification of the gene responsible for Best macular dystrophy. Nat Genet 19:241–247 [DOI] [PubMed] [Google Scholar]
- Rivolta C, Sweklo EA, Berson EL, Dryja TP (2000) Missense mutation in the USH2A gene: association with recessive retinitis pigmentosa without hearing loss. Am J Hum Genet 66:1975–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozet J-M, Gerber S, Ghazi I, Perrault I, Ducroq D, Souied E, Cabot A, Dufier J-L, Munnich A, Kaplan J (1999) Mutations of the retinal specific ATP binding transporter gene (ABCR) in a single family segregating both autosomal recessive retinitis pigmentosa RP19 and Stargardt disease: evidence of clinical heterogeneity at this locus. J Med Genet 36:447–451 [PMC free article] [PubMed] [Google Scholar]
- Rozet J-M, Gerber S, Souied E, Perrault I, Châtelin S, Ghazi I, Leowski C, Dufier J-L, Munnich A, Kaplan J (1998) Spectrum of ABCR gene mutations in autosomal recessive macular dystrophies. Eur J Hum Genet 6:291–295 [DOI] [PubMed] [Google Scholar]
- Shroyer NF, Lewis RA, Allikmets R, Singh N, Dean M, Leppert M, Lupski JR (1999) The rod photoreceptor ATP-binding cassette transporter gene, ABCR, and retinal disease: from monogenic to multifactorial. Vision Res 39:2537–2544 [DOI] [PubMed] [Google Scholar]
- Shroyer NF, Lewis RA, Lupski JR (2000) Complex inheritance of ABCR mutations in Stargardt disease: linkage disequilibrium, complex alleles, and pseudodominance. Hum Genet 106:244–248 [DOI] [PubMed] [Google Scholar]
- Simonelli F, Testa F, de Crecchio G, Rinaldi E, Hutchinson A, Atkinson A, Dean M, D'Urso M, Allikmets R (2000) New ABCR mutations and clinical phenotype in Italian patients with Stargardt disease. Invest Ophthalmol Vis Sci 41:892–897 [PubMed] [Google Scholar]
- Stone EM, Webster AR, Vandenburgh K, Streb LM, Hockey RR, Lotery AJ, Sheffield VC (1998) Allelic variation in ABCR associated with Stargardt disease but not age-related macular degeneration. Nat Genet 20:328–329 [DOI] [PubMed] [Google Scholar]
- Sun H, Molday RS, Nathans J (1999) Retinal stimulates ATP hydrolysis by purified and reconstituted ABCR, the photoreceptor-specific ATP-binding cassette transporter responsible for Stargardt disease. J Biol Chem 274:8269–8281 [DOI] [PubMed] [Google Scholar]
- Szlyk JP, Fishman GA, Alexander KR, Peachey NS, Derlacki DJ (1993) Clinical subtypes of cone-rod dystrophy. Arch Ophthalmol 111:781–788 [DOI] [PubMed] [Google Scholar]
- Valle D, Simell O (1989) The hyperornithinemias. In: Scriver CR, Beaudet AL, Sly WS, Valle D (eds) The metabolic basis of inherited disease, 6th ed. McGraw-Hill, New York, pp 599–627 [Google Scholar]
- van Driel MA, Maugeri A, Klevering BJ, Hoyng CB, Cremers FPM (1998) ABCR unites what ophthalmologists divide(s). Ophthalmic Genet 19:117–122 [DOI] [PubMed] [Google Scholar]
- Weng J, Mata NL, Azarian SM, Tzekov RT, Birch DG, Travis GH (1999) Insights into the function of rim protein in photoreceptors and etiology of Stargardt's disease from the phenotype in abcr knockout mice. Cell 98:13–23 [DOI] [PubMed] [Google Scholar]